Introduction
During the last decade, automated diagnostic image
analysis methods have been developed to improve the standardization
and reproducibility of the results of histopathological evaluation.
Immunohistochemistry (IHC) has been of particular interest, as an
objective quantitative assessment is often required. Currently,
there are different computer-assisted software tools for automatic
IHC analysis based on whole-slide imaging, including Aperio
software (Aperio Technologies; Vista, CA, USA), an integrated
scanning and image analysis system. This tool incorporates positive
pixel count nuclear, membrane and cytoplasmic algorithms,
therefore, can be employed to measure areas and staining properties
(1). Numerous studies have been
conducted regarding the employment of Aperio software analysis in
clinical practice, as well as in research models (2–4).
Objective and reproducible automated analysis has been shown to
facilitate diagnosis, prognosis and management of oncologic
diseases (5–6). However, to the best of our knowledge,
there are no studies concerning automated analysis using the Aperio
algorithms in the cases of ocular surface squamous neoplasia
(OSSN).
OSSN (including intraepithelial and invasive
neoplasia) is a non-pigmented malignant tumor of the conjunctiva
that results in significant ocular surface destruction and visual
dysfunction. The reported incidence ranges from 0.02 to 3.5
cases/100,000 individuals/year and exhibits an increasing trend in
developing countries with a high prevalence of the human
immunodeficiency virus (7). OSSN is
commonly a slow growing, low-grade malignancy, however,
occasionally it acts aggressively and may result in recurrences,
invasion, metastases and even mortality, particularly in
immunocompromised patients (8).
Previous studies described the following prognostic factors for
OSSN recurrence: Older age, greater lesion size, tumor invasion,
non-radical excision, absence of cryotherapy and an increased level
of positive expression of the biomarker, Ki-67 (8–10).
Ki-67 is a nuclear antigen, which is expressed in the proliferating
cells of healthy and neoplastic tissue, and is evaluated using IHC
(11,12). It was hypothesized in the present
study that automated image analysis using the Aperio nuclear V9
algorithm may aid with the quantitative assessment of the Ki-67
value in OSSN.
The aim of the present study was to evaluate the
Aperio nuclear V9 algorithm as an image analysis tool for the
histopathological changes of OSSN. The results of the automated,
stereological and visual IHC assessments of tumor conjunctiva (TC)
and healthy conjunctiva (HC) tissues were compared. In addition,
the nuclear area in the TC and HC cells was quantified. To the best
of our knowledge this is the first study to present the results of
this algorithm when applied to the analysis of OSSN.
Materials and methods
Patients
The current retrospective pilot study was conducted
at the Centre of Eye Diseases, Vilnius University Hospital
Santariškių Klinikos (Vilnius, Lithuania) between April 2002 and
June 2012. The study consisted of six conjunctival specimens from
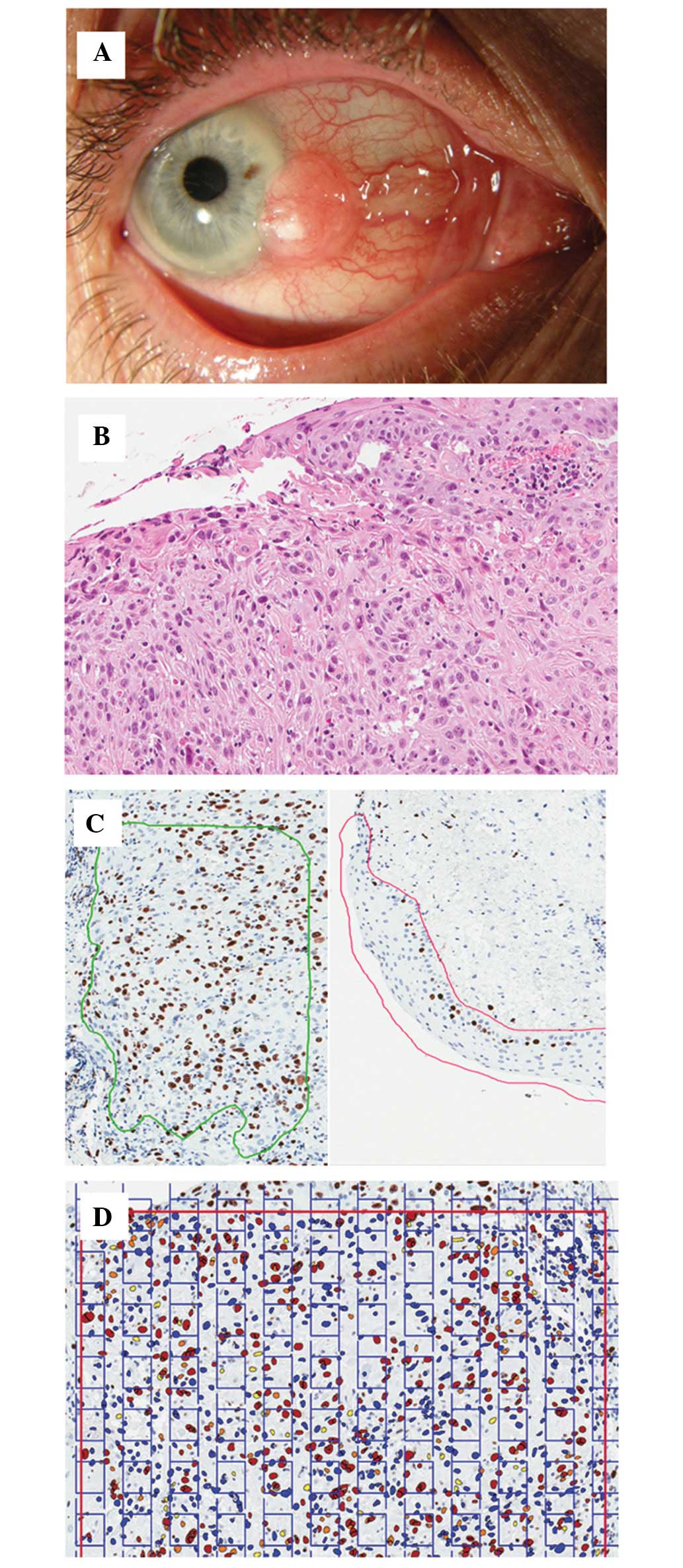
six patients with a histopathological diagnosis of OSSN (Fig. 1A and B). In three patients (three
eyes) the TC was excised, cryo-application and an amniotic membrane
transplantation was performed. In two patients (two eyes) excision
of the tumor without any additional surgical manipulation was
performed. The data regarding the treatment of one patient is
missing. The study was approved by the Ethics Committee of Vilnius
University Hospital Santariškių Klinikos, Vilnius, Lithuania
(EK-38) Patients provided written informed consent.
Tissue preparation
Excised tissues for histological analysis were fixed
in 10% neutral buffered formalin for 6–24 h and paraffin embedded
in a tissue processor (Shandon Pathcentre® Tissue
Processor; Thermo Shandon Ltd., Runcorn, UK). Formalin-fixed and
paraffin-embedded (FFPE) tissues were sectioned (2-μm thick) and
processed for subsequent staining with Hematoxylin and Eosin
(H&E). The Ki-67-immunostained slides were prepared according
to the manufacturer’s instructions, using the mouse anti-human
monoclonal Mib-1 clone antibody (dilution, 1:200; DAKO,
Carpinteria, CA, USA) and the Ventana BenchMark XT staining
platform (Ventana Medical Systems, Tucson, AZ, USA). Digital images
of H&E- and IHC-stained glass slides were obtained using a
ScanScope Digital slide scanner (Aperio Technologies) at a
magnification of ×20 (Fig. 1B and
C).
Quantification techniques
Various Ki-67 proliferative index (PI)
quantification techniques were adopted, including the following: i)
Visual evaluation by one pathologist; ii) digital image analysis
(DIA) using an LG L226WTQ-SF Flatron monitor (LG Electronics,
Seoul, South Korea) and the nuclear V9 algorithm (Aperio ScanScope
XT System; Aperio Technologies) counting >1,252 cells in the TC
tissue and 242 cells in the HC tissue; and iii) DIA using a
stereology module (Stereology Toolkit 4.2.0 [ADCIS, www.adcis.net]; Fig.
1D). Cells exhibiting questionable nuclear staining were
discounted (visually and stereologically). Ki-67 PI values from the
visual, stereological and automated analyses were compared. In
addition, the Aperio nuclear V9 algorithm was used to quantify the
area of epithelial nuclei in the TC and HC tissues.
Statistical analysis
Collinearity in the linear regression analysis was
performed with the SAS® 4.2 Enterprise Guide software
(SAS Institute Inc., Cary, NC, USA). P<0.05 was considered to
indicate a statistically significant difference.
Results
Visual, stereological and automated
analysis
OSSN was diagnosed in six patients (three males and
three females) with a mean age of 69 years (range, 54–84 years).
Table I summarizes the clinical,
histopathological and management data of these patients. The
results of IHC staining for the Ki-67 PI, which was visually
assessed by the pathologist, the Aperio nuclear V9 algorithm and
using the stereology module are presented in Table II. The visual scoring of Ki-67 PI
ranged from 22 to 60% (mean, 38.5%) in TC tissue and from 5 to 20%
(mean, 9.5%) in HC tissue. The computer-aided analysis using the
Aperio nuclear V9 algorithm demonstrated that the Ki-67 PI ranged
from 21.5 to 43.5% (mean, 33.6%) and from 1.9 to 21.0% (mean,
11.8%) in TC and HC tissues, respectively. The stereological method
identified a Ki-67 PI range of 30.1 to 51.5% (mean, 41.0%) and from
3.2 to 30.1 (mean, 15.1%) in TC and HC tissues, respectively.
 | Table IClinical, histopathological and
management data of the patients exhibiting ocular surface squamous
neoplasia. |
Table I
Clinical, histopathological and
management data of the patients exhibiting ocular surface squamous
neoplasia.
| Patient | Age at diagnosis
(years)/gender | Histopathological
diagnosis | Location of the
tumor | Management | Excisional
margins |
|---|
| 1 | 60/Female | Keratinizing invasive
CSCC (pT1) | Nasal bulbar,
conjunctiva limbus | Excision | No data due to poor
orientation of the specimen |
| 2 | 74/Female | Non-keratinizing
invasive CSCC (≥pT1) | Nasal bulbar
conjunctiva, limbus | Excision | Involved |
| 3 | 54/Male | Non-keratinizing
invasive CSCC (≥pT1) | Nasal bulbar
conjunctiva, limbus | Excision + Cryo +
AMT | Clear |
| 4 | 76/Female | Keratinizing invasive
CSCC (pT2) | Nasal bulbar
conjunctiva, limbus | Excision + Cryo +
AMT | Involved |
| 5 | 66/Male | Invasive CSCC
(pT2) | No data | No data | No data |
| 6 | 84/Male | Keratinizing CSCC
in situ (CIN 3) | Temporal bulbar
conjunctiva, limbus, cornea | Excision + Cryo +
AMT | Clear |
 | Table IIResults of visual, automated (Aperio
nuclear V9 algorithm) and stereological analysis of the Ki-67
PI. |
Table II
Results of visual, automated (Aperio
nuclear V9 algorithm) and stereological analysis of the Ki-67
PI.
| Ki-67 PI of tumor
conjunctiva (%) | Ki-67 PI of healthy
conjunctiva (%) |
|---|
|
|
|
|---|
| Patient | Visual | Aperio V9 | Stereological | Visual | Aperio V9 | Stereological |
|---|
| 1 | 34 | 34.0 | 42.0 | 7 | 13.2 | 22.8 |
| 2 | 40 | 33.4 | 45.7 | 7 | 13.6 | 15.7 |
| 3 | 60 | 41.0 | 51.5 | 20 | 21.0 | 30.1 |
| 4 | 22 | 28.0 | 30.1 | 6 | 7.2 | 9.7 |
| 5 | 35 | 21.5 | 30.9 | 5 | 1.9 | 3.2 |
| 6 | 40 | 43.5 | 45.8 | 12 | 14.0 | 9.0 |
| Mean | 38.5 | 33.6 | 41.0 | 9.5 | 11.8 | 15.1 |
Collinearity in the regression
analysis
The strongest association in collinearity of the
regression analysis was observed between the Aperio nuclear V9
algorithm/stereological analysis models in the TC tissue
(r2=0.7, P=0.04) and in the HC tissue
(r2=0.7, P=0.03), as well as in the visual/stereological
models in the TC tissue (r2=0.7, P=0.04) and
visual/Aperio nuclear V9 algorithm in the HC tissue
(r2=0.7, P=0.04). A weak and statistically insignificant
association was identified between the Aperio nuclear V9
algorithm/visual analysis in the TC tissue and the
visual/stereological analysis in the HC tissue; r2=0.4
(P=0.2) and r2=0.5 (P=0.13), respectively. No
significant difference was observed between the nuclear area of the
TC tissue (mean, 36.5 μm2 and range, 33.6–38.2
μm2) and the HC tissue (mean, 35.7 μm2 and
range, 33.0–40.0 μm2; P=0.88) (Table III).
 | Table IIIMean nuclear area of the tumor and
healthy conjunctiva as assessed using the Aperio nuclear V9
algorithm. |
Table III
Mean nuclear area of the tumor and
healthy conjunctiva as assessed using the Aperio nuclear V9
algorithm.
| Mean nuclear area
(μm2) |
|---|
|
|
|---|
| Patient | Tumor
conjunctiva | Healthy
conjuctiva |
|---|
| 1 | 38.2 | 34.7 |
| 2 | 33.6 | 36.7 |
| 3 | 35.3 | 33.0 |
| 4 | 36.7 | 34.1 |
| 5 | 38.0 | 35.4 |
| 6 | 37.4 | 40.0 |
| Mean | 36.5 | 35.7 |
Discussion
In the last decade, progress in information and
communication technologies has offered novel possibilities for
increasing the level of objectiveness and effectiveness of
histopathological examination using automated image analysis.
However, until recently, the insufficient quality obtained during
image acquisition, and of program software, prevented the routine
use of automated histopathological analysis in the clinical and
research settings. As a result of technological improvements,
virtual microscopy and DIA are currently widely available, and
there are numerous studies regarding the diagnostic and prognostic
value of computer-assisted histopathological analysis (2,3,5,1,13,14).
In the present study the experience of adopting automated
histopathological analysis for the analysis of OSSN is described
and, to the best of our knowledge, this is the first study to apply
this analysis method to OSSN.
There are various solutions available for
computer-assisted image analysis, ranging from general-purpose
software to fully- or semi-automated commercial packages (15). All of these systems aim to achieve
an accurate, reproducible, increasingly efficient and economical
method of histopathological diagnosis. It is particularly important
in quantitative analysis where variability and subjectivity
significantly influences the accuracy. Therefore, the different
methods of evaluating the IHC staining intensity, including direct
observation under a microscope, stereological analysis (by
superimposing grids on images) and computer-aided analysis, are
often compared. In the present study, the strongest association in
regression analysis was observed between the results of digital
evaluation using the Aperio nuclear V9 algorithm and the
stereological method, with the latter considered to be the gold
standard (an independent variable in statistical analysis). The
association between the results of automated DIA and the visual
method in the HC tissue revealed a similar r2-value,
however, was identified to be weaker in the TC tissue. This may
result from a more regular arrangement of cells (thus facilitating
cell counting), and fewer lymphocytes and reactive stromal
fibroblasts (which could be mistaken for tumorous cells) in the HC
tissue, when using automated analysis. Previous studies have
reported a good correlation between the results of visual IHC and
automated analysis using Aperio nuclear V9 algorithms, and
concluded that the latter method is reliable in histopathological
examination (3,13,14,16,17).
Visual analysis is considered to be a subjective,
and time- and human resource-consuming process; its weakest point
being inter- and intra-observer variability. Differences in the
competency of pathologists, and preconceptions, expectations and
fatigue are considered to be the primary causes of the
abovementioned disadvantages and may lead to inaccurate results and
erroneous managerial decisions (15). By contrast, automated image analysis
offers a more rapid and effective approach and demonstrates
improved reproducibility (14,16).
The elimination of subjectivity risk is another advantage of
automated analysis, which aids with obtaining increasingly
objective qualitative and, importantly, quantitative results. In
addition, automated image analysis simplifies the collection,
sharing and visualization of data. However, the limitations of
computer-assisted image analysis are artifacts in histopathology,
exceptional variations in the sample and non-uniform staining,
which result in significant issues with the accuracy of the
results. Thus, certain studies emphasize the necessity of
standardization, strict quality control of sample slides and
supervision by a pathologist in order to obtain precise and
accurate results (1,2).
The use of the Aperio nuclear V9 algorithm for
quantification of the expression level of various biomarkers is
considered to be valuable in numerous types of malignancy.
Automated Aperio analysis of estrogen and progesterone expression
in breast cancer was identified to be valuable for prognostic and
therapeutic strategies in previous studies (2,5).
Chabot-Richards et al (3)
demonstrated a significant correlation between the results
estimated by a pathologist and those obtained using automated image
analysis of the percentage of Ki-67 positivity in diffuse large
B-cell lymphoma. Singh et al (6) adopted the Aperio analysis method and
demonstrated that ovarian cancer cells express chemokine receptor,
CCR9 and its ligand, CCL25 and indicated that this
chemokine-receptor axis may be involved in ovarian cancer
progression.
The Ki-67 proliferation biomarker is widely used in
histopathology to measure the population of actively cycling cells,
and has been identified as a significant index for the diagnosis
and prognosis of different types of malignancy (18–20).
Certain studies reported Ki-67 to be an independent prognostic
marker for conjunctival squamous cell carcinoma (9,21). The
percentage of Ki-67 positivity, as assessed by the Aperio nuclear
V9 algorithm in the current study, was comparable with the data
obtained by Jung et al (11). McKelvie et al (9) reported smaller values of Ki-67
staining as a result of visual analysis. However, an association
between the Ki-67 positivity and the disease outcome was not
determined due to the limited sample size.
According to Wolberg et al (22) the nuclear area of the cells of
benign breast masses is smaller compared with that of malignant
masses. Therefore, in the present study, it was hypothesized that
the nuclear area of HC cells may be smaller than that of the TC
cells of OSSN. However no significant difference was identified
between the nuclear area of the TC and HC cells.
In conclusion, the Aperio nuclear V9 algorithm is
considered to be a useful tool for the reliable analysis of
histopathological changes of OSSN. The results of this type of DIA
algorithm correlate strongly with the stereoscopic method when
assessing the Ki-67 PI and may facilitate the accurate quantitative
evaluation of IHC-stained biomarkers. Future studies are required
to further evaluate the potential of the Aperio nuclear V9
algorithm for the analysis of OSSN.
References
|
1
|
Rojo MG, Bueno G and Slodkowska J: Review
of imaging solutions for integrated quantitative
immunohistochemistry in the Pathology daily practice. Folia
Histochem Cytobiol. 47:349–354. 2009.
|
|
2
|
Lloyd MC, Allam-Nandyala P, Purohit CN, et
al: Using image analysis as a tool for assessment of prognostic and
predictive biomarkers for breast cancer: How reliable is it? J
Pathol Inform. 1:292010.
|
|
3
|
Chabot-Richards DS, Martin DR, Myers OB,
Czuchlewski DR and Hunt KE: Quantitative image analysis in the
assessment of diffuse large B-cell lymphoma. Mod Pathol.
24:1598–1605. 2011.
|
|
4
|
Klapczynski M, Gagne GD, Morgan SJ, et al:
Computer-assisted imaging algorithms facilitate histomorphometric
quantification of kidney damage in rodent renal failure models. J
Pathol Inform. 3:202012.
|
|
5
|
Laurinavicius A, Laurinaviciene A,
Ostapenko V, et al: Immunohistochemistry profiles of breast ductal
carcinoma: factor analysis of digital image analysis data. Diagn
Pathol. 7:272012.
|
|
6
|
Singh R, Stockard CR, Grizzle WE, Lillard
JW Jr and Singh S: Expression and histopathological correlation of
CCR9 and CCL25 in ovarian cancer. Int J Oncol. 39:373–381.
2011.
|
|
7
|
Yang J and Foster CS: Squamous cell
carcinoma of the conjunctiva. Int Ophthalmol Clin. 37:73–85.
1997.
|
|
8
|
Kiire CA, Srinivasan S and Karp CL: Ocular
surface squamous neoplasia. Int Ophthalmol Clin. 50:35–46.
2010.
|
|
9
|
McKelvie PA, Daniell M, McNab A, Loughnan
M and Santamaria JD: Squamous cell carcinoma of the conjunctiva: a
series of 26 cases. Br J Ophthalmol. 86:168–173. 2002.
|
|
10
|
Tunc M, Char DH, Crawford B and Miller T:
Intraepithelial and invasive squamous cell carcinoma of the
conjunctiva: analysis of 60 cases. Br J Ophthalmol. 83:98–103.
1999.
|
|
11
|
Jung SM, Lin HC, Chu PH, et al: Expression
of cell cycle-regulatory proteins, MIB-1, p16, p53, and p63, in
squamous cell carcinoma of conjunctiva: not associated with human
papillomavirus infection. Virchows Arch. 448:301–305. 2006.
|
|
12
|
Schlüter C, Duchrow M, Wohlenberg C, et
al: The cell proliferation-associated antigen of antibody Ki-67: a
very large, ubiquitous nuclear protein with numerous repeated
elements, representing a new kind of cell cycle-maintaining
proteins. J Cell Biol. 123:513–522. 1993.
|
|
13
|
Alvarenga AW, Coutinho-Camillo CM,
Rodrigues BR, et al: A comparison between manual and automated
evaluations of tissue microarray patterns of protein expression. J
Histochem Cytochem. 61:272–282. 2013.
|
|
14
|
Brazdziute E and Laurinavicius A: Digital
pathology evaluation of complement C4d component deposition in the
kidney allograft biopsies is a useful tool to improve
reproducibility of the scoring. Diagn Pathol. 6(Suppl 1):
S52011.
|
|
15
|
Prasad K and Prabhu GK: Image analysis
tools for evaluation of microscopic views of immunohistochemically
stained specimen in medical research - a review. J Med Syst.
36:2621–3261. 2012.
|
|
16
|
Fasanella S, Leonardi E, Cantaloni C, et
al: Proliferative activity in human breast cancer: Ki-67 automated
evaluation and the influence of different Ki-67 equivalent
antibodies. Diagn Pathol. 6(Suppl 1): S72011.
|
|
17
|
Słodkowska J, Filas V, Buszkiewicz E, et
al: Study on breast carcinoma Her2/neu and hormonal receptors
status assessed by automated images analysis systems: ACIS III
(Dako) and ScanScope (Aperio). Folia Histochem Cytobiol. 48:19–25.
2010.
|
|
18
|
Salek D, Vesela P, Boudova L, et al:
Retrospective analysis of 235 unselected patients with mantle cell
lymphoma confirms prognostic relevance of Mantle Cell Lymphoma
International Prognostic Index and Ki-67 in the era of rituximab:
long-term data from the Czech Lymphoma Project Database. Leuk
Lymphoma. 55:802–810. 2014.
|
|
19
|
Liu Y, Yin W, Yan T, et al: The clinical
significance of Ki-67 as a marker of prognostic value and
chemosensitivity prediction in hormone-receptor-positive breast
cancer: a meta-analysis of the published literature. Curr Med Res
Opin. 29:1453–1461. 2013.
|
|
20
|
Otto W, Denzinger S, Fritsche HM, et al:
Introduction and first clinical application of a simplified
immunohistochemical validation system confirms prognostic impact of
KI-67 and CK20 for stage T1 urothelial bladder carcinoma:
single-center analysis of eight biomarkers in a series of three
hundred six patients. Clin Genitourin Cancer. 11:537–544. 2013.
|
|
21
|
Ohara M, Sotozono C, Tsuchihashi Y and
Kinoshita S: Ki-67 labeling index as a marker of malignancy in
ocular surface neoplasms. Jpn J Ophthalmol. 48:524–529. 2004.
|
|
22
|
Wolberg WH, Street N, Heisey DM and
Mangasarian OL: Computer-derived nuclear features distinguish
malignant from benign breast cytology. Hum Pathol. 26:792–796.
1995.
|