Introduction
Chromosomal translocations and gene mutations are
common genetic abnormalities observed in leukemia patients
(1). In total, ~50% of patients
with acute myeloid leukemia (AML) carry a distinct chromosomal
translocation, such as t(8;21) (q22;q22) or t(8;21), the latter of
which ~10% of all AMLs exhibit and is considered to be an AML group
that is associated with a favorable prognosis. The t(8;21)(q22;q22)
and inv(16)(p13.1;q22)/t(16;16)(p13.1;q22)
chromosomal alterations are the most common genetic abnormalities
and give rise to the AML1-ETO and CBFB-MYH11 fusion
genes, respectively. As AML1 encodes the α subunit of the
core-binding factor (CBF) and CBFB encodes its β subunit,
these two gene fusions interfere with normal CBF function.
Therefore, AML with AML1-ETO or CBFB-MYH11 is termed
CBF-AML and accounts for 15% of AML cases worldwide (2,3).
The c-kit gene is located on chromosome
4q11–12 and encodes a 145-kDa type III receptor tyrosine kinase.
c-kit has five extracellular immunoglobulin-like domains, a
juxtamembrane domain and an intracellular kinase domain.
c-kit mutations have been identified in ≥70% of
gastrointestinal stromal tumors, ≥90% of mastocytosis and ~10% of
germ cell tumors (4,5). In addition, c-kit mutations
have been found in 12–25% of CBF-AML cases (6). It has also been reported that CBF-AML
cases exhibiting a c-kit mutation are associated with a
higher rate of relapse and a poor prognosis (7,8). Thus,
the c-kit mutation may be a prognostic factor for
CBF-AML.
Various methods have been used to detect
c-kit mutations and one of the most common methods is the
amplification refractory mutation system (9). However, its application is limited due
to the requirement for high primer concentrations, its ability to
only detect a small quantity of mutation sites and the complexity
of the detection process. High-resolution melting analysis
(10) detects DNA mutations based
on the melting characteristics of the DNA molecules. It is an
additional method that is relatively simple, however, it may be too
sensitive as the ion concentrations in the samples may affect the
results. Currently available hybridization probes (11) only detect mutations around the hot
spot at D816 and, although frequently used at present, denaturing
high-performance liquid chromatography combined with direct
sequencing (12) requires the
polymerase chain reaction (PCR) products to be post-processed,
which may result in contamination. Furthermore, this method is
complex and not applicable for mutation detection in clinical
samples. Therefore, a simple, accurate and highly efficient method
is required for detecting c-kit mutations.
Our previous study established a novel melting
curve-based method for detecting gene mutations (13). In the present study, a unique probe
arrangement was designed to establish a novel melting curve-based
method for detecting c-kit mutations. The results
demonstrated that this method detected the majority of mutations at
the exon 17 hot spot. Furthermore, this method is advantageous due
to its simplicity combined with its high sensitivity and
specificity.
Materials and methods
Clinical samples
Bone marrow (2 ml) or peripheral blood (5 ml)
samples were collected from 107 patients with leukemia at the
Zhongshan Hospital of Xiamen University (Xiamen, China), between
July 2008 and January 2010. All patients were diagnosed in
accordance with the leukemia diagnostic standards (14), which was confirmed by morphological
and immunophenotypic analyses of the bone marrow. Of the samples,
12 were from CBF-AML patients who were positive for AML-ETO.
The patients provided written informed consent for the collection
of the bone marrow and blood samples for the diagnostic and study
purposes in accordance with the principles outlined in the Code of
Ethics of the World Medical Association (Declaration of Helsinki).
The experimental procedures were performed following the guidelines
of the Xiamen University Medical Research Council and were approved
by the ethics committee of the Zhongshan Hospital of Xiamen
University.
DNA extraction
Genomic DNA was extracted using a Qiagen genomic DNA
extraction kit [Tiangen Biotech (Beijing) Co., Ltd., Beijing,
China] within 24 h of the collection of the blood samples. The DNA
concentration was measured using spectrophotometry (UV-2450/2550;
Shimadzu Corp., Kyoto, Japan); the absorbance was measured at 260
nm and the DNA samples were diluted to a concentration of 10
ng/ml.
Primer and probe design
Primer Premier v5.00 (Premier Biosoft, Palo Alto,
CA, USA) and Tm Utility v1.3 (Sangon Biotech (Shanghai) Co., Ltd.
(Shanghai, China) software packages were used to design the primers
and probes. The primers and probes were synthesized by Sangon
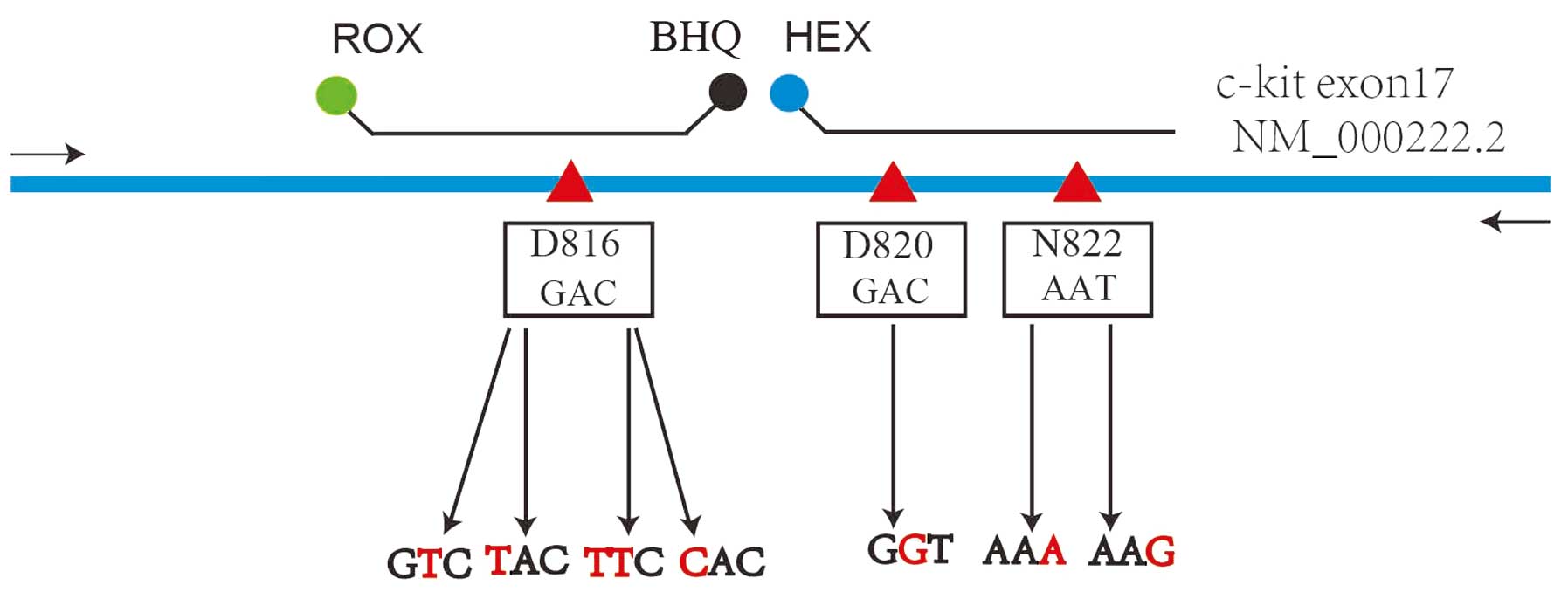
Biotech (Shanghai) Co., Ltd. The probes contained the following two
segments: i) A self-quenched probe segment labeled with a
carboxyrhodamine (ROX) fluorophore at its 5′ end (the first three
basic groups were thiophosphorylated to prevent shearing of the
fluorophore-carrying basic group by the DNA polymerase) and a black
hole quencher (BHQ) at its 3′ end; and ii) a probe segment labeled
with a hexachlorofluorescein (HEX) fluorophore at its 5′ end and a
NH2 group at its 3′ end to prevent probe extension by
the DNA polymerase (Fig. 1,
Table I). There were three basic
groups between the two sequences; therefore, the quenching group of
the first sequence was able to function with the two fluorophores.
This resulted in the formation of a single probe in the first
sequence and the formation of a hybridization probe when combined
with the second sequence. In the combined probe that contained the
two segments, the sequence of the first segment was designed to
detect mutations around D816, and the sequence of the second
segment was designed to detect mutations at N820 and N822. The
hybridization of the probe to the target sequence that contained
sequences around D816 alone, N820/N822 alone, or D816 and N820/N822
together enabled the signal from ROX alone, HEX alone, or ROX and
HEX together to be detected. Furthermore, the melting curve
analysis indicated the presence of unique sequences (a single peak,
which was unique to the wild-type (WT) or mutant sequence) or
mixtures of the sequences (multiple peaks, each corresponding to
the WT or mutant sequence).
 | Table IPrimer and probe seqences. |
Table I
Primer and probe seqences.
| A, Primers |
|---|
|
|---|
| Description | Sequence (5′ to
3′) |
|---|
| d-Kit17-F1 |
ACAGAGACTTGGCAGCCAGAA |
| d-Kit17-R |
TTGCAGGACTGTCAAGCAGAG |
|
| B, Probes |
|
| Description | Sequence (5′ to
3′) |
|
| D816-ROXa |
ROX-TGGTCTAGCCAGAGaCATCAA-BHQ |
| N822-HEXa |
HEX-TGATTCTAATTATGTGGTTAAA-NH2 |
Construction of mutation-positive
plasmids
Using genomic DNA from 293T human embryonic kidney
cells as the template, mutation-positive control plasmids were
constructed using the overlap extension PCR method (15,16).
The plasmids contained the following c-kit WT or mutant
sequences: D816WT, D816V, D816Y, D816H, D816F, N822K(A), N822K(G)
and N820G. D816WT contained the WT sequence, while in D816V, the
GAC codon for amino acid 816 was mutated to GTC, resulting in a D
(aspartic acid) to V (valine) change. The relevant plasmid
sequences are listed in Table
II.
 | Table IISequences of the different
plasmids. |
Table II
Sequences of the different
plasmids.
| Mutation | Sequence |
|---|
| D816WT |
…gtgattttggtctagccagagacatcaagaatgattctaattatgtggttaaa… |
| D816V |
…GtgattttggtctagccagagTcatcaagaatgattctaattatgtggttaaa… |
| D816Y |
…gtgattttggtctagccagaTacatcaagaatgattctaattatgtggttaaa… |
| D816H |
…gtgattttggtctagccagaCacatcaagaatgattctaattatgtggttaaa… |
| D816F |
…gtgattttggtctagccagaTTcatcaagaatgattctaattatgtggttaaa… |
| N820G |
…gtgattttggtctagccagagacatcaagaatgGttctaattatgtggttaaa… |
| N822K(A) |
…gtgattttggtctagccagagacatcaagaatgattctaaAtatgtggttaaa… |
| N822K(G) |
…gtgattttggtctagccagagacatcaagaatgattctaaGtatgtggttaaa… |
PCR amplification and mutation
detection
The PCR reactions contained 1× sequence-specific
primer buffer [67 mM Tris-HCl, 16.6 mM
(NH4)2SO4, 6.7 μM EDTA and 0.085
mg/ml bovine serum albumin], 4 mM Mg2+, 0.2 mM dNTPs, 1
pmol upstream primer, 10 pmol downstream primer, 2 pmol D816-ROX
probe, 2 pmol N822-HEX probe, 1 unit of Taq HS, 5 μl of template
DNA and ddH2O in a final volume of 25 μl.
Amplification was conducted using a Gene-pro Gene
Amplifier (Bioer Biotechnology Co., Ltd., Hangzhou, China) with 50
cycles of 95°C for 20 sec, 52°C for 30 sec and 72°C for 30 sec. The
melting curves were analyzed using a CFX96 Real-Time PCR detection
system (Bio-Rad, Hercules, CA, USA) and measurement of fluorescence
(HEX and ROX channels) at 0.5°C increments was performed between 35
and 80°C.
Sensitivity testing
The mutation-positive control plasmids were diluted
to 2×103 copies/μl. The WT and control plasmids were
used as templates to produce mixtures with 50, 25, 10, 5 and 1% of
the plasmids that contained the individual mutations. The plasmid
mixtures were used as templates for amplification and mutation
detection. The samples were tested in duplicate, together with a
WT-positive control and template-free negative control.
Sample detection and sequencing
In total, 5-μl aliquots of DNA samples from patients
were used for amplification and melting curve analysis. In
addition, the 12 PCR products from the CBF-AML patients were
sequenced using a commercial sequencing service (Major Biosystem
Co., Ltd., Shanghai, China). The results of the sequencing analysis
of the patient DNA samples were compared with those of the
mutation-positive control plasmids.
Results
Sensitivity of the mutation detection
system
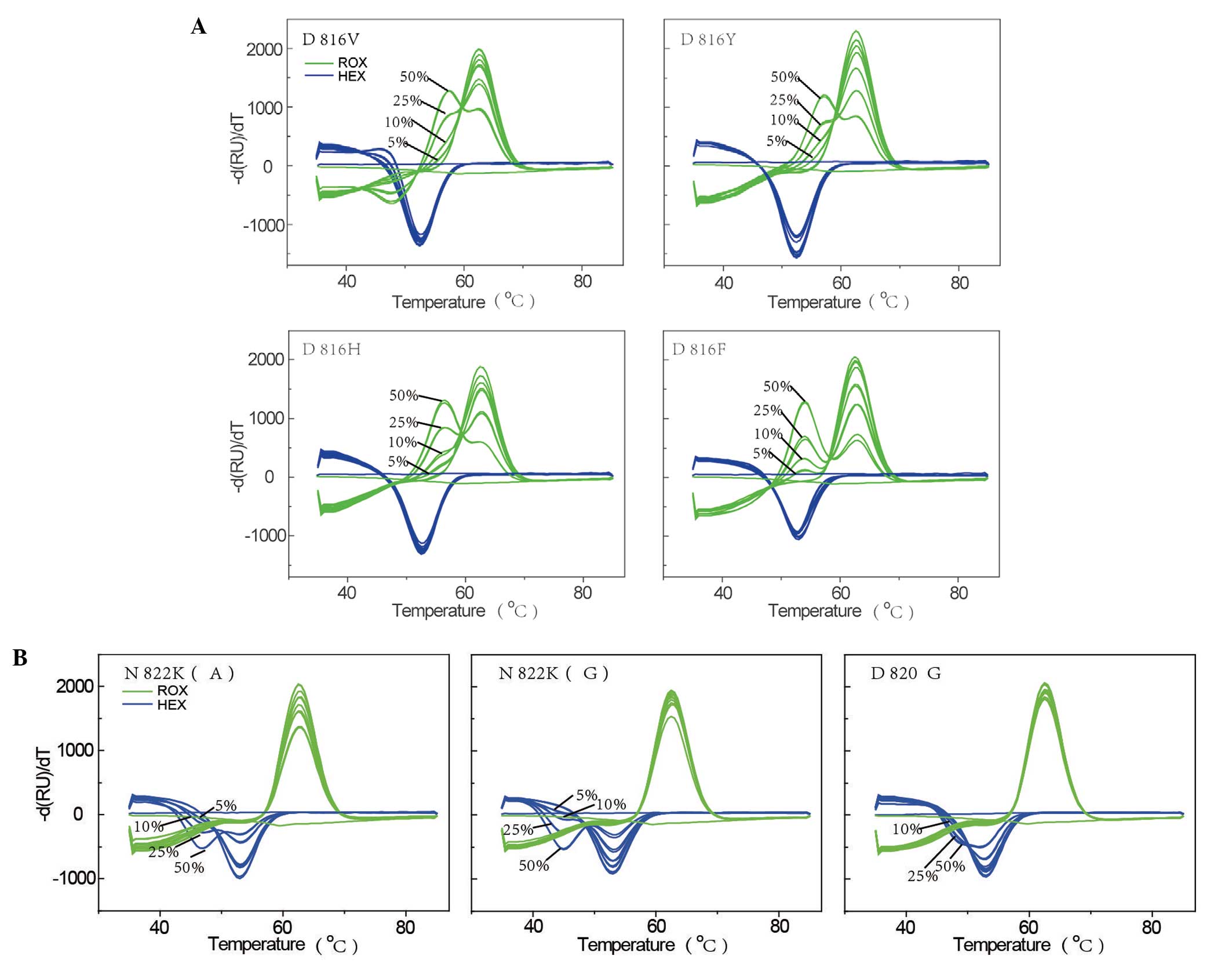
To test the sensitivity of the novel system, the
plasmid mixtures containing the plasmid with the WT c-kit
sequence and each of the seven plasmids carrying c-kit
mutations were examined. The mutations included four D816 mutations
(D816V, D816Y, D816H, and D816F), two N822 mutations [N822K(A) and
N822K(G)], and a N820 mutation (N820G). The results of the
sensitivity analysis are shown in Fig.
2. For the four D816 mutations, the signal from the ROX channel
for the WT plasmid exhibited only one melting peak (at ~62.5°C),
whereas the 50, 25, 10 and 5% plasmid mixtures exhibited double
peaks that clearly differed from that of the WT plasmid. Double
peaks were not evident for the 1% mixture, indicating that the
detection sensitivity for the four D816 mutations was ~5%. For the
remaining mutations, the signal from the HEX channel for the WT
plasmid exhibited only one melting peak (at ~52.5°C), while the
mixed plasmids exhibited double peaks. The detection sensitivity
for N822K(A) and N822K(G) was 5%, while that of N820G was 10%.
Melting curve and sequencing analyses of
CBF-AML samples
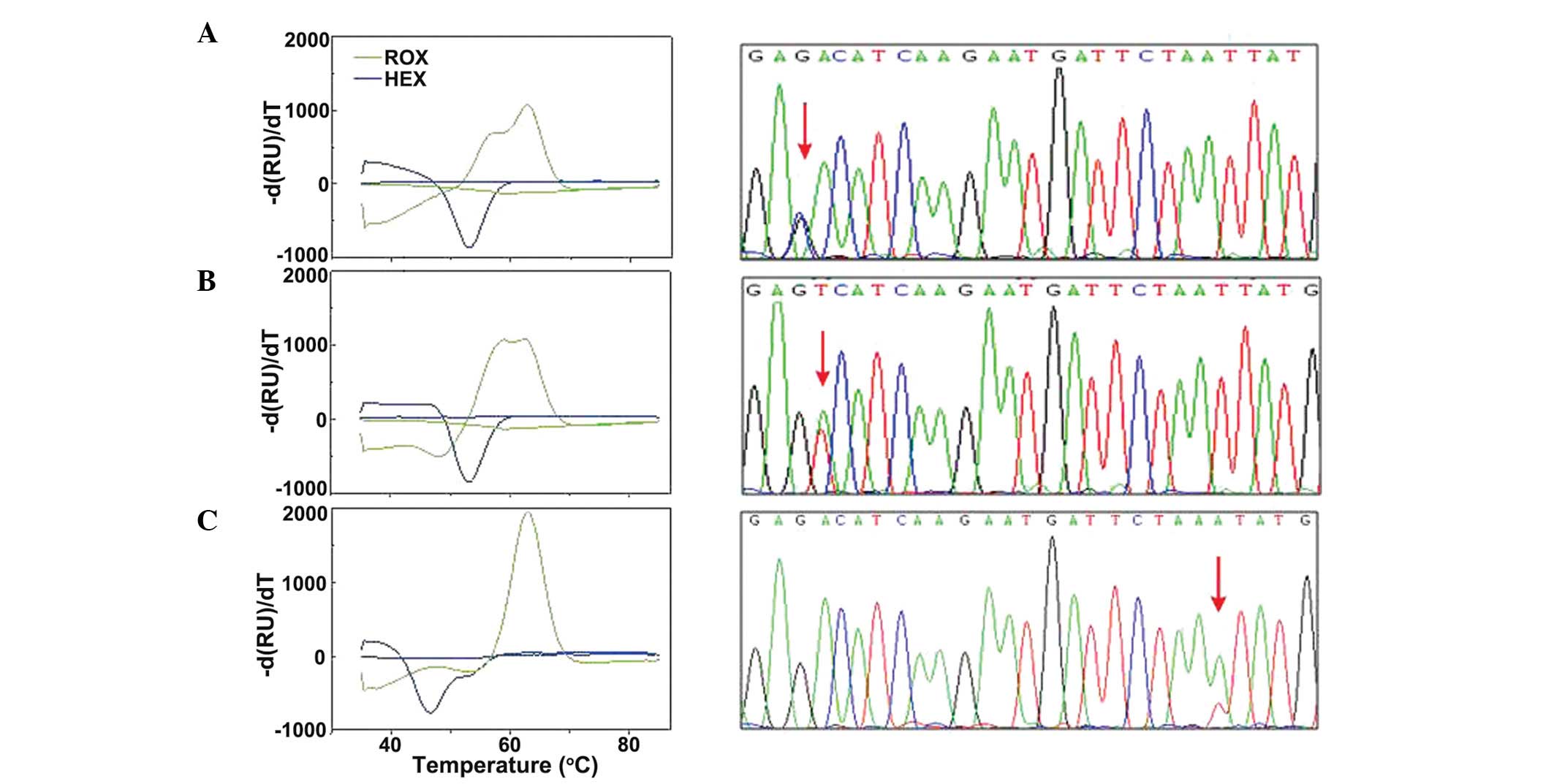
The results of the melting curve and sequencing
analyses for the 12 CBF-AML patient-derived samples are shown in
Fig. 3. The ROX signal identified
four samples for which the melting curve was different from that of
the WT sequence, indicating a mutation at D816. Three samples
exhibited a single peak at 57.5°C, however, they were clearly
different from the WT peak at 62°C. The final sample that was
different exhibited a melting peak at 62°C (WT) and an additional
peak at 56.5°C.
The HEX signal identified one sample with an
abnormal HEX melting peak, with a melting peak at 54°C (WT) and an
additional peak at 46°C, indicating the presence of a mutation at
N820 or N822. Sequencing analysis of the 12 samples supported the
melting curve data. Among the five c-kit mutation-positive
samples, three different mutations were identified: A D816H
mutation (one sample), a D816V mutation (three samples) and a
N822K(A) mutation (one sample; Fig.
3). No mutations were detected in the remaining six CBF-AML
cases.
Melting curve analysis of non-CBF-AML
samples
To assess the c-kit mutation rate in samples
from patients with other types of leukemia, the novel method was
used to analyze 95 non-CBF-AML samples, including 58 AML (negative
for AML1-ETO and CBFB-MYH11), 25 acute lymphoblastic
leukemia, 10 chronic myelocytic leukemia and two chronic
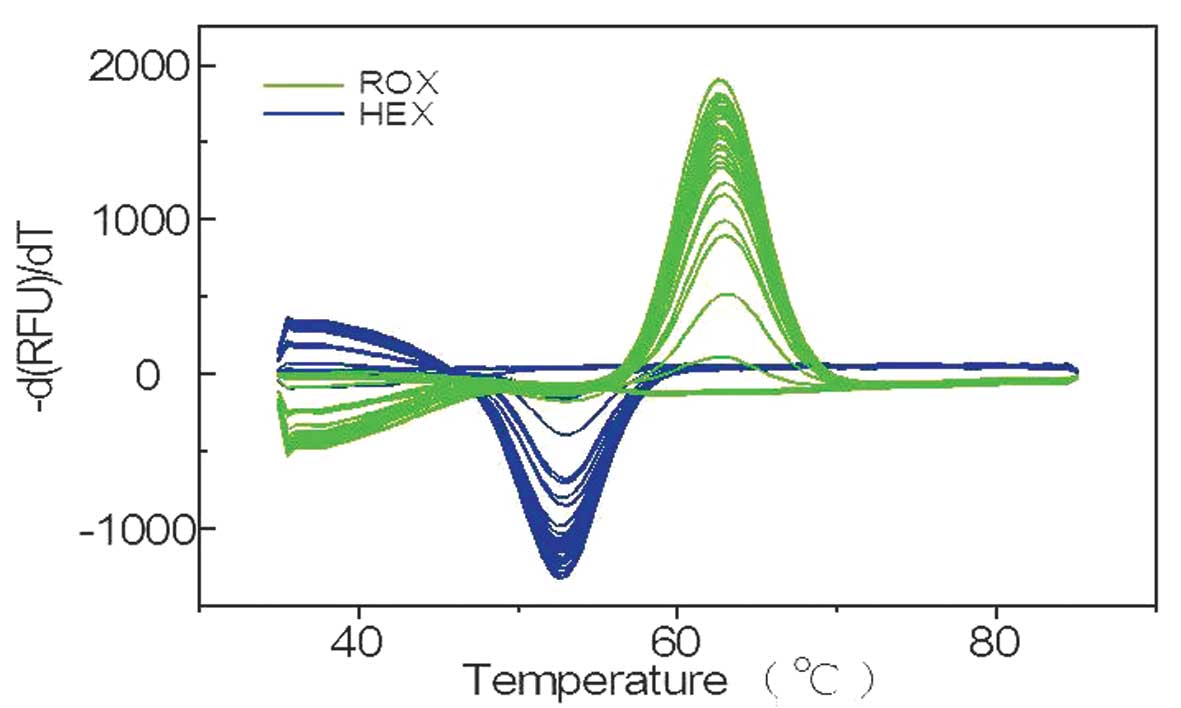
eosinophilic leukemia samples. Representative data for the 31
samples are shown in Fig. 4. Two
samples exhibited no amplification signals, while the remaining 29
ROX and HEX signals exhibited single melting peaks, indicating that
the signals were negative for c-kit mutations.
Discussion
Aberrant c-kit in t(8;21) AML has been
reported in the extracellular domain (encoded in exon 8), the
juxtamembrane domain (encoded in exons 10 and 11) and the A-loop
domain with tyrosine kinase activity (encoded in exon 17). Certain
previous studies reported that the D816V mutation (in exon 17)
confers increased tumor growth and antiapoptotic potential compared
with mutations in the extracellular or juxtamembrane domains
(17,18). Therefore, it was hypothesized that
the development of a highly sensitive method for detecting
c-kit mutations at exon 17 is required and may facilitate
the appropriate management of AML.
The current study modified a previously described
hybridization probe technique (13), where a single quencher was used to
quench two fluorophores on the probe. In this modified probe
method, the anterior segment of the probe was an independent
self-quenching probe labeled with a fluorophore (ROX) at its 5′ end
and a BHQ at its 3′ end. In addition, the first three basic groups
at the 5′ end were thiophosphorylated to prevent the shearing
effects that are caused by the excision step of DNA polymerase on
the fluorophore-carrying basic group. In the annealing step, the
probe hybridized to the amplification product and product-specific
unique sequence information was obtained from the melting curve
analysis. The posterior segment of the probe was an oligonucleotide
labeled with a different fluorophore (HEX) at its 5′ end and the
sequence of this oligonucleotide allowed for hybridization with the
front half of the probe, enabling the hybridization of the
amplification products during annealing. The melting curve analysis
of the probe-covered regions directly reflected the sequence of the
region. This modified probe is advantageous as it provides sequence
information by overlapping the two segments of the probe, whereas
the original hybridization probe only reveals sequence information
in the region that is overlapped by the fluorescent probe. However,
as the region that is covered by two segments of the probe is long,
the detection of one self-quenched or molecular probe may not
provide sufficiently high fluorescence signals or may fail to
detect mutations due to reduced sensitivity.
It is known that WT DNA may interfere with the
detection of mutant DNA. Therefore, it is important to analyze
sensitivity. The gold standard sensitivity for c-kit
mutation detection has been set at 20% (11). The method used in the current study
exceeded this threshold for sensitivity for all the mutations
analyzed; the sensitivity was 10% for N820G and 5% for the other
six mutations tested.
In the present study, c-kit mutations were
identified in six of the 12 AML-ETO-positive samples,
yielding a positivity rate (50%) comparable with those previously
reported; 12.8–46.8% (19–21). Furthermore, to evaluate c-kit
mutations in non-CBF-AML cases, c-kit mutations were also
analyzed in 95 samples obtained from non-CBF-AML patients. As
predicted, no c-kit mutations were identified, which
indicates that the c-kit mutation is rare in non-CBF-AML
cases.
In conclusion, the method described in the present
study is simple and rapid, and exhibits high sensitivity and
specificity. This modified probe method may facilitate the
classification and individual treatment of patients with
CBF-AML.
Acknowledgements
The present study was partially supported by funds
from the National Nature Science Fund (No 81172246)
References
|
1
|
Döhner K and Döhner H: Molecular
characterization of acute myeloid leukemia. Haematologica.
93:976–982. 2008.
|
|
2
|
Beghini A, Peterlongo P, Ripamonti CB, et
al: C-kit mutations in core binding factor leukemias. Blood.
95:726–727. 2000.
|
|
3
|
Dombret H, Preudhomme C and Boissel N:
Core binding factor acute myeloid leukemia (CBF-AML): is high-dose
Ara-C (HDAC) consolidation as effective as you think? Curr Opin
Hematol. 16:92–97. 2009.
|
|
4
|
Heinrich MC, Blanke CD, Druker BJ and
Corless CL: Inhibition of KIT tyrosine kinase activity: a novel
molcular approach to the treatment of KIT-positive malignancies. J
Clin Oncol. 20:1692–1703. 2002.
|
|
5
|
Roskoski R Jr: Structure and regulation of
Kit protein-tyrosine kinase - the stem cell factor receptor.
Biochem Biophys Res Commun. 338:1307–1315. 2005.
|
|
6
|
Mrózek K and Bloomfield CD: Chromosome
aberrations, gene mutations and expression changes, and prognosis
in adult acute myeloid leukemia. Hematology Am Soc Hematol Educ
Program. 169–177. 2006.
|
|
7
|
Nanri T, Matsuno N, Kawakita T, et al:
Mutations in the receptor tyrosine kinase pathway are associated
with clinical outcome in patients with acute myeloblastic leukemia
harboring t(8;21)(q22;q22). Leukemia. 19:1361–1366. 2005.
|
|
8
|
Schnittger S, Kohl TM, Haferlach T, Kern
W, Hiddemann W, Spiekermann K and Schoch C: KIT-D816 mutations in
AML1-ETO-positive AML are associated with impaired event-free and
overall survival. Blood. 107:1791–1799. 2006.
|
|
9
|
Corless CL, Harrell P, Lacouture M, et al:
Allele-specific polymerase chain reaction for the
imatinib-resistant KIT D816V and D816F mutations in mastocytosis
and acute myelogenous leukemia. J Mol Diagn. 8:604–612. 2006.
|
|
10
|
Fuster O, Barragán E, Bolufer P, et al:
Rapid detection of KIT mutations in core-binding factor acute
myeloid leukemia using high-resolution melting analysis. J Mol
Diagn. 11:458–463. 2009.
|
|
11
|
Sotlar K, Escribano L, Landt O, et al:
One-step detection of c-kit point mutations using peptide nucleic
acid-mediated polymerase chain reaction clamping and hybridization
probes. Am J Pathol. 162:737–746. 2003.
|
|
12
|
Paschka P, Marcucci G, Ruppert AS, et al;
Cancer and Leukemia Group B. Adverse prognostic significance of KIT
mutations in adult acute myeloid leukemia with inv(16) and t(8;21):
a Cancer and Leukemia Group B Study. J Clin Oncol. 24:3904–3911.
2006.
|
|
13
|
Huang Q, Liu Z, Liao Y, et al: Multiplex
fluorescence melting curve analysis for mutation detection with
dual-labeled, self-quenched probes. PloS One. 6:e192062011.
|
|
14
|
Vardiman JW, Thiele J, Arber DA, et al:
The 2008 revision of the World Health Organization (WHO)
classification of myeloid neoplasms and acute leukemia: rationale
and important changes. Blood. 114:937–951. 2009.
|
|
15
|
Higuchi R, Krummel B and Saiki RK: A
general method of in vitro preparation and specific mutagenesis of
DNA fragments: study of protein and DNA interactions. Nucleic Acids
Res. 16:7351–7367. 1988.
|
|
16
|
Heckman KL and Pease LR: Gene splicing and
mutagenesis by PCR-driven overlap extension. Nat Protoc. 2:924–932.
2007.
|
|
17
|
Kohl TM, Schnittger S, Ellwart JW,
Hiddemann W and Spiekermann K: KIT exon 8 mutations associated with
core-binding factor (CBF)-acute myeloid leukemia (AML) cause
hyperactivation of the receptor in response to stem cell factor.
Blood. 105:3319–3321. 2005.
|
|
18
|
Frost MJ, Ferrao PT, Hughes TP and Ashman
LK: Juxtamembrane mutant V560GKit is more sensitive to Imatinib
(STI571) compared with wild-type c-kit whereas the kinase domain
mutant D816VKit is resistant. Mol Cancer Ther. 1:1115–1124.
2002.
|
|
19
|
Wang YY, Zhou GB, Yin T, et al: AML1-ETO
and C-KIT mutation/overexpression in t(8;21) leukemia: implication
in stepwise leukemogenesis and response to Gleevec. Proc Natl Acad
Sci USA. 102:1104–1109. 2005.
|
|
20
|
Lück SC, Russ AC, Du J, et al: KIT
mutations confer a distinct gene expression signature in core
binding factor leukaemia. Br J Haematol. 148:925–937. 2010.
|
|
21
|
Paschka P, Marcucci G, Ruppert AS, et al;
Cancer and Leukemia Group B. Adverse prognostic significance of KIT
mutations in adult acute myeloid leukemia with inv(16) and t(8;21):
a Cancer and Leukemia Group B Study. J Clin Oncol. 24:3904–3911.
2006.
|