Introduction
Worldwide, hepatitis B virus (HBV) is an important
pathogen causing both acute and chronic liver diseases. Chronic HBV
carriers have an increased risk of developing hepatocellular
carcinoma (HCC). The mechanism of HBV-related carcinogenesis is
poorly understood. Almost all HBV-associated HCCs studied thus far
harbor chromosomally integrated HBV DNA (1–3). The
integrated HBV DNA can encode two types of transcriptional
transactivators: The well studied HBxAg and the preS2
transactivators, large hepatitis B virus surface proteins (LHBs)
and C-terminally truncated middle size surface proteins
(MHBst) (4–6). The wild-type middle hepatitis B virus
surface protein (MHBs) consists of a 55-amino acid (aa)
preS2-domain and a 226 aa S-domain. To generate the transactivating
forms of MHBs, deletion of at least 87 C-terminal aa is required,
and the transcriptional transactivator function is based on the
cytoplasmic orientation of the preS2-domain (7). Considerable advances have been made in
the fields of transcriptional transactivation of MHBst;
however, the molecular basis of MHBst-dependent
transcriptional transactivation remains enigmatic. Studies in this
area will provide a better understanding of the association between
HBV and host hepatocytes, and pave the way for elucidating the
pathogenesis of HBV-related HCC.
In the present study, the transactivating potential
of MHBst C-terminally truncated at aa position 167
(MHBst167) on Simian virus (SV40) early promoter was
analyzed and, subsequently, genes transactivated by
MHBst167 were screened using suppression subtractive
hybridization (SSH). SSH was designed to generate a cDNA library
that is enriched in differentially expressed sequences and, more
importantly, equalized for the number of individual cDNA species,
thus allowing the detection of rare transcripts. The full-length
genes from the library were searched for homologs in GenBank.
Finally, the association between the human proto-oncogene c-Myc and
MHBst167 was discussed.
Materials and methods
Construction of vectors
For construction of eukaryotic expression vector
pcDNA3.1(−)-MHBst167, the MHBst167 fragment
was PCR-amplified from pCP10 containing two copies of HBV DNA
subtype ayw (GenBank accession: U95551), with the forward primer
(5′-GCCGGGCCCATGCAGTGGAATTCCACAAC) containing an ApaI site
and reverse primer (5′-GGAAAGCTTCTATCCTGGAATTAGAGGACAAAC)
containing a HindIII site. The fragment was inserted into
the cloning vector pGEM-T (Promega, Madison, WI, USA), resulting in
pGEM-T-MHBst167. An ApaI-HindIII fragment
was isolated from the vector and inserted into
ApaI-HindIII-digested pcDNA3.1(−) (Invitrogen Life
Technologies, Carlsbad, CA, USA), resulting in
pcDNA3.1(−)-MHBst167. The pcDNA3.1(−)-MHBs coding for
intact HBV middle surface protein was constructed previously in the
laboratory (8). To construct the
reporter vector pCAT3-c-Myc, the promoter of human c-Myc was
amplified from the genomic DNA of the HepG2 human HCC cell line
(HBsAg-negative) by PCR with a pair of forward
(5′-GGTACCATCCTCTCTCGCTAATCTCC) and reverse (5′-AGATCTATGGG
CAGAATAGCCTCCCC) primers containing a KpnI and a
BglII site, respectively. The promoter fragment was inserted
into pGEM-T, yielding pGEM-T-c-Myc. A KpnI-BglII
fragment was isolated from the plasmid and inserted at the
KpnI-BglII sites of pCAT3-basic (promoterless;
Promega), resulting in the pCAT3-c-Myc reporter vector. DL2000 DNA
marker (Takara Biotechnology (Dalian) Co., Ltd, Dalian, China) was
used as a DNA molecular weight marker. All the vectors were
sequenced and digested with corresponding restriction enzymes to
confirm the sequence accuracy.
Cell culture and transient
transfection
HepG2 cells were purchased from the Cell Resource
Center, Shanghai Institutes for Biological Sciences, Chinese
Academy of Sciences (Shanghai, China) and were cultivated in
Dulbecco’s modified Eagle’s medium (Invitrogen Life Technologies)
containing 100 IU penicillin and 100 μg streptomycin per
milliliter, supplemented with 10% (v/v) heat-inactivated fetal
bovine serum (Hyclone Laboratories, Inc., Logan, UT, USA), at 37°C
in 5% CO2 and 90% relative humidity. The cells were
seeded on the day before transfection at a density of
8×105 cells per 35-mm dish and reached 50% confluence at
the time of transfection. All transfections were performed with
FuGene® 6 Transfection Reagent (Roche Applied Science,
Indianapolis, IN, USA) according to the manufacturer’s
instructions. The medium was changed 5 h after transfection and
cells were harvested 40–48 h following transfection. All
transfections and assays were repeated independently three times in
triplicate.
Detection of MHBst167
expression
mRNA from HepG2 cells transfected with
pcDNA3.1(−)-MHBst167 and pcDNA3.1(−) was isolated using
a QuickPrep Micro mRNA purification kit (Amersham Biosciences,
Little Chalfont, UK), and cDNA was reverse-transcribed from the
mRNA. MHBst167 expression was detected by reverse
transcription-polymerase chain reaction (RT-PCR) with
MHBst167-specific primers by using 35 amplification
cycles, and by western blotting using lysates of the HepG2 cells.
The extracts were boiled for 5 min and separated by
SDS-polyacrylamide gel electrophoresis (SDS-PAGE), and then
transferred to nitrocellulose membranes (Pierce Biotechnology,
Inc., Rockford, IL, USA). The membranes were reacted with
anti-pre-S2 mouse anti-human monoclonal antibody and HRP-labeled
goat anti-mouse polyclonal IgG as the primary and secondary
antibodies, respectively and then with a SuperSignal West Pico
Chemiluminescent Substrate working solution (Pierce Protein Biology
Products, Thermo Fisher Scientific, Inc., Rockford, IL, USA)
according to the manufacturer’s instructions. The immunoreactive
bands were visualized after exposure to X-ray film.
Detecting the effect of
MHBst167 on SV40 early promoter and c-Myc promoter
To detect the effect of MHBst167 on the
SV40 promoter, various quantities of reporter vector pCAT3-promoter
(Promega) containing CAT reporter gene controlled by the SV40
immediate early promoter element were transiently transfected into
HepG2 cells. Co-transfections were made with
pcDNA3.1(−)-MHBst167 (2.0 μg) + pCAT3-promoter (0.4 μg)
as the test group, and pcDNA3.1(−) (2.0 μg) + pCAT3-promoter (0.4
μg) and pcDNA3.1(−)-MHBs (2.0 μg) + pCAT3-promoter (0.4 μg) as the
control groups, respectively. To detect the effect of
MHBst167 on the c-Myc promoter, various quantities of
pCAT3-c-Myc were transiently transfected into HepG2 cells. The
co-transfections were made with 1.0 μg reporter vector pCAT3-c-Myc
and 0.5, 1.0, 1.5 and 2.0 μg effector vector
pcDNA3.1(−)-MHBst167. The relative CAT activity was
measured using enzyme-linked immunosorbent assay according to the
manufacturer’s instructions (CAT ELISA kit; Roche Applied
Science).
Generation and analysis of a subtracted
cDNA library
SSH was performed with the PCR-Select™ cDNA
subtraction kit (Clontech Laboratories, Inc., Mountain View, CA,
USA) according to the manufacturer’s instructions. In brief, 2.0 μg
of poly A+ mRNA, each from the pcDNA3.1(−)-MHBst167
tester group and the pcDNA3.1(−) driver group was subjected to cDNA
synthesis, respectively. Following restriction with RsaI,
small sizes of cDNAs were obtained. The tester cDNAs were then
subdivided into two parts, ligated with the specific adaptor 1 and
adaptor 2, respectively. After two subtractive hybridization
reactions and two suppression PCR amplifications, differentially
expressed cDNAs were selectively amplified. Subsequently, the
second PCR products were used as templates for PCR amplification of
G3PDH (a housekeeping gene) at 18, 23, 28, 33 cycles, respectively,
to analyze subtraction efficiency. The second PCR products were
directly purified using the Wizard® PCR-Preps DNA
Purification system (Promega), and inserted into pGEM-T Easy
(Promega) to construct the subtracted library. Colony PCRs were
conducted to confirm that the size of the cDNA inserts ranged
between 200 and 1,000 bp by using T7/SP6 specific primers localized
in pGEM-T Easy. Following DNA sequencing of the positive colonies,
nucleotide homology searches were performed using the BLAST program
at NCBI (http://blast.st-va.ncbi.nlm.nih.gov/Blast.cgi).
Detecting the effect of
MHBst167 on c-Myc expression
To detect the effect of MHBst167 on c-Myc
mRNA, total RNA was extracted from HepG2 cells transiently
transfected by pcDNA3.1(−) and pcDNA3.1(−)-MHBst167
using TRIzol reagent (Invitrogen Life Technologies) according to
the manufacturer’s instructions, and was used for RT-PCR. The
following primers (sense, 5′-TTCGGGTAGTGGAAAACCAG and antisense,
5′-CAGCAGCTCGAATTTCTTC) were used to amplify the c-Myc cDNA.
β-actin specific primers (sense, 5′-TGACGGGGTCACCCACACTGTGCCCATCTA
and antisense, 5′-CTAGAAGCATTTGCGGTGGACGATGGAGGG) was used as
internal reference. To detect the effect of MHBst167 on
c-Myc protein, total soluble proteins were extracted in
radioimmunoprecipitation assay buffer (Pierce Biotechnology, Inc.)
from the transfected HepG2 cells and separated on 12.5% SDS-PAGE
gels for immunoblotting assay (prepared in house, Institute of
Infectious Diseases, Beijing Ditan Hospital, Capital Medical
University, Beijing, China). The expression of c-Myc was probed by
mouse monoclonal antibody against human c-Myc derived from a cell
line from American Type Culture Collection, Manassas, VA, USA.
Mouse anti-human monoclonal β-actin antibody (Santa Cruz
Biotechnology, Inc., Santa Cruz, CA, USA) was used as internal
reference.
Results
Transient expression of
MHBst167 in HepG2 cells
Digestion of recombinant vector
pcDNA3.1(−)-MHBst167 with ApaI/HindIII,
EcoRI, XbaI and XhoI yielded the expected
bands (data not shown). DNA sequencing results indicated that the
recombinant vector contained HBV DNA fragment encoding the
truncated middle surface protein in-frame and the sequence was
completely correct. MHBst167 mRNA and protein expression
in HepG2 cells was successfully detected by RT-PCR (Fig. 1A) and western blotting (Fig. 1B), respectively.
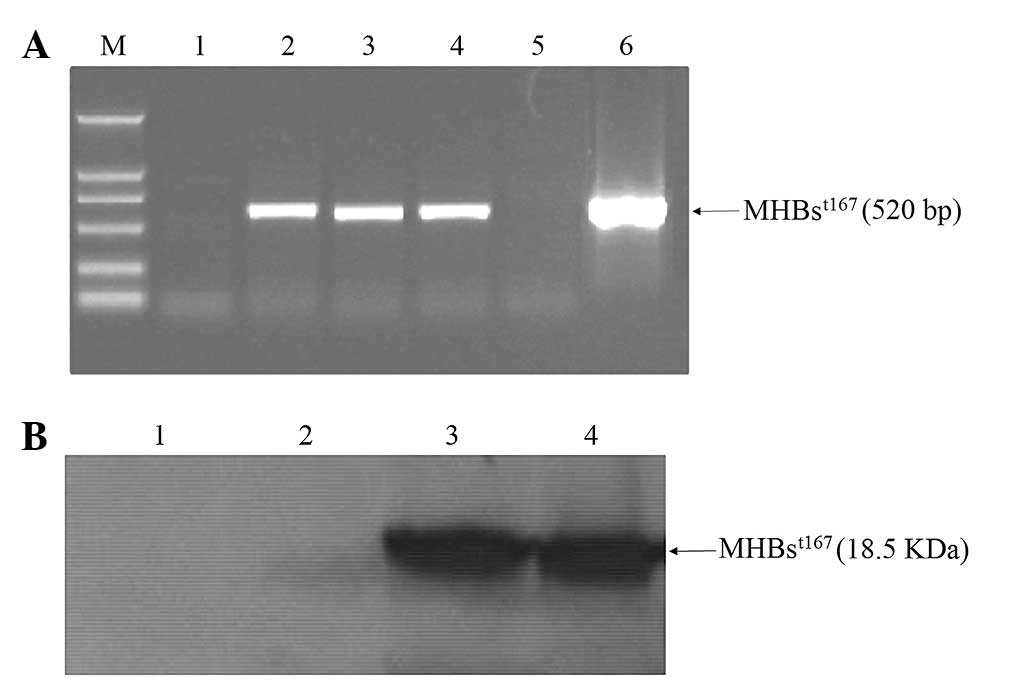 | Figure 1Transient expression of
MHBst167 in HepG2 cells. (A) Products of reverse
transcription-polymerase chain reaction amplification of
MHBst167 mRNA. Lane 1, mRNA from HepG2 cells; lanes 2–4,
mRNA from HepG2 cells transfected with
pcDNA3.1(−)-MHBst167; lane 5, mRNA from HepG2 cells
transfected with pcDNA3.1(−); lane 6,
pcDNA3.1(−)-MHBst167 vector positive control; M, DL2000
DNA marker. (B) Western blot analysis of MHBst protein.
Lane 1, lysates from HepG2 cells; lane 2, lysates from HepG2 cells
transfected with pcDNA3.1(−); lanes 3 and 4, lysates from HepG2
cells transfected with pcDNA3.1(−)-MHBst167.
MHBst167, hepatitis B virus middle size surface protein
C-terminally truncated at amino acid position 167; M, molecular
weight marker. |
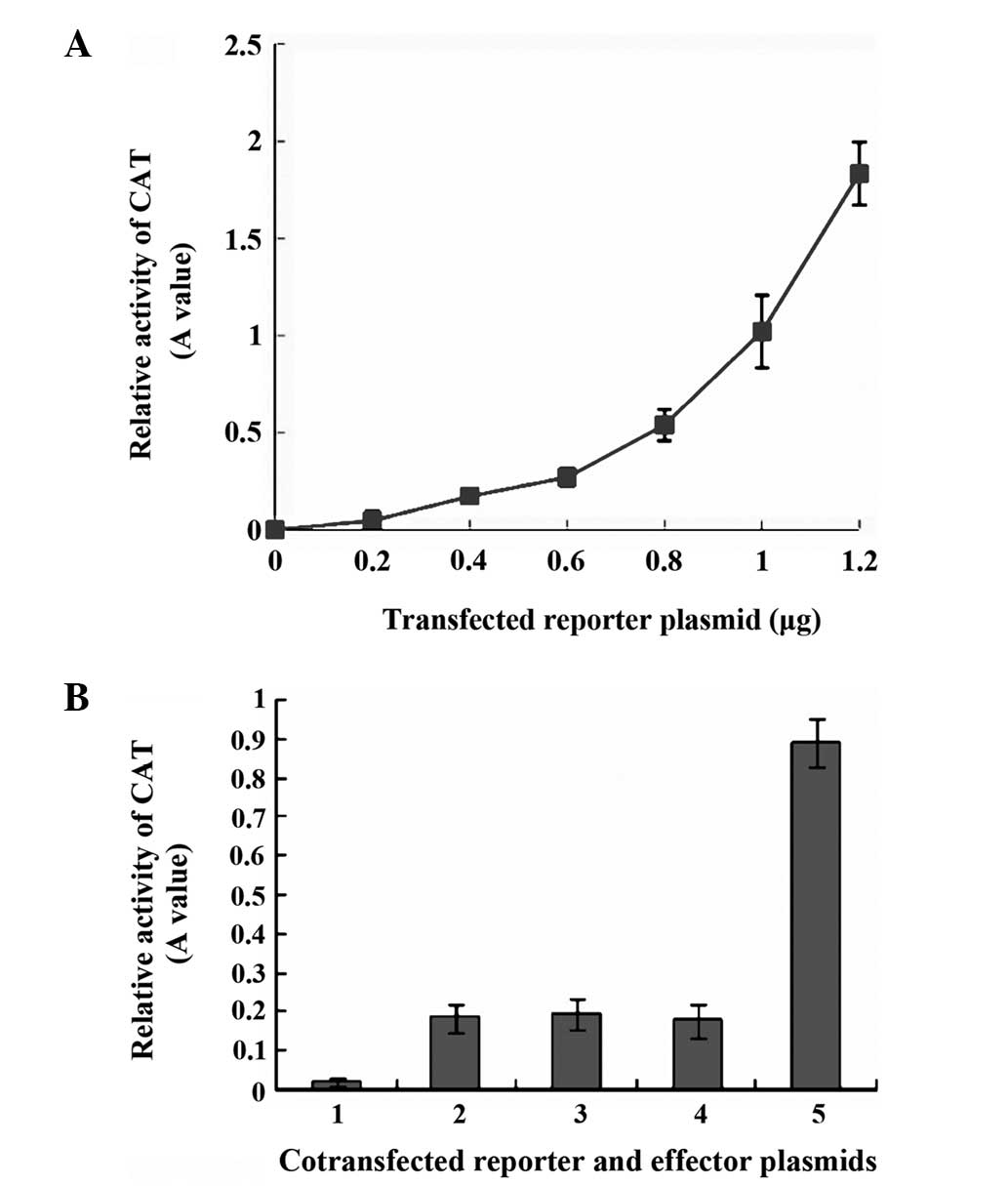
Transactivation of MHBst167 on
SV40 immediate early promoter
To determine the sensitivity of the kit for CAT
measurement, we first evaluated the CAT activities in total cell
lysates of HepG2 cells transfected with different quantities of
pCAT3-promoter. The results showed that CAT gene expression
exhibited an approximately linear association with the quantity of
reporter vector used in transfection (Fig. 2A). Subsequently, 0.4 μg of reporter
vector was selected to be used in the transfection experiments, so
that it was easy to detect the reporter activity with room for
further increase if a greater quantity of reporter vector was used.
In the transient co-transfection assays, CAT gene expression from
the pCAT3-promoter was ~4.5-fold higher following co-transfection
with pcDNA3.1(−)-MHBst167 compared with that after
co-transfection with pcDNA3.1(−) or pcDNA3.1(−)-MHBs. The marked
increase in CAT gene expression may be attributed to the
transactivating effect of the truncated HBV MHBst167 on
the SV40 early promoter element, leading to the observed increase
in CAT expression, whereas the intact MHBs was not transactive
(Fig. 2B).
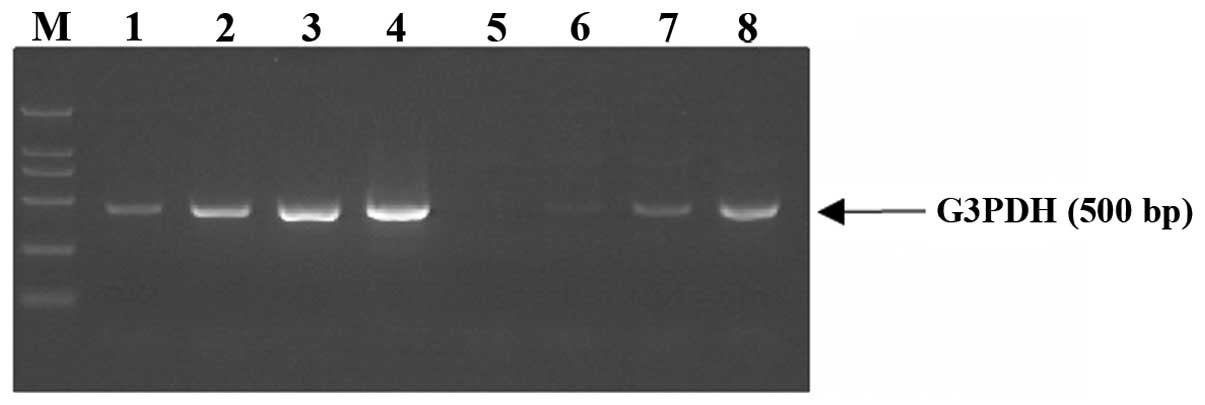
Analysis of the cDNA subtracted library
transactivated by MHBst167
To gain a general view of the genes which may be
involved in the pathogenesis of HBV, genes that were upregulated in
HepG2 cells expressing MHBst167 were identified by the
generation of a subtracted cDNA library. Subtraction efficiency
analysis showed that PCR products of the housekeeping gene G3PDH in
the unsubtracted library were obviously visible after 18 cycles;
however, 28 cycles were required in the subtracted library
(Fig. 3), indicating that the
abundance of non-differentially expressed genes was effectively
reduced and the subtraction method had a high subtraction
efficiency. Using SSH, a total of 94 positive clones were obtained.
These clones were prescreened by using PCR amplification to ensure
that they had different inserts before sequencing. Among these
clones, 77 contained inserts of 200–1,000 bp. A total of 50 clones
from the cDNA library were randomly chosen and sequenced, and their
nucleotide sequence homology searches were performed using the
BLAST program at NCBI. The analysis results showed that there were
22 coding sequences, of which 18 were known and 4 were unknown
genes. Some of the proteins coded by these genes have been shown to
be involved in cell cycle regulation, cell apoptosis, signal
transduction pathways and tumor development. Notably, the cell
proto-oncogene c-Myc was up-regulated by MHBst167. A
summary of the data is presented in Table I.
 | Table ISequence analysis of 42 clones
isolated from subtracted cDNA library transactivated by
MHBst167. |
Table I
Sequence analysis of 42 clones
isolated from subtracted cDNA library transactivated by
MHBst167.
| GenBank
accession | Gene description | Number of clones | Homology (%) |
|---|
| NM_001402 | Homo sapiens
eukaryotic translation elongation factor 1 alpha 1 (EEF1A1) | 8 | 98 |
| NM_001025 | Homo sapiens
ribosomal protein S23 (RPS23) | 6 | 100 |
| NM_212482 | Homo sapiens
fibronectin 1 (FN1) | 4 | 100 |
| NM_000477 | Homo sapiens albumin
(ALB) | 3 | 99 |
| NM_005004 | Homo sapiens NADH
dehydrogenase (ubiquinone) 1 beta subcomplex, 8, 19kDa
(NDUFB8) | 3 | 100 |
| NM_000014 | Homo sapiens
alpha-2-macroglobulin (A2M) | 2 | 100 |
| NM_000300 | Homo sapiens
phospholipase A2, group IIA (PLA2G2A) | 2 | 100 |
| NM_001354 | Homo sapiens
aldo-keto reductase family 1, member C2 (AKR1C2) | 2 | 96 |
| NM_002970 | Homo sapiens
spermidine/spermine N1-acetyltransferase (SSAT) | 2 | 100 |
| NM_002467 | Homo sapiens v-myc
avian myelocytomatosis viral oncogene homolog (MYC) | 2 | 98 |
| NM_001032281 | Homo sapiens tissue
factor pathway inhibitor (TFPI) | 1 | 99 |
| NM_002128 | Homo sapiens high
mobility group box 1 (HMGB1) | 1 | 100 |
| NM_000126 | Homo sapiens
electron-transfer-flavoprotein, alpha polypeptide (ETFA) | 1 | 99 |
| NM_000482 | Homo sapiens
apolipoprotein A-IV (APOA4) | 1 | 99 |
| NM_012073 | Homo sapiens
chaperonin containing TCP1, subunit 5 (CCT5) | 1 | 99 |
| NM_000687 | Homo sapiens
adenosylhomocysteinase (AHCY) | 1 | 99 |
| NM_000582 | Homo sapiens secreted
phosphoprotein 1 (SPP1) | 1 | 95 |
| NM_005141 | Homo sapiens
fibrinogen beta chain (FGB) | 1 | 100 |
MHBst167 upregulates the c-Myc
promoter activity
To examine the relationship between
MHBst167 and c-Myc, the effect of MHBst167 on
the promoter activity of c-Myc was investigated. The promoter
activity was evaluated using a CAT assay in HepG2 cells transfected
with pcDNA3.1(−)-MHBst167. As shown in Fig. 4, transiently expressed
MHBst167 was found to markedly increase the promoter
activity of c-Myc in HepG2 cells in a dose-dependent manner. The
results suggested that MHBst167 protein could
transactivate c-Myc expression by transcriptionally activating its
promoter element.
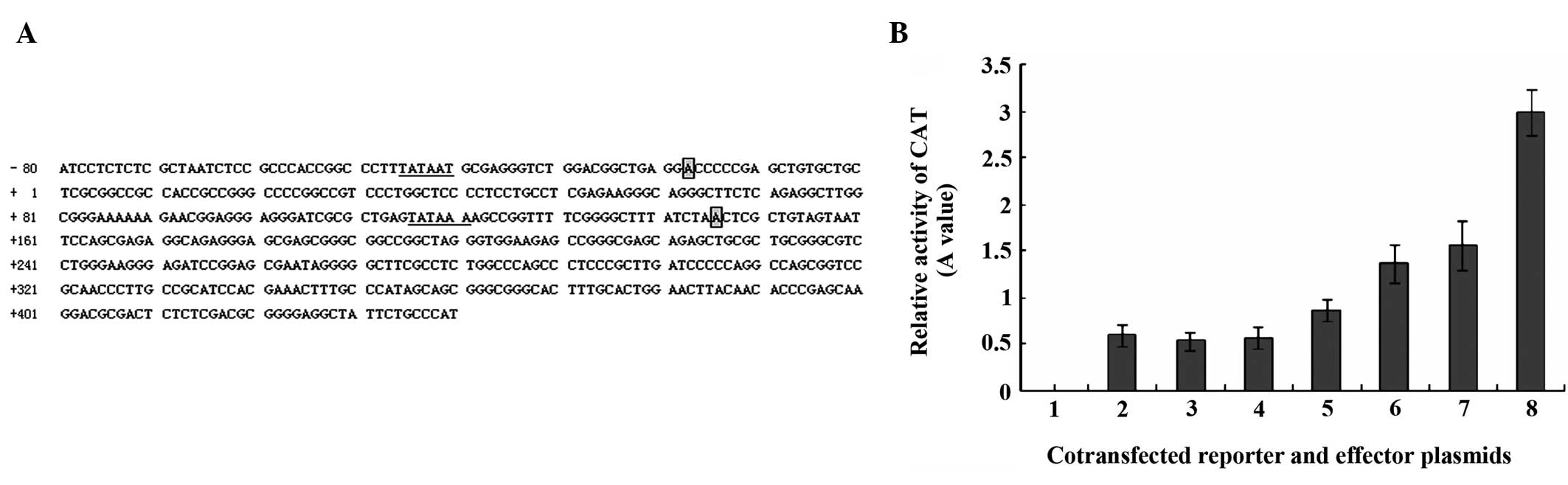 | Figure 4(A) Nucleotide sequence of human
oncogene c-Myc promoter. The classical ‘TATA’ boxes are underlined
and ‘A’ denotes the start sites of c-Myc promoters 1 and 2,
respectively. The first nucleotide of exon 1 acts as +1. (B)
Transactivation of human oncogene c-Myc promoter element by
MHBst167 in HepG2 cells in a dose-dependent manner. Lane
1, 1.0 μg of pCAT3-basic (promoterless); lane 2, 1.0 μg of
pCAT3-promoter (positive control); lane 3, 1.0 μg of pCAT3-c-Myc;
lane 4, 1.0 μg of pCAT3-c-Myc and 1.0 μg of pcDNA3.1(−); lanes 5–8,
1.0 μg of pCAT3-c-Myc and 0.5, 1.0, 1.5 and 2.0 μg
pcDNA3.1(−)-MHBst167, respectively. The standard
deviation is shown in the diagram. MHBst167, hepatitis B
virus middle size surface protein C-terminally truncated at amino
acid position 167; CAT, chloramphenicol acetyltransferase. |
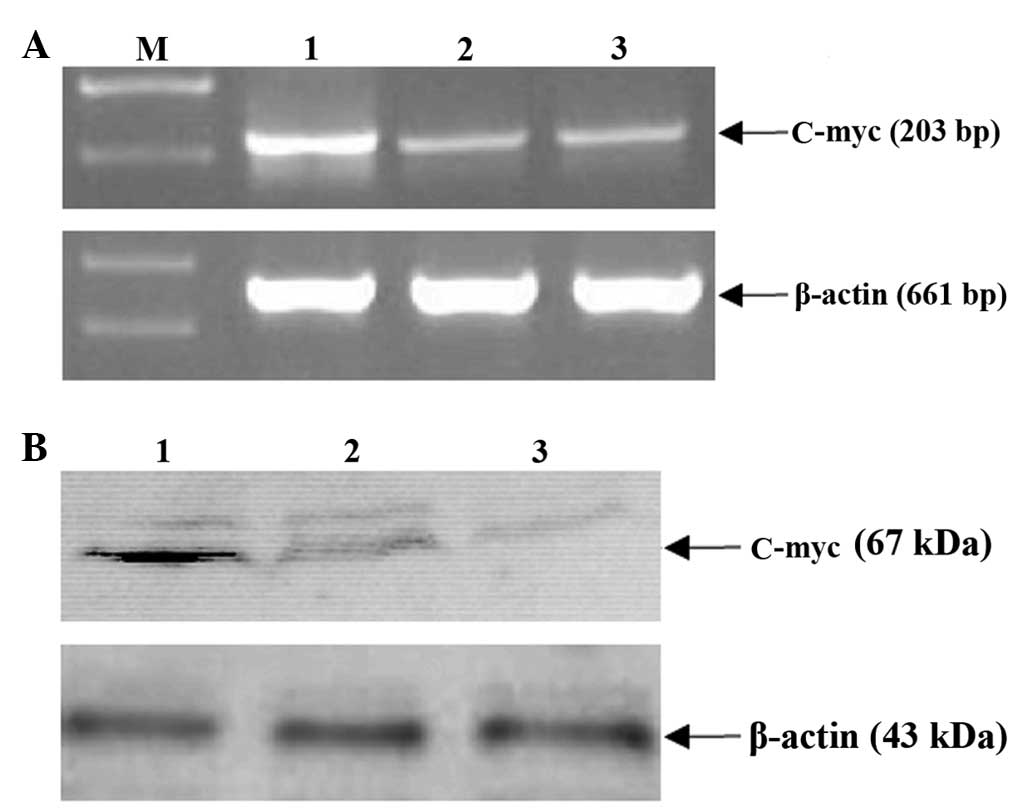
MHBst167 upregulates the c-Myc
expression
To further elucidate the mechanisms of
MHBst167 on c-Myc expression at the transcription and
translation levels, the effect of MHBst167 on expression
of the gene was investigated. As shown in Fig. 5A, level of mRNA of c-Myc markedly
increased following transient transfection with
pcDNA3.1(−)-MHBst167. The western blot analysis
indicated that the expression of the gene was low in the control
groups, whereas in the experiment group, its expression was
markedly enhanced (Fig. 5B). These
results indicated that MHBst167 could transactivate the
expression of c-Myc at both the transcription and translation
levels.
Discussion
The molecular mechanism of HBV-related
carcinogenesis is poorly understood. A possibility could be
activation of the expression of cellular genes involved in cell
growth regulation by viral proteins. For example, HBx has been
shown to act as a transcriptional transactivator, stimulating
various viral and cellular enhancer and promoter elements (9). Transactivating functions have also
been attributed to viral proteins translated from 3′-truncated HBV
preS/S sequences that are frequently found in human HCCs (10). Lauer et al found there are
three hydrophobic regions in the S-domain, located at MHBs residues
62–78, 135–53 and 224–281, respectively, which are separated by two
highly hydrophilic regions (at MHBs residues 79–134 and 154–223).
The authors showed that if truncation occurred beyond the third
hydrophobic region with the first hydrophobic region being
complete, the truncated protein acquired transactivation functions
(11). MHBst-encoding sequences are
found in numerous integrates subcloned from HBV-associated HCC, and
previous studies showed there was truncated-form S protein in the
circulation of patients with chronic hepatitis B virus infection
(12,13).
To evaluate the putative relevance of
MHBst167 in the process in HBV-associated HCC
development, a detailed analysis of MHBst167 activator
function is of great biological significance. In the present study,
the authors demonstrated that MHBst167 was successfully
expressed in the transiently transfected HepG2 cells. In the
co-transfection experiments, the relative CAT expression in the
cells transfected with pcDNA3.1(−)-MHBst167 +
pCAT3-promoter was approximately 4.5-fold higher than that with
pcDNA3.1(−) + pCAT3-promoter or pcDNA3.1(−)-MHBs + pCAT3-promoter.
This indicated that MHBst167 had a significant
transactivating function on the SV40 early promoter, leading to the
observed increase in the expression of the downstream gene CAT,
while the intact MHBs did not show transactivation. This suggests
that HBV MHBst167 transiently expressed in HepG2 cells
retains its biological activity in transcriptional activation,
which is consistent with previous reports (14).
To gain further insights into the genes
transactivated by MHBst167, SSH was used to clone the
genes transactivated by MHBst167. Sequencing of the
genes obtained from the subtracted library revealed 22 different
coding sequences, of which18 were known and 4 were unknown
genes.
The genes with known functions can be divided into
five groups, namely genes related to cell transcription and protein
synthesis, cell energy and substance metabolism, the formation
mechanism of hepatic fibrosis, cell signal transduction and
apoptosis, and tumor development (15). Notably, upregulated expression of
proto-oncogene c-Myc was observed. Yuen et al demonstrated
that 74% of HCC tissues had a high level of c-Myc expression
(16). Overexpression of c-Myc has
been implicated in liver regeneration and hepatocarcinogenesis. It
is also an indicator of malignant potential and poor prognosis
(17). The biological significance
of c-Myc gene upregulation by the truncated middle surface protein
of HBV in human hepatocellular carcinoma, however, has not been
confirmed.
To further elucidate the regulatory mechanisms of
MHBst167 on c-Myc expression, a reporter vector
pCAT3-c-Myc was generated where the CAT gene was placed under the
control of the c-Myc promoter, which contains the partly
5′-flanking region and the majority of the exon 1 regions of the
c-Myc gene (18). The upregulated
expression of proto-oncogene c-Myc in HepG2 cells has been
confirmed by cell transient transfection at the mRNA and protein
levels. It is reasonable to believe that the transformation effect
of MHBst167 is involved in the upregulation of the
expression of proto-oncogene c-Myc.
In conclusion, the present study analyzed the
transactivator function of MHBst167 and constructed a
subtracted cDNA library of genes transactivated by
MHBst167. Furthermore, it was confirmed that
MHBst167 could transactivate the expression of c-Myc at
the transcriptional and translational levels. These findings
provide new insights into the biological functions of
MHBst167 and new directions to elucidate the
hepatocarcinogenesis mechanisms of HBV infection.
Acknowledgements
The authors thank the technical staff of the Viral
Hepatitis Research Center, Institute of Infectious Diseases,
Beijing PLA 302 Hospital (Beijing, China), for excellent technical
assistance.
References
|
1
|
Humphries JC and Dixon JS: Antivirals for
the treatment of chronic hepatitis B: current and future options.
Intervirology. 46:413–420. 2003.
|
|
2
|
Seeger C and Mason WS: Hepatitis B virus
biology. Microbiol Mol Biol Rev. 64:51–68. 2000.
|
|
3
|
Glebe D and Bremer CM: The molecular
virology of hepatitis B virus. Semin Liver Dis. 33:103–112.
2013.
|
|
4
|
Bruss V, Lu X, Thomssen R and Gerlich WH:
Post-translational alterations in transmembrane topology of the
hepatitis B virus large envelope protein. EMBO J. 13:2273–2279.
1994.
|
|
5
|
Kekulé AS, Lauer U, Meyer M, Caselmann WH,
Hofschneider PH and Koshy R: The preS2/S region of integrated
hepatitis B virus DNA encodes a transcriptional transactivator.
Nature. 343:457–461. 1990.
|
|
6
|
Hildt E, Urban S and Hofschneider PH:
Characterization of essential domains for the functionality of the
MHBst transcriptional activator and identification of a
minimal MHBst activator. Oncogene. 11:2055–2066.
1995.
|
|
7
|
Hildt E, Urban S, Lauer U, Hofschneider PH
and Kekulé AS: ER-localization and functional expression of the HBV
transactivator MHBst. Oncogene. 8:3359–3367. 1993.
|
|
8
|
Li ZQ, Ma YJ and Cheng J: Screening
proteins in hepatocytes interacting with the middle surface protein
of hepatitis B virus using the yeast-two hybrid technique. Zhonghua
Gan Zang Bing Za Zhi. 15:111–113. 2007.(In Chinese).
|
|
9
|
Henkler FF and Koshy R: Hepatitis B virus
transcriptional activators: mechanisms and possible role in
oncogenesis. J Viral Hepat. 3:109–121. 1996.
|
|
10
|
Caselmann WH: Transactivation of cellular
gene expression by hepatitis B viral proteins: a possible molecular
mechanism of hepatocarcinogenesis. J Hepatol. 22(1 Suppl): 34–37.
1995.
|
|
11
|
Lauer U, Weiss L, Hofschneider PH and
Kekulé AS: The hepatitis B virus pre-S/S(t) transactivator is
generated by 3′ truncations within a defined region of the S gene.
J Virol. 66:5284–5289. 1992.
|
|
12
|
Hildt E, Urban S, Eckerskorn C and
Hofschneider PH: Isolation of highly purified, functional
carboxy-terminally truncated hepatitis B virus middle surface
protein activators from eucaryotic expression systems. Hepatology.
24:502–507. 1996.
|
|
13
|
Hildt E, Munz B, Saher G, Reifenberg K and
Hofschneider PH: The PreS2 activator MHBst of hepatitis
B virus activates c-raf-1/Erk2 signaling in transgenic mice. EMBO
J. 21:525–535. 2002.
|
|
14
|
Caselmann WH, Renner M, Schluter V,
Hofschneider PH, Koshy R and Meyer M: The hepatitis B virus
MHBst167 protein is a pleiotropic transctivator
mediating its effect via ubiquitous cellular transcription factors.
J Gen Virol. 78:1487–1495. 1997.
|
|
15
|
Badapanda C: Suppression subtractive
hybridization (SSH) combined with bioinformatics method: an
integrated functional annotation approach for analysis of
differentially expressed immune-genes in insects. Bioinformation.
9:216–221. 2013.
|
|
16
|
Yuen MF, Wu PC, Lai VC, Lau JY and Lai CL:
Expression of c-Myc, c-Fos, and c-jun in hepatocellular carcinoma.
Cancer. 91:106–112. 2001.
|
|
17
|
Cheng J: Molecular mechanisms of hepatitis
virus-hepatocyte interactions. J Gastroenterol Hepatol. 17(Suppl
3): S342–S343. 2002.
|
|
18
|
Guo F, Song F, Zhang J, Li J and Tang Y:
Study of transcription activity of X-box binding protein 1 gene in
human different cell lines. J Genet Genomics. 34:790–798. 2007.
|