Introduction
It has been widely accepted that tumor growth is
sustained by a rare subpopulation of putative cancer stem cells
(CSCs)/progenitor-like cells that share specific characteristics
with normal stem cells, namely self-renewal, clonogenicity and
multipotency (1–3). Previous investigations have shown that
a number of tumors may actually arise from the transformation of
progenitor cells rather than stem cells (4,5).
Normal stem cells and CSCs share significant properties, such as
heterogeneity and plasticity. Maturation and differentiation are
important in cancer cell heterogeneity, and tumor cell
heterogeneity may result from clonal evolution driven by genetic
instability of stem-like cells, frequently called CSCs or
tumor-initiating cells (6). Cells
in this heterogeneous population exist in various stages throughout
their lifetime. During early tumor development or in unperturbed
tumor conditions, CSCs mainly undergo one-way maturation by
developing into tumor progenitor cells and even differentiated
tumor cells (7). It is possible to
assume that these differentiated cells may arise from CSCs, which
have self-renewal capacity and/or phenotypically differentiated
tumor cells that functionally possess low or no tumor-regenerating
capacity (non-CSCs/bulk population). CSCs are the cell
subpopulation that are most likely responsible for treatment
failure and cancer recurrence, while the bulk population of tumor
cells exhibit low self-renewal capacity and a higher probability of
terminal differentiation (i.e. transit-amplifying cancer progenitor
cells) (8).
CSCs have been previously isolated and identified
using common cell surface markers in the majority of cancer types,
including brain (9,10), kidney (11), liver (12,13),
colon (14), pancreas (15) and prostate (16). CD133, also known as prominin-1 or
AC133 (a glycoprotein comprising of five transmembrane
domains), has been described as a marker of stem cells in several
organs and appears to be the CSC marker for a number of tumor types
(17). However, there have been
accumulating results demonstrating that CD133+ and
CD133− subpopulations are tumorigenic in metastatic
glioblastoma and colon cancer (18–20).
CD44 is a member of the cell adhesion protein family
and the expression of several CD44 proteins has been found to
correlate with aggressive stages of various types of human cancer
(21). An evident function of the
CD44 family members is their alternative splicing. Previously,
Ponta et al demonstrated that CD44 family members differ in
the extracellular domain by the insertion of variable regions
through alternative splicing (22).
A small subset of CD44+ cells in prostate cell cultures
and xenograft tumors are more tumorigenic, proliferative,
clonogenic and metastatic as compared with the CD44−
subpopulation. This CD44+ subset expresses higher mRNA
levels of several genes that are characteristic of embryonic stem
cells (23). In addition, Collins
et al have shown that prostate cancer tumorigenic cells have
a CD44+/1α2β1high/CD133+ phenotype
(24).
A challenge has been encountered with regard to the
enrichment of CSCs from the established cell lines of a variety of
solid tumors that develop as three-dimensional (3D) cell cultures.
The 3D spheroid model is a new technique for the propagation of
cells in vitro using serum-free medium and cultured under
low-adherence conditions (25). An
additional usage of spheroids constitutes the liquid overlay
technique, namely multicellular tumor spheroids (26)
The 3D spheroid model presents a convenient model to
investigate cancer cells and has been increasingly used for this
purpose. It reproduces in vitro results in accordance with
in vivo results and generates significant in vitro
characteristics not observed in monolayers or suspension
cultures.
The present study hypothesized that the structure of
CSCs may show differentiation when compared with non-CSCs, and
differentiation of stem cell markers may aid therapeutic strategies
of cancer. Therefore, the current study describes approaches to
present and analyze the differentiation properties of human
prostate CSCs within 3D spheroids, which may serve as the basis for
defining the gene and protein trace of CSCs.
Materials and methods
Cell culture conditions and reagents
The DU145 human prostate cancer cell line was
supplied by the American Type Culture Collection (Manassas, VA,
USA) and was grown in monolayer culture in Dulbecco’s modified
Eagle’s medium-F12 (DMEM-F12; Biological Industries Israel
Beit-Haemek Ltd., Kibbutz Beit-Haemek, Israel) supplemented with
10% heat-inactivated fetal calf serum (Gibco, Invitrogen Life
Science, Paisley, UK), 100 U/ml penicillin and 100 μg/ml
streptomycin (Sigma-Aldrich, St Louis, MO, USA). Cells in
semi-confluent flasks were harvested using 0.05% trypsin
(Sigma-Aldrich), centrifuged (Nüve NF200, Laboratory and
Sterilization Technology, Ankara, Turkey) following the addition of
DMEM-F12 for trypsin inactivation and then resuspended in culture
medium. The antibodies used consisted of C-kit (sc-168),
Notch1 (sc-6014), Jagged1 (sc-6011) and Delta1
(sc-8155) (all 1:100; Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA), Sox2 (1:300; Abnova, Taipei, Taiwan), c-Myc
(1:300; Santa Cruz Biotechnology, Inc.), Oct4 and
Klf4 (1:300, Abcam, Cambridge UK), CD90 (THY-1;
1:300, Abcam) and SSEA1 (1:300, Abcam), secondary antibody
(sc-2053; Histostain®-Plus Streptavidin-Peroxidase;
Gibco, Invitrogen Life Technologies and Santa Cruz Biotechnology,
Inc.).
Fluorescence-activated cell sorting
(FACS) and experimental groups
For FACS (Facs Aria, BD Biosciences, San Jose, CA,
USA), cells were detached using non-enzymatic cell dissociation
solution (Sigma-Aldrich) and ~5×104 cells were incubated
with antibody [dilution of 1:100 in FACS wash (0.5% bovine serum
albumin, 2 mM NaN3 and 5 mM EDTA)] for 15 min at 4°C. An
isotype and concentration-matched PE-labeled control antibody
(Miltenyi Biotech, Bisley, UK) was used and the samples were
labeled with PE-labeled CD133/1 (clone AC133/1; Miltenyi Biotech)
and FITC-labeled CD44 (clone G44-26; BD Pharmingen, San Diego,
USA). After 3–5 min, the cells were washed with FACS wash and
resuspended. The cells were sorted into
CD133high/CD44high (CSC) and non-CSC
subpopulations. The two subpopulations were cultured in two
different settings, monolayer 2D culture or 3D multicellular tumor
spheroid. Briefly, the experimental groups comprised of monolayer
CSC (M+) and non-CSC (M−) and spheroid CSC
(S+) and non-CSC (S−) subpopulations.
Constitution of spheroids and sphere
formation assay
For spheroid cultures, the tumor cells grown as
monolayer were resuspended with trypsin and the clonogenic
potential of various phenotypic populations was analyzed in a 3D
non-adherent culture condition (plates coated with 3% Noble agar;
Difco Laboaratories Inc., BD Diagnostic Systems, Detroit, MI, USA).
The cells were counted, resuspended and plated with
1×103 cells per well in a six-well plate. Two weeks
following initiation, the plates were inspected for colony (sphere)
growth. The number of colonies within each well was counted under
the microscope (Olympus BX-51, Olympus, Hamburg, Germany) and
images of representative fields were captured. First passage
floating spheres were removed and gently disaggregated with a new
3% Noble agar-coated well.
Polymerase chain reaction (PCR) array
assay
Total RNA was extracted from CSCs and non-CSCs
(miRNeasy kit; Qiagen, Hilden, Germany) and synthesis of cDNA was
performed using the SuperArray kit (C-03; SABiosciences, Frederick,
MD, USA). Stem cell-specific gene expression profiles were studied
with the PCR array assay (SABiosciences) according to the
manufacturer’s instructions. Briefly, total RNA was isolated from
monolayer cell populations or whole floating spheroids. In total,
≤1 μg of total RNA was treated with DNase and cDNA was prepared
using the RT2 first-strand kit. For each analysis, pairs
of the test and control cDNA samples were mixed with RT2
qPCR master mix and distributed across the 96-well plate of the PCR
array, each of which contained 84 stem cell-related probes and
control housekeeping genes. After cycling with qPCR (LightCycler
480; Roche Diagnostics GmbH, Mannheim, Germany), the obtained
amplification data (fold-changes in Ct values of all the genes)
were analyzed using SABiosciences software and ≥1.5 fold-change was
used for filtering criteria. Detailed analyses of telomerase
reverse transcriptase (TERT); cyclin A2; cyclin D1
(CCND1); CCND2; cyclin E1 (CCNE1);
cyclin-dependent kinase 1 (CDK1); GTP-binding protein
(CDC42); E1A-binding protein (EP300);
myelocytomatosis viral oncogene homolog (MYC);
retinoblastoma 1; forkhead box A2 (FOXA2); actin, α, cardiac
muscle 1 (ACTC1); achaete-scute complex homolog 2
(ASCL2); ISL LIM homeobox 1 (ISL1); keratin15
(KRT15); msh homebox 1 (MSX1); myogenic
differentiation 1 (MYOD1); T, brachyury homolog (T);
NOTCH1; NOTCH2; jagged1 (JAG1); Delta-like 1
(DLL1); DLL3; deltex homolog 1 (DTX1);
DTX2; dishevelled, dsh homolog 1 (DVL1); K (lysine)
acetyltransferase 2A (KAT2A); histone deacetylase 2
(HDAC2); numb homolog (NUMB); sex determining region
Y, box 1 (SOX1); SOX2; aggrecan (ACAN); alkaline
phosphatase, intestinal (ALPI); bone γ-carboxyglutamate
protein (BGLAP); collagen, type I, α 1 (COL1A1);
COL2A1; COL9A1; and peroxisome proliferator-activated
receptor γ (PPARG) were performed.
Immunohistochemical analysis
Immunohistochemistry was adapted and modified from
our previous protocols (27).
Briefly, monolayer cells were maintained in 24-well plates and
fixed with paraformaldehyde. The spheroids were processed in
routine histological processing for embedding in paraffin wax.
Cells were incubated with primary antibodies overnight at 40°C in a
humidity chamber. The modified Streptavidin-Peroxidase technique
was then used. Following incubation with 3,3′-diaminobenzidine
(Invitrogen Life Technologies), sections were counterstained with
Mayer’s hematoxylin (Sigma-Aldrich). Immunoreactivity of molecules
was assessed by light microscopy using Olympus BX-51 and C-5050
digital cameras. Staining was graded independently by two
observers, blinded to the groups, who evaluated semi-quantitatively
using the following scale: Mild, +; moderate, ++; and strong,
+++.
Results
CD133high/CD44 high
CSC and non-CSC subpopulation purity and sorting rates
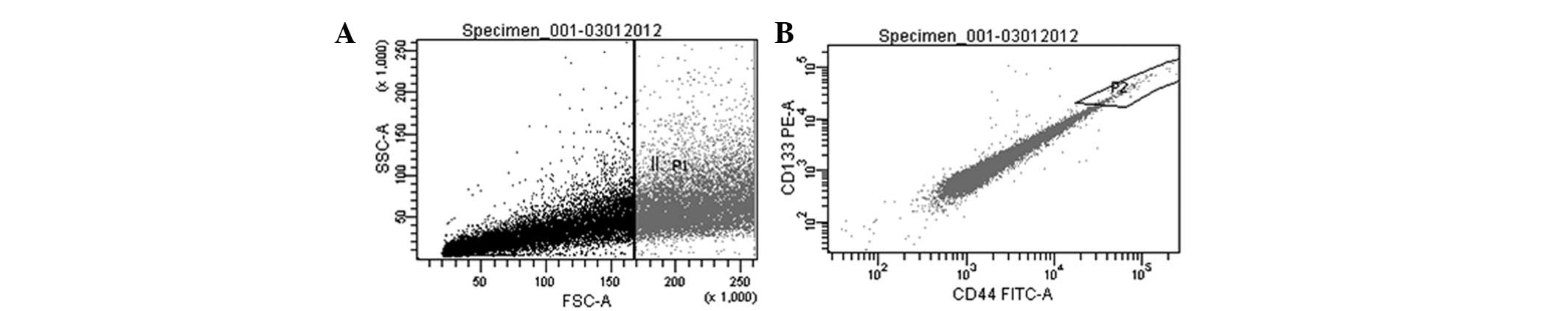
Prior to performing the microarray, the purity of
CSC and non-CSC samples was tested with CD133 and CD44. CSC sorting
is performed 300 times per year in our laboratory (Molecular
Embyology Laboratory, Department of Histology and Embryology,
Faculty of Medicine, Ege University, Bornova, Turkey). Sorting rate
analysis and purity of cells were evaluated sequentially and
statistical analysis was performed using SPSS 15 (SPSS, Inc.,
Chicago, IL, USA). Rates were 9.67±5.4% for CSCs and 90.33±5.4 for
non-CSCs. In order to confirm the flow cytometry analysis, cells
were re-evaluated following sorting, and this analysis was repeated
after one passage. Results showed that the purity of the cells was
85% and immunofluorescence staining yielded a cell purity of
>85% in all samples.
Analysis of TERT and cell cycle
regulation gene products
Following cell separation with FACS (Fig. 1), the differentially expressed genes
of the DU145 human prostate cell line were analyzed in
CD133high/CD44high (CSCs) and their bulk
counterpart (non-CSCs) cultured as monolayer cells or 3D spheroids.
In general, notable differences were observed between the CSCs
spheroid (S+) and monolayer (M+) groups.
These two groups constituted of CSCs and TERT expression was
increased significantly in the S+ group when compared
with the M+ group (Fig.
2A). An additional large population of genes related to cell
cycle regulation were analyzed, including CCND1,
CCND2, CCNE1, CDK1, CDC42, EP300 and MYC.
These genes were upregulated in the S+ group versus the
M+ group. CCND2 expression increased in the non-CSC
counterpart of the S− group compared with the
S+ group. On the other hand, CCND2 was significantly
reduced in the M+ group compared with the M−
group (Fig. 2A).
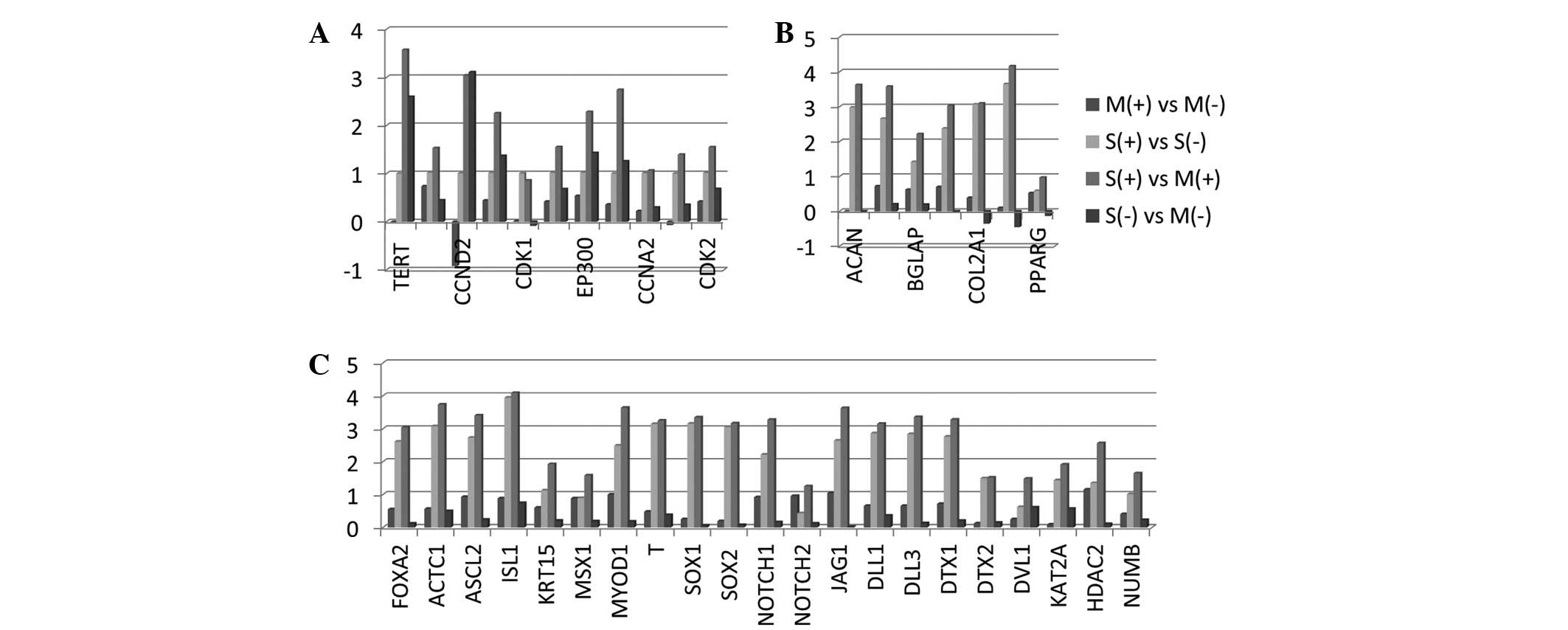 | Figure 2Microarray analysis in DU145 human
prostate cell line in monolayer cells (M) and 3D spheroids (S), as
well as in CD133high/CD44high CSCs
(S+ and M+) and their bulk counterpart
non-CSCs (S− and M−) cells was performed. (A)
TERT and cell cycle regulation, (B) embryonic cell lineage and
Notch signaling and (C) mesenchymal cell linage-related genes were
demonstrated. These microarrays were analyzed to calculate the
log-ratios. CSCs, cancer stem cells; TERT, telomerase reverse
transcriptase; CCND2, cyclin D2; CDK1, cyclin-dependent kinase 1;
EP300, E1A-binding protein; CCNA2, cyclin A2; CDK2,
cyclin-dependent kinase 2; ACAN, aggrecan; BGLAP, bone
γ-carboxyglutamate protein; COOL2A1, collagen, type II, α 1; PPARG,
peroxisome proliferator-activated receptor γ; FOXA2, forkhead box
A2; ACTC1, actin, α, cardiac muscle 1; ASCL2, achaete-scute complex
homolog 2; ISL1, ISL LIM homeobox 1; KRT15, keratin15; MSX1, msh
homebox 1; MYOD1, myogenic differentiation 1; T, T, brachyury
homolog; SOX1/2, sex determining region Y, box 1/2; JAG1, jagged1;
DLL1/3, Delta-like 1/3; DTX1/2, deltex homolog 1/2; DVL1,
dishevelled, dsh homolog 1; KAT2A, K(lysine) acetyltransferase 2A;
HDAC2, histone deacetylase 2; NUMB, numb homolog. |
Analysis of embryonic cell lineage and
Notch signaling gene products
The differentiation of the embryonic cell linage
genes of prostate CSC-enriched CD133high/CD44
high cells were determined and compared with the bulk
counterparts in the monolayer and spheroid subpopulations. With
respect to the embryonic cell lineages, FOXA2, ACTC1, ASCL2,
ISL1, KRT15, MSX1, MYOD1, T, SOX1 and SOX2, expression
profiles were investigated. These genes were commonly upregulated
in the CSC spheroid versus CSC monolayer cultures (S+
vs. M+], with significantly higher levels of ISL1,
ACTC1, MYOD1, ASCL2, SOX1, T and SOX2. These genes were
expressed at extremely low levels in the non-CSCs. The lowest
expression was observed when comparing the S− group with
the M− group.
The Notch signaling pathway is important in
cell-to-cell communications that regulate multiple cell
differentiation processes during embryonic and adult life (28). In the present study, the expression
of NOTCH1, NOTCH2, JAG1, DLL1, DLL3, DTX1, DTX2, DVL1, KAT2A,
HDAC2 and NUMB was investigated. The expression of these
genes significantly increased in the S+ versus the
M+ populations compared with the other groups. The Notch
signaling genes, JAG1, DLL3, NOTCH1, DTX1 and
DLL1, were of higher levels when compared with other
upregulated genes (Fig. 2B).
Analysis of mesenchymal cell linage gene
products
The ACAN, ALPI, BGLAP, COL1A1, COL2A1, COL9A1
and PPARG genes were evaluated. Significantly, the
COL9A1 gene was upregulated in the CSC spheroid as compared
with the CSC monolayer. The expression of COL2A1 and
COL9A1 genes was reduced in the non-CSC spheroids when
compared with the monolayers (S− vs. M−)
(Fig. 2C).
Immunohistochemical analysis of stem cell
markers
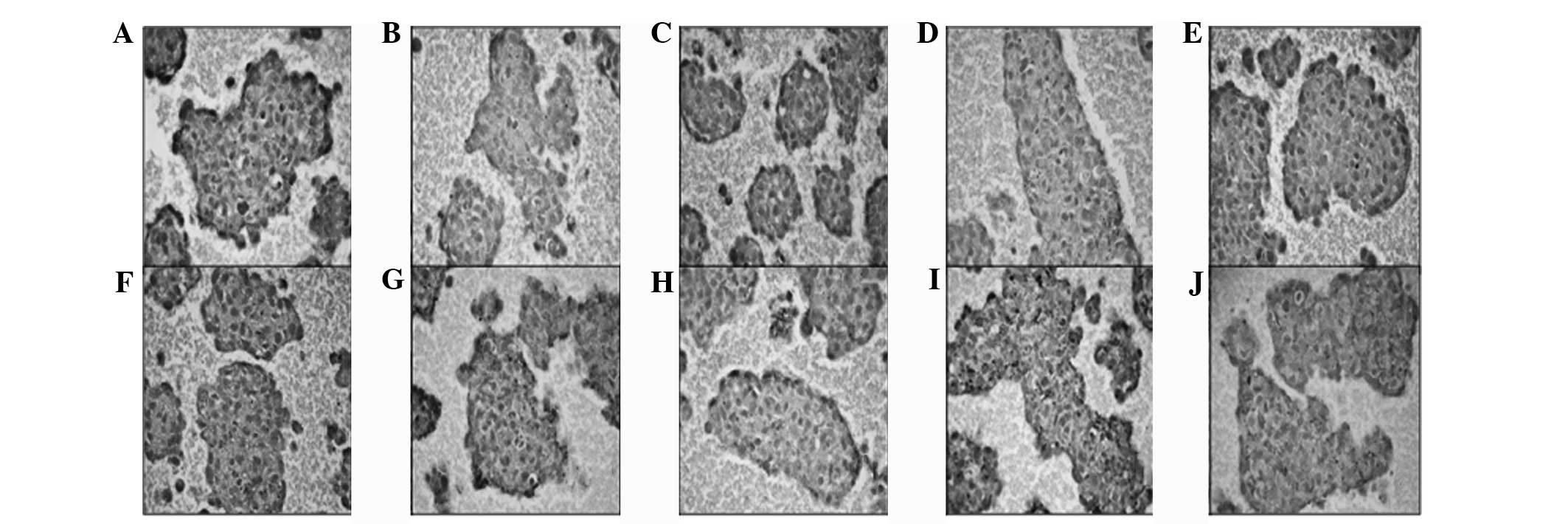
Results of the immunohistochemical analyses revealed
that embryonic stem cell markers increased following the
differentiation of CSCs when the cells constituted a spheroid
formation. Immunohistochemistry of CD117, Notch1,
Jagged1, Delta1, Sox2, c-Myc,
Oct4, KLF4, CD90 and SSEA1 was
determined in the various groups. Positive immunoreactivity was
observed in CSCs and non-CSCs whether the cells were maintained in
monolayer culture or as spheroid. The monolayer CSCs showed low (+)
immunoreactivity scores (Fig. 3),
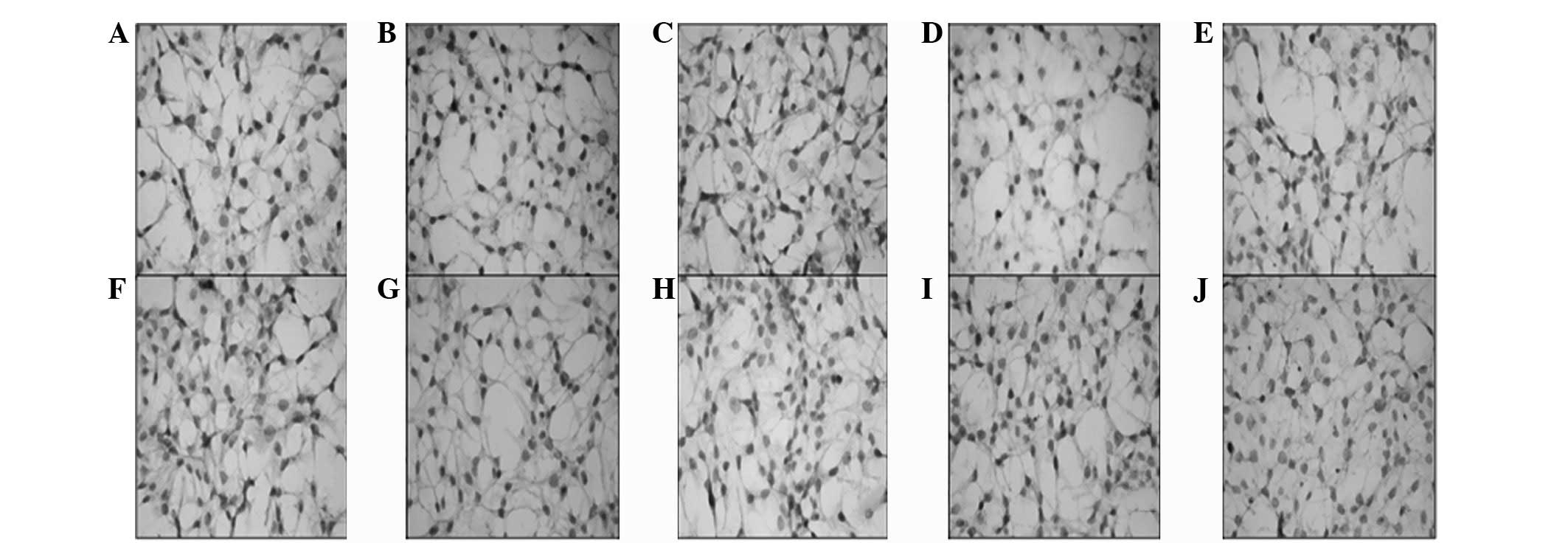
while the monolayer non-CSCs (Fig.
4) showed moderate (++) immunoreactivity. Increased
nucleus/cytoplasm ratios, decreased cell diameter and enhanced
immunoreactivity were observed in the CD133high/CD44
high population. The staining density of Jagged1,
Sox2, Oct4 and Klf-4 increased significantly
in this monolayer CSCs population. On the other hand, strong (+++)
immunoreactivity was observed in the CSC spheroids (Fig. 5) when compared with the non-CSC
spheroids (Fig. 6). Among these
spheroids, a moderate (++) immunoreactivity score was observed for
Notch1, Jagged1 and Delta1 in the non-CSCs
spheroids while strong (+++) immunoreactivity was observed in the
other groups. Moreover, the highest immunoreactivity was observed
in the CSC spheroid group when compared with the monolayer CSCs,
monolayer non-CSCs or spheroid non-CSCs group.
 | Figure 3Immunohistochemistry of (A) CD117,
(B) Notch1, (C) Jagged1, (D) Delta1, (E) Sox2, (F) c-Myc, (G) Oct4,
(H) KLF4, (I) CD90 and (J) SSEA1 was determined in monolayer CSCs.
Increased nucleus/cytoplasm ratio, decreased cell diameter and
enhanced immunoreactivity were observed in the
CD133high/CD44high population. Although
positive immunoreactivity was determined in all cells in all
groups, the staining density of Jagged1, Sox2, Oct4 and Klf-4
increased significantly in the
CD133high/CD44high CSC population. CSCs,
cancer stem cells. |
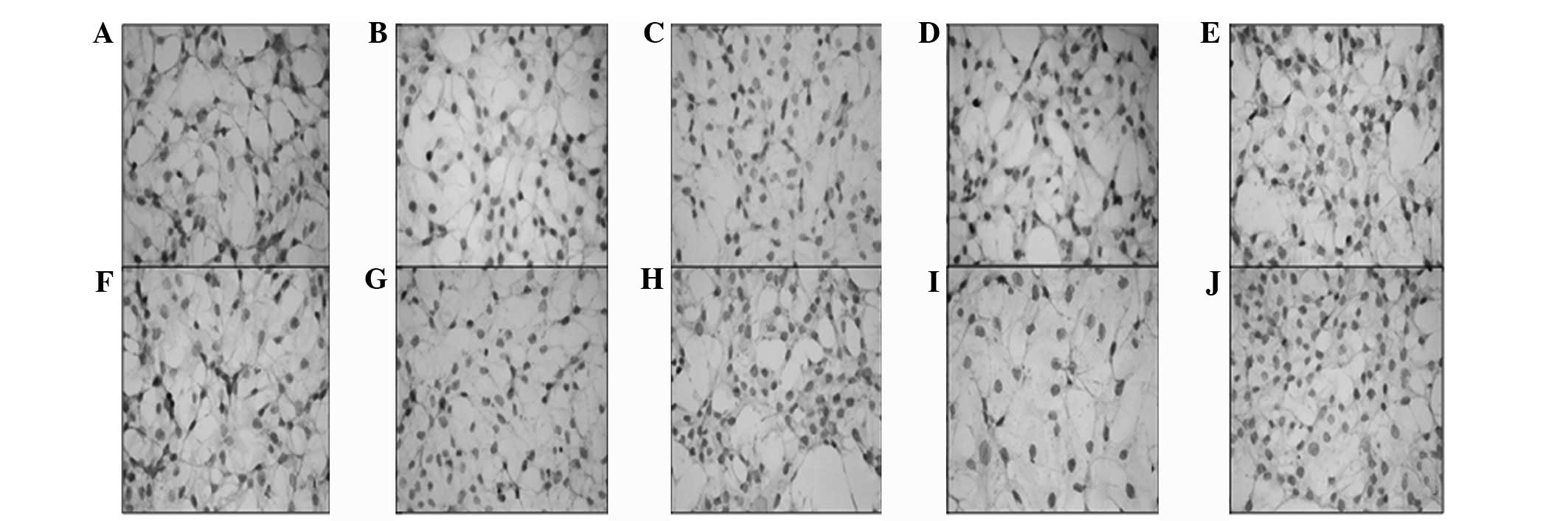 | Figure 4Immunohistochemistry of (A) CD117,
(B) Notch1, (C) Jagged1, (D) Delta1, (E) Sox2, (F) c-Myc, (G) Oct4,
(H) KLF4, (I) CD90 and (J) SSEA1 was determined in monolayer
non-CSCs. Decreased nucleus/cytoplasm ratio, increased diameter and
positive immunoreactivity was observed in the cells; however, the
staining density of this non-CSC population was significantly
decreased compared with the monolayer non-CSCs. CSCs, cancer stem
cells. |
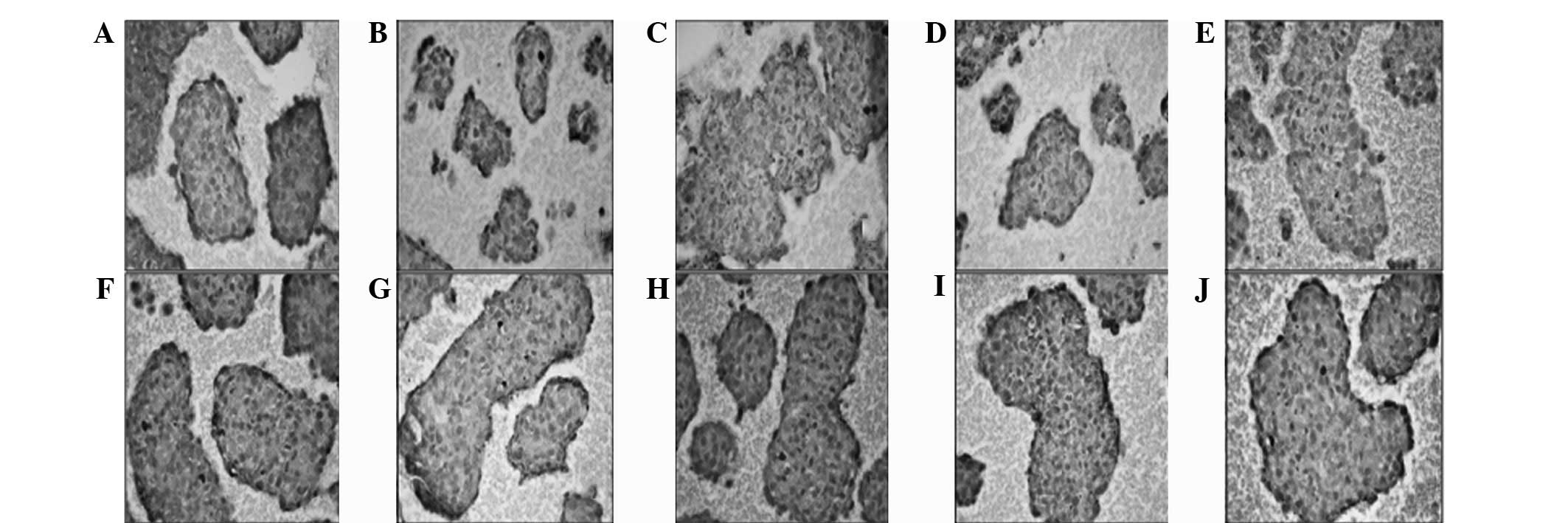 | Figure 6Immunohistochemistry of (A) CD117,
(B) Notch1, (C) Jagged1, (D) Delta1, (E) Sox2, (F) c-Myc, (G) Oct4,
(H) KLF4, (I) CD90 and (J) SSEA was determined in non-CSC
spheroids. Notch1, Jagged1 and Delta1 immunoreactivity was moderate
compared with the marked immunoreactivity of the other groups. CSC,
cancer stem cell. |
Discussion
Despite limited data in the previous literature, the
differentiation of CSCs may be investigated. Cancer cells capable
of undergoing proliferation have the ability to self-renew and
their differentiation properties are unique to CSCs. In the present
study, this differentiation hypothesis was examined by using an
in vitro 3D-tumor differentiation spheroid model. The cells
were found to alter their gene expression profiles during this
process. Our hypothesis was supported by the observation that
significant gene alterations were observed in the
CD133high/CD44 high population when the
monolayer cells were allowed to grow as spheroid. In this group, a
marked upregulation was determined in COL9A1 and ISL1
compared with other genes. Type IX collagen is covalently bound to
the surface of type II collagen fibrils within the cartilage
extracellular matrix (29).
Collagen IX is required for the integrity of collagen II fibrils
and the regulation of vascular plexus formation (30). Additionally, Piotrowski et al
previously demonstrated that complete loss of methylation affected
COL9A1 in tumors (31).
However, no previous literature is available with regard to the
role of COL9A1 in cancer. In the current study, it was
possible to assume that the upregulation of COL9A1
correlates with the arrangement of the extrafibrillar proteoglycan,
glycoprotein matrix and vascular development. ISL1 is a
LIM-homeodomain transcription factor that marks cell population and
establishes endothelial, myocardial and smooth muscle cells.
Previously, Schmitt et al demonstrated that ISL1 is a
reliable marker of pancreatic endocrine tumors and metastases
(32). The present study reported,
for the first time, that ISL1 is an additional significantly
upregulated gene in prostate spheroid CSCs. ISL1+
multipotent precursors have the potential of self-renewal and
differentiation into endothelial, cardiomyocyte and smooth muscle
lineages. These features highlight postnatal angiogenesis and
vasculogenesis by improving the angiogenic properties of
endothelial cells and mesenchymal stem cells (33). Angiogenesis is critical for tumor
growth, and the VEGF pathway and Notch signaling are
perhaps two of the most important mechanisms in the regulation of
embryonic vascular development and tumor angiogenesis (34). According to our recent study,
Notch signaling affects ovarian carcinomas and Notch1
expression correlates with metastasis, while Jagged1
expression correlates with tumor grade (27). However, it was demonstrated that in
spheroids, all genes in Notch signaling are significantly
upregulated, particularly Jagged1, DLL3 and
Notch1. High Jagged1 expression has been demonstrated
to predict a worse outcome in breast cancer (35,36),
renal cell carcinoma (37) and
colon adenocarcinoma (38). It has
also been reported that high Jagged1 expression is
associated with prostate cancer recurrence (39). Furthermore, Jagged1 signaling
regulates hemangioma stem cell-to-pericyte/vascular smooth muscle
cell differentiation (40). The
abovementioned observations indicate that cellular organizations in
CSCs accompany vascular development or extracellular structuring
with the possible tendency of epithelial mesenchymal transition.
The most upregulated cyclin was CCND2, which is implicated
in cell differentiation and malignant transformation and is
inactivated by promoter hypermethylation in several types of human
cancer. High DNA methylation levels of CCND2 cause
deregulation of the G1/S checkpoint and correlate with
clinicopathological features of tumor aggressiveness in breast and
types of prostate cancer (41,42).
In conclusion, isolated CSCs in human tumors may
alter their cellular characterization with time and exhibit
differentiation by maintaining their former surface antigens at the
level of transcription or translation. This differentiation may be
a principal mechanism that is responsible for the malignant process
and tumor growth. As demonstrated in the current study, upregulated
genes of angiogenesis and mesenchymal transition or cellular
tendency to the vascular development appear to be due to malignancy
and tumor progression. Overall, these determinations indicated the
differentiation of CSCs, but must be further validated with a
series of patient samples derived from primary and/or metastatic
lesions of prostate cancer.
Acknowledgements
The authors would like to thank Professor Benjamin
Bonavida for his editorial expertise; Assistant Professor Ogun
Sercan for statistical analysis; Ummu Guven and Turker Cavusoglu
for their support in Cell Culture Laboratory maintenance and
inter-laboratory correlations; and Tayyibe Yeni and Merve Kurşuner
for flow cytometry and cell-sorting experiments in AREL. The
current study was funded by a grant from the Ege University
Scientific Research Project Fund (no. Ege-BAP 2008 Tıp-019).
The abstract for this study was previously
presented at the American Association for Cancer Research Annual
Meeting April 6–10, 2013 in Washington DC, USA and published as
abstract no. 255 in Cancer Research, Volume 73, Supplement 1,
2013.
References
|
1
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar
|
|
2
|
Vermeulen L, Sprick MR, Kemper K, Stassi G
and Medema JP: Cancer stem cells - old concepts, new insights. Cell
Death Differ. 15:947–958. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hill RP and Perris R: ‘Destemming’ cancer
stem cells. J Natl Cancer Inst. 99:1435–1440. 2007.
|
|
4
|
Miki J and Rhim JS: Prostate cell cultures
as in vitro models for the study of normal stem cells and cancer
stem cells. Prostate Cancer Prostatic Dis. 11:32–39. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Botchkina GI, Zuniga SZ, Das M, Wang Y,
Wang H, Zhu S, Savitt AG, Rowehl RA, Leyfman Y, Ju J, et al:
New-generation taxoid SB-T-1214 inhibits stem cell-related gene
expression in 3D cancer spheroids induced by purified colon
tumor-initiating cells. Mol Cancer. 9:1922010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shackleton M, Quintana E, Fearon ER and
Morrison SJ: Heterogeneity in cancer: cancer stem cells versus
clonal evolution. Cell. 138:822–829. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tang DG: Understanding cancer stem cell
heterogeneity and plasticity. Cell Res. 22:457–472. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y and Laterra J: Cancer stem cells:
distinct entities or dynamically regulated phenotypes? Cancer Res.
72:576–580. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Singh SK, Clarke ID, Terasaki M, Bonn VE,
Hawkins C, Squire J and Dirks PB: Identification of a cancer stem
cell in human brain tumors. Cancer Res. 63:5821–5828.
2003.PubMed/NCBI
|
|
10
|
Liu G, Yuan X, Zeng Z, Tunici P, Ng H,
Abdulkadir IR, Lu L, Irvin D, Black KL and Yu JS: Analysis of gene
expression and chemoresistance of CD133+ cancer stem
cells in glioblastoma. Mol Cancer. 5:672006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bussolati B, Bruno S, Grange C,
Buttiglieri S, Deregibus MC, Cantino D and Camussi G: Isolation of
renal progenitor cells from adult human kidney. Am J Pathol.
166:545–555. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suetsugu A, Nagaki M, Aoki H, Motohashi T,
Kunisada T and Moriwaki H: Characterization of CD133+
hepatocellular carcinoma cells as cancer stem/progenitor cells.
Biochem and Biophys Res Commun. 351:820–824. 2006.
|
|
13
|
Yin S, Li J, Hu C, Chen X, et al: CD133
positive hepatocellular carcinoma cells posess high capacity for
tumorigenicity. Int J Cancer. 120:1444–1450. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
O’Brien CA, Pollett A, Gallinger S and
Dick J: A human colon cancer cell capable of initiating timor
growth in immunodeficient mice. Nature. 445:106–110.
2007.PubMed/NCBI
|
|
15
|
Hermann PC, Huber SL, Herrler T, Aicher A,
Ellwart JW, Guba M, Bruns CJ and Heeschen C: Distinct populations
of cancer stem cells determine tumor growth and metastatic activity
in human pancreatic cancer. Cell Stem Cell. 1:313–323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Maitland NJ and Collins AT: Prostate
cancer stem cells: a new target for therapy. J Clin Oncol.
26:2862–2870. 2008. View Article : Google Scholar
|
|
17
|
Shmelkov SV, St Clair R, Lyden D and Rafii
S: AC133/CD133/Prominin-1. Int J Biochem Cell Biol. 37:715–719.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shmelkov SV, Butler JM, Hooper AT, Hormigo
A, Kushner J, Milde T, St Clair R, et al: CD133 expression is not
restricted to stem cells, and both CD133+ and
CD133− metastatic colon cancer cells initiate tumors. J
Clin Invest. 118:2111–2120. 2008.PubMed/NCBI
|
|
19
|
Beier D, Hau P, Proescholdt M, Lohmeier A,
Wischhusen J, Oefner PJ, Aigner L, Brawanski A, Bogdahn U and Beier
CP: CD133+ and CD133− glioblastoma-derived
cancer stem cells show differential growth characteristics and
molecular profiles. Cancer Res. 67:4010–4015. 2007.PubMed/NCBI
|
|
20
|
Joo KM, Kim SY, Jin X, Song SY, Kong DS,
Lee JI, Jeon JW, Kim MH, Kang BG, Jung Y, et al: Clinical and
biological implications of CD133-positive and CD133-negative cells
in glioblastomas. Lab Invest. 808:808–815. 2008.PubMed/NCBI
|
|
21
|
Orian-Rousseau V: CD44, a therapeutic
target for metastasising tumours. Eur J Cancer. 46:1271–1277. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ponta H, Sherman L and Herrlich PA: CD44:
from adhesion molecules to signaling regulators. Nat Rev Mol Cell
Biol. 4:33–45. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ugolkov AV, Eisengart LJ, Luan C and Yang
XJ: Expression analysis of putative stem cell markers in human
benign and malignant prostate. Prostate. 71:18–25. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Collins AT, Berry PA, Hyde C, Stower MJ
and Maitland NJ: Prospective identification of tumorigenic prostate
cancer stem cells. Cancer Res. 65:10946–10951. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Robertson FM, Ogasawara MA, Ye Z, Chu K,
Pickei R, Debeb BG, Woodward WA, Hittelman WN, Cristofanilli M and
Barsky SH: Imaging and analysis of 3D tumor spheroids enriched for
a cancer stem cell phenotype. J Biomol Screen. 15:820–829. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Acker H: Microenvironmental conditions in
multicellular spheroids grown under liquid-overlay tissue culture
conditions. Recent Results Cancer Res. 95:116–133. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Oktem G, Sanci M, Bilir A, Yildirim Y,
Kececi SD, Ayla S and Inan S: Cancer stem cell and embryonic
development-associated molecules contribute to prognostic
significance in ovarian cancer. Int J Gynecol Cancer. 22:23–29.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lai EC: Notch signaling: control of cell
communication and cell fate. Development. 131:965–973. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parsons P, Gilbert SJ, Vaughan-Thomas A,
Sorrell DA, Notman R, Bishop M, Hayes AJ, Mason DJ and Duance VC:
Type IX collogen interacts with fibronectin providing an important
molecular bridge in articular cartilage. J Biol Chem.
286:34986972011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang CC, Wang TC, Lin BH, Wang YW,
Johnson SL and Yu J: Collagen IX is required for the integrity of
the collagen II fibrils and the regulation of vascular plexus
formation in zebrafish caudalfins. Dev Biol. 332:360–370. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Piotrowski A, Benetkiewicz M, Menzel U, et
al: Microarray-based survey of CpG islands identifies concurrent
hyper- and hypomethylation patterns in tissues derived from
patients with breast cancer. Genes Chromosomes Cancer. 45:656–667.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Schmitt AM, Riniker F, Anlauf M, Schmid S,
Soltermann A, Moch H, Heitz PU, Klöppel G, Komminoth P and Perren
A: Islet 1 (Isl1) expression is a reliable marker for pancreatic
endocrine tumorsand their metasteses. Am J Surg Pathol. 32:420–425.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Barzelay A, Ben-Shoshan J, Entin-Meer M,
Maysel-Auslender S, Afek A, Barshack Keren G and George J: A
potential role for islet-1 in post-natal angiogenesis and
vasculogenesis. Thromb Haemost. 103:188–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li JL and Harris AL: Crosstalk of VEGF and
Notch pathways in tumour angiogenesis: therapeutic implications.
Front Biosci. 14:3094–3110. 2009.PubMed/NCBI
|
|
35
|
Sethi N, Dai X, Winter CG and Kang Y:
Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast
cancer by engaging notch signaling in bone cells. Cancer Cell.
19:192–205. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Reedijk M, Odorcic S, Chang L, Zhang H,
Miller N, McCready DR, et al: High-level coexpression of JAG1 and
NOTCH1 is observed in human breast cancer and is associated with
poor overall survival. Cancer Res. 65:8530–8537. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Wu K, Xu L, Zhang L, Lin Z and Hou J: High
Jagged1 expression predicts poor outcome in clear cell renal cell
carcinoma. Jpn J Clin Oncol. 41:411–416. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Gao J, Liu J, Fan D, Xu H, Xiong Y, Wang
Y, Xu W, Wang Y, Cheng Y and Zheng G: Up-regulated expression of
Notch1 and Jagged1 in human colon adenocarcinoma. Pathol Biol
(Paris). 59:298–302. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Santagata S, Demichelis F, Riva A,
Varambally S, Hofer MD, Kutok JL, et al: JAGGED1 expression is
associated with prostate cancer metastasis and recurrence. Cancer
Res. 64:6854–6857. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Boscolo E, Stewart CL, Greenberger S, Wu
JK, Durham JT, Herman IM, Mulliken JB, Kitajewski J and Bischoff J:
JAGGED1 signaling regulates hemangioma stem cell-to
pericyte/vascular smooth muscle cell differentiation. Arterioscler
Thromb Vasc Biol. 31:2181–2192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Sharma G, Mirza S, Prasad CP, Srivastava
A, Gupta SD and Ralhan R: Promoter hypermethylation of p16INK4A,
p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast
cancer patients. Life Sci. 80:1873–1881. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Henrique R, Costa VL, Cerveira N, Carvalho
AL, Hoque MO, Ribeiro FR, Oliveira J, Teixeira MR, Sidransky D and
Jeronimo C: Hypermethylation of Cyclin D2 is associated with loss
of mRNA expression and tumor development in prostate cancer. J Mol
Med. 84:911–918. 2006. View Article : Google Scholar : PubMed/NCBI
|