Introduction
Cisplatin (CDDP) is one of the most commonly used
drugs in chemotherapy for gastric cancer. However, when gastric
cancer cells develop resistance to CDDP, the chemotherapeutic
effect of CDDP is reduced and this may even result in the failure
of chemotherapy. Therefore, the analysis of CDDP resistance in
gastric cancer cells has important implications. The Survivin gene
is a member of the inhibitor of apoptosis protein family (1). Since Survivin inhibits apoptosis and
is beneficial to the growth of tumor cells, the Survivin gene is
also known as ‘survival factor’ (2). The Survivin gene is overexpressed in
gastric cancer cells (3), which may
be associated with the resistance of CDDP to gastric cancer cells
and this was investigated in the present study.
Materials and methods
SGC7901 cell culture
Human gastric cancer SGC7901 cells were obtained
from the cell bank of the Chinese Academy of Sciences (Shanghai,
China). The SGC7901 cells were cultured in a humidified atmosphere
of 5% CO2 and 95% air using RPMI 1640 (Invitrogen Life
Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine
serum, 100 U/ml penicillin and 100 μg/ml streptomycin (both North
China Pharmaceutical Group Corporation, Shijiazhuang, China) in
75-cm2 flasks at 37°C. The RPMI 1640 was adjusted to pH
7.2 with 5.6% sterile NaHCO3. The cell culture medium
was changed every 2–3 days. The cells were subcultured when 80%
confluence was reached.
SGC7901/CDDP cell culture
RPMI 1640 with 100 ng/ml CDDP (lyophilized type;
batch number, 6040122DC; Qilu Pharmaceutical Co., Ltd., Jinan,
China) was added to the culture medium and the medium was changed
every 2–3 days. When 80% confluence was reached, the cells were
subcultured with RPMI 1640 to achieve a good adhesive condition. As
the cells became adherent to the bottom of cell culture flasks,
RPMI 1640 with 200 ng/ml CDDP was added to the medium and the
medium was changed every 2–3 days. When the cells has reached 80%
confluence, the cells were subcultured with RPMI 1640 to achieve a
good adhesive condition. As the cells became adherent to the bottom
of the cell culture flasks, RPMI 1640 with 500, 700 or 1,000 ng/ml
CDDP was added to the medium and the medium was changed every
two-three days.
Survivin mRNA detection
Total RNA was extracted from the cells using TRIzol
reagent (Invitrogen Life Technologies) and cDNA synthesized from
RNA (1 μg) was used as a template for the RT reaction (Invitrogen
Life Technologies). The 447-bp Survivin DNA fragment was amplified
using the following two primers synthesized by Invitrogen Life
Technologies: Forward, 5′-GCATGGGTGCCCCGACGTTG-3′ and reverse,
5′-GCTCCGGCCAGAGGCCTCAA-3′. Polymerase chain reaction (PCR) was
performed in a solution containing 2 μl 10X PCR buffer (Invitrogen
Life Technologies), 0.8 μl MgCl2, 1.0 μl dNTPs
(Invitrogen Life Technologies), 0.2 μl of each primer, 2.0 μl cDNA
and 1.0 μl Taq DNA polymerase (Promega Corporation, Madison, WI,
USA), to obtain a total volume of 20 μl. The amplification was
performed in a microcentrifuge tube under the following conditions:
Denaturation at 94°C for 30 sec, annealing at 55°C for 60 sec and
elongation at 72°C for 60 sec, for 30 cycles. The 241-bp β-actin
fragment was amplified using the following two primers synthesized
by Invitrogen Life Technologies: Forward,
5′-TAAAGACCTCTATGCCAACACAGT-3′ and reverse,
5′-CACCATGGAGGGGCCGGACTCTTC-3′. PCR was performed in a solution
containing 2 μl 10X PCR buffer (Invitrogen Life Technologies), 1.6
μl MgCl2, 1.0 μl dNTPs (Invitrogen Life Technologies),
0.2 μl of each primer, 2.0 μl cDNA and 1.0 μl Taq DNA polymerase
(Promega Corporation) to obtain a total volume of 20 μl. The
amplification conditions were as follows: Denaturation at 94°C for
30 sec, annealing at 58°C for 40 sec and elongation at 72°C for 40
sec, for 28 cycles. The PCR products were separated on a 1% agarose
gel containing ethidium bromide. The gel images were digitally
recorded and analyzed by computer assisted image analyzer with
Lab-work 4.5 analysis software (Ultra Violet Products, Upland, CA,
USA). The relative content of Survivin mRNA was indicated by the
absorbance ratio of the Survivin mRNA band and the β-actin
band.
Survivin protein detection
The SGC7901 and SGC7901/CDDP cells were washed twice
with 4°C phosphate-buffered saline (PBS). RIPA buffer (Invitrogen
Life Technologies) was added and the cells were lysed on ice for 30
min, then clarified by centrifugation at 10,000 × g for 10 min at
4°C. The supernatants were used to assay protein concentrations.
Subsequently, 25 μg protein was loaded and separated by
polyacrylamide gel electrophoresis and then transferred to a
polyvinylidene difluoride (PVDF) membrane. The PVDF membranes were
incubated for 2 h at room temperature with 5% skimmed powdered milk
in 500 mm NaCl, 20 mm Tris-HCL (pH 7.5) and 0.5% PBS-Tween 20, and
then for 24 h at 4°C with the following dilutions of primary
antibody: 1:2,000 anti-human Survivin antibody (catalog number,
AF6471; immunoglobulin-type, human Survivin specific goat IgG;
R&D Systems, Minneapolis, MN, USA) and 1:500 anti-β-actin
antibody (Wuhan Boster Biological Technology, Ltd., Wuhan, China).
Subsequent to being washed with Tris-buffered saline-Tween 20, the
PVDF membranes were incubated with 1:3,000 peroxidase-conjugated
rabbit anti-goat secondary antibodies (Wuhan Boster Biological
Technology, Ltd.) for 2 h at room temperature. Proteins were
visualized using chemiluminescent peroxidase substrate (Pierce
Biotechnology, Inc., Rockford, USA), and the protein blots were
quantified and analyzed by computer-assisted image analyzer with
Lab-work 4.5 analysis software. The relative content of Survivin
protein was indicated by the absorbance ratio of the Survivin
protein band to the β-actin band.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Student’s t-test was used for comparisons involving two groups. All
statistically analyses were performed using SPSS 19.0 software
(IBM, Armonk, NY, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
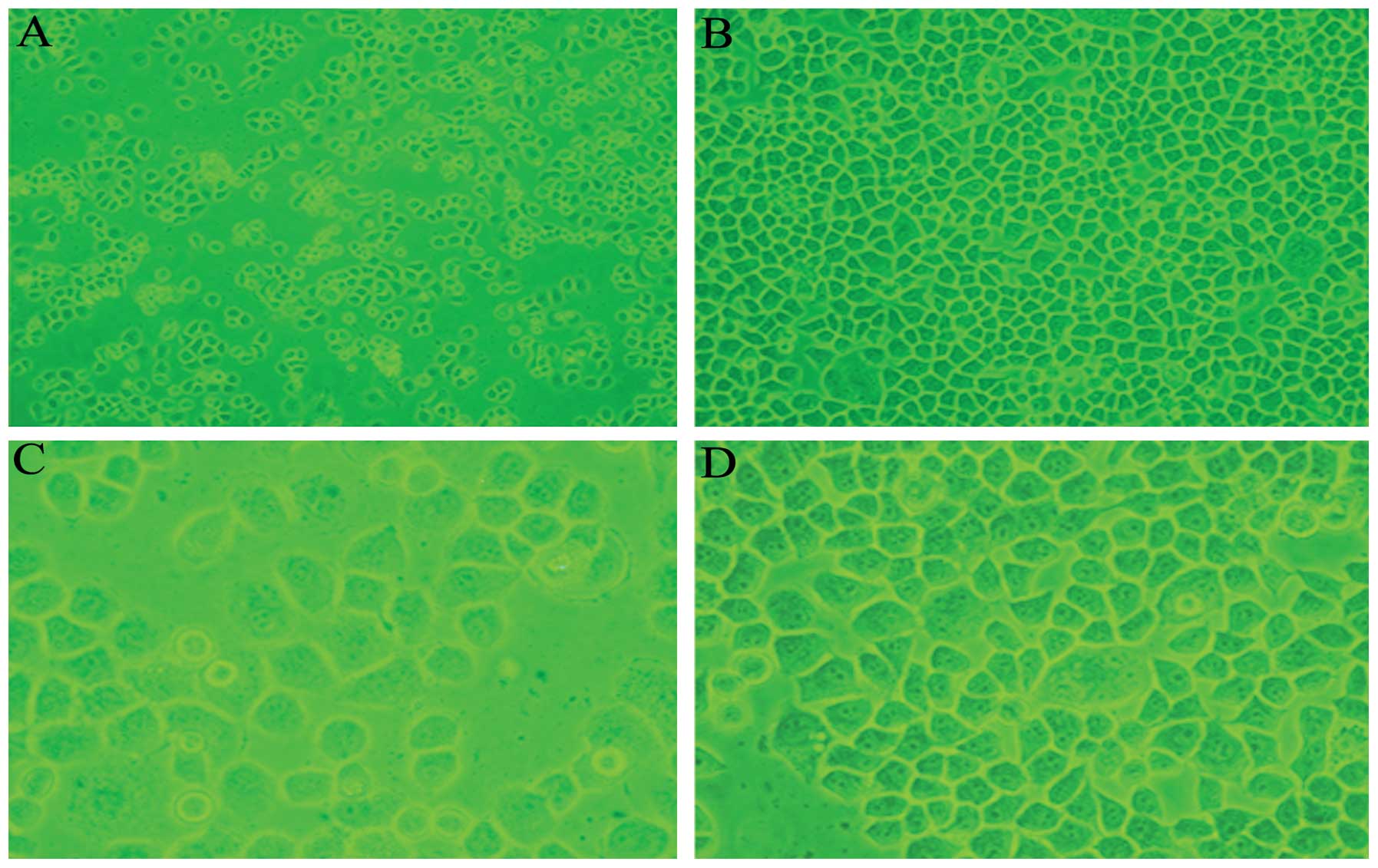
Morphology of SGC7901 and SGC7901/CDDP
cells
The SGC7901 cells appeared polygonal,
cobblestone-like and tightly adherent to the flask, and were highly
refractive and proliferative. The SGC7901/CDDP cells were
relatively reduced in number, only marginally refractive and weakly
adherent to the flask, and increased space was observed between the
cells. A few SGC7901/CDDP cells were deformed, increased in size or
floating in the culture medium (Fig.
1).
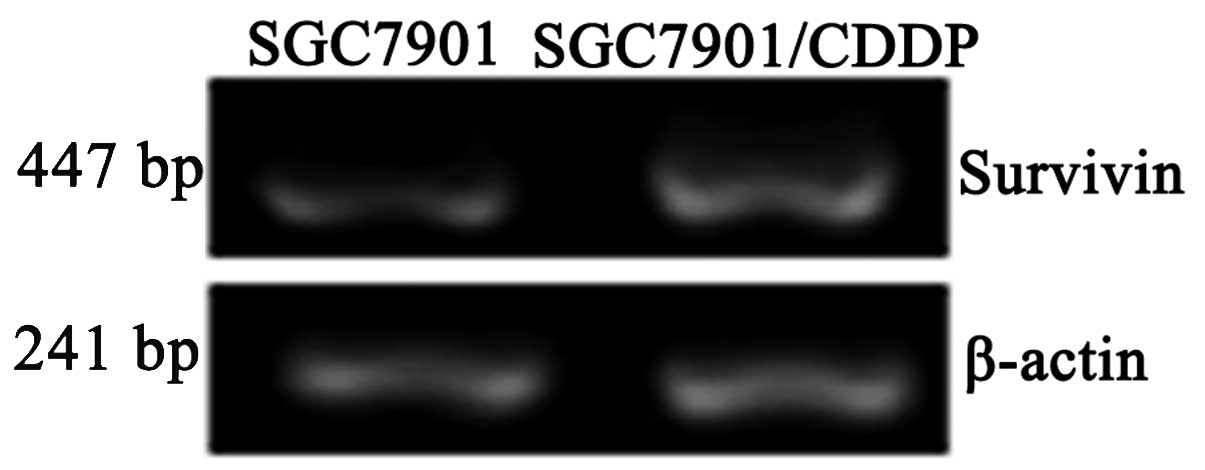
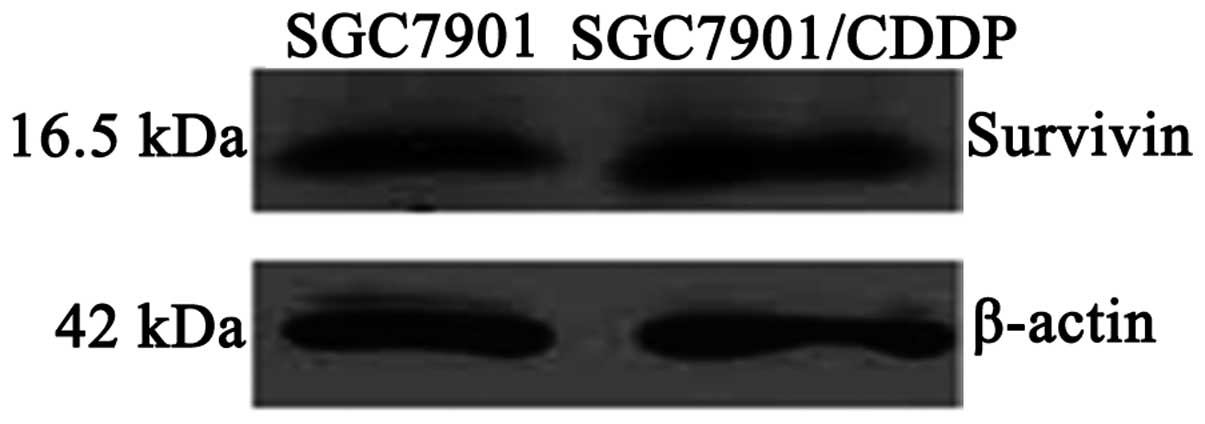
Survivin mRNA and protein expression
levels in SGC7901 and SGC7901/CDDP cells
Survivin mRNA expression levels were significantly
higher in the SGC7901/CDDP cells compared with the SGC7901 cells
(P<0.05; Table I; Fig. 2). Survivin protein expression levels
were also significantly higher in the SGC7901/CDDP cells compared
with the SGC7901 cells (P<0.05; Table I; Fig.
3).
 | Table ISurvivin mRNA and protein expression
levels in SGC7901 and SGC7901/CDDP human gastric cancer cells. |
Table I
Survivin mRNA and protein expression
levels in SGC7901 and SGC7901/CDDP human gastric cancer cells.
| Cell type | |
|---|
|
| |
|---|
| Parameter | SGC7901 | SGC7901/CDDP | P-value |
|---|
| mRNA | 0.748±0.011 | 1.010±0.068 | <0.05 |
| Protein | 1.430±0.234 | 2.565±0.382 | <0.05 |
Discussion
Gastric cancer is a malignant disease with high
morbidity and mortality. Since gastrointestinal tumors are
sensitive to chemotherapy, this form of therapy is an indispensable
element in the comprehensive treatment of gastric cancer. In recent
years adjuvant chemotherapy and neoadjuvant chemotherapy have been
increasingly used in clinical practice, but the survival times of
patients with gastric cancer have not been significantly prolonged
(3). One reason for this is the
resistance of gastric cancer cells to chemotherapeutic drugs. CDDP
remains a commonly used classical drug for patients with gastric
cancer. CDDP kills tumor cells by binding the DNA, forming
cross-links with the DNA, and subsequently inhibiting DNA synthesis
and the division of the cancer cells (4–6). In
tumor cell resistance to CDDP, the efficacy of CDDP is reduced,
even resulting in the failure of chemotherapy in patients with
gastric cancer. However, the mechanism of chemotherapy resistance
of gastric cancer cells to CDDP is unclear.
To clarify the underlying mechanism of the
chemotherapeutic resistance of gastric cancer cells to CDDP, a
CDDP-resistant gastric cancer cell strain was established in
vitro. At present, there are three methods to establish
resistant tumor cell strains, consisting of the stepwise exposure
of cells to increasing concentrations of the drug (7–9), low
doses of the drug (intermittent induced method) (10) and high doses of the drug
(intermittent induced method) (11). In the present study, SGC7901/CDDP
cells were established by the stepwise exposure of SGC7901 cells to
increasing CDDP concentrations.
Goldie and Coldman (12) considered there to be two types of
drug resistance in tumor cells, namely endogenous drug resistance
and acquired drug resistance. Acquired drug resistance in tumor
cells indicates that the sensitivity of tumor cells to drugs is
progressively reduced, thus the drug becomes less effective or even
ineffective; a mechanism similar to the development of antibiotic
resistance. Conversely, endogenous drug resistance does not undergo
a gradual desensitization process; the resistance is already
present prior to the initiation of drug treatment. The SGC7901/CDDP
strain produced in the present study underwent the former process.
Drug resistance may involve the mutation of tumor cells during cell
growth and proliferation. Resistant cells are produced in each
mutation. The cells that adapt to the mutation during this time may
have a novel mutation which has been induced by the change in drug
concentrations. Thus, drug resistance is not an all-or-nothing
phenomenon, but a gradual process (13). The establishment of the SGC7901/CDDP
cells was also a gradual process in the present study. As the drug
concentration was continuously increased in the culture medium, the
CDDP resistance strain, SGC7901/CDDP, was developed.
The established SGC7901/CDDP cells exhibited stable
growth and proliferation following cryopreservation, and long-term
culture indicated that the resistance of the cells to CDDP was
relatively stable. Therefore, the SGC7901/CDDP cell line was
reliable and an ideal cell model for analyzing the mechanism of
CDDP resistance. The majority of drug-resistant strains exhibit
multidrug-resistant characteristics. Whether the SGC7901/CDDP cells
are resistant to other commonly used chemotherapy drugs, such as
5-fluorouracil, paclitaxel and hydroxycampothecin, requires further
investigation.
Comparing the morphology of SGC7901 and SGC7901/CDDP
cells by microscopy, the SGC7901/CDDP cells were reduced in number,
with certain cells deformed or increased in size. Giant cells were
also visualized, demonstrating that the cells were damaged. The
degree of refraction had declined, the space between the cells was
increased, the cells were less adherent to the flask and a few
cells were floating in the culture medium. Reduced cell
proliferation was also observed. Survivin mRNA and protein were
expressed in the SGC7901 and SGC7901/CDDP cells, but the expression
levels were significantly higher in the SGC7901/CDDP cells compared
with the SGC7901 cells. The results suggest that the induction of
increased Survivin gene expression levels is a cause of CDDP
resistance in gastric cancer cells.
One study has shown that the knockdown of Survivin
expression enhances the sensitivity of gastric cancer cells to CDDP
(12). Furthermore, overexpression
of the Survivin gene in gastric cancer cells has been associated
with the resistance to docetaxel-based chemotherapy in patients
with advanced gastric cancer (15),
and the overexpression of the Survivin gene induced by CDDP has
been demonstrated to aid cancer cells in overcoming the apoptosis
checkpoint at the G2/M phase of the cell cycle (16). Prior to chemotherapy, the analysis
of Survivin gene expression does not indicate whether the tumor is
sensitive or resistant to chemotherapeutic drugs. However, during
chemotherapy, the assessment of Survivin gene expression may
provide novel information regarding tumor drug sensitivity
(17), although the mechanism is
unclear (18). Therefore, Survivin
gene expression levels may add significant prognostic value to the
current tumor-node-metastasis staging system (19), and the correlation between the
expression of Survivin and overall survival time for patients with
gastric cancer is evident (20).
In the intraocular environment of the body, the
metabolic changes and drug resistance induced by CDDP are difficult
to explain clearly, and the underlying mechanism may not be
elucidated simply by establishing a cell model in vitro.
Thus, the underlying mechanism with regard to the development of
chemotherapeutic resistance to CDDP in gastric cancer cells
requires further investigation.
References
|
1
|
Johnson ME and Howerth EW: Survivin: a
bifunctional inhibitor of apoptosis protein. Vet Pathol.
41:599–607. 2004.
|
|
2
|
Ambrosini G, Adida C and Altieri DC: A
novel anti-apoptosis gene, survivin, expressed in cancer and
lymphoma. Nat Med. 3:917–921. 1997.
|
|
3
|
Fujitani K: Overview of adjuvant and
neoadjuvant therapy for resectable gastric cancer in the East. Dig
Surg. 30:119–129. 2013.
|
|
4
|
Raffo AJ, Kim AL and Fine AL: Formation of
nuclear Bax/p53 complexes is associated with chemotherapy induced
apotosis. Oncogene. 19:6216–6228. 2000.
|
|
5
|
Blanc C, Deveraux QL, Krajewski S, et al:
Caspase-3 is essential for procaspase-9 processing and
cisplatin-induced apoptosis of MCF-7 breast cancer cells. Cancer
Res. 60:4386–4390. 2000.
|
|
6
|
Sasaki CY, Lin HC and Passaniti A:
Expression of E-cadherin reduces bcl-2 expression and increases
sensitivity to etoposide-induced apoptosis. Int J Cancer.
86:660–666. 2000.
|
|
7
|
Yang LY and Trujillo JM: Biological
Characterisation of Multidrug-resistant Human Colon Carcinoma
Sublines Induced/Selected by Two Methods. Cancer Res. 50:3218–3225.
1990.
|
|
8
|
Vandier D, Calvez V, Massade L, et al:
Transactivation of the metallothionein promoter in
cisplatin-resistant cancer cells: a specific gene therapy strategy.
J Natl Cancer Inst. 92:642–647. 2000.
|
|
9
|
Kotoh S, Naito S, Yokomizo A, et al:
Increased expression of DNA topoisomerase I gene and collateral
sensitivity to camptothecin in human cisplatin-resistant bladder
cancer cells. Cancer Res. 54:3248–3252. 1994.
|
|
10
|
Godwin AK, Meister A, O’Dwyer PJ, et al:
High resistance to cisplatin in human ovarian cancer cell lines is
associated with marked increase of glutathione synthesis. Proc Natl
Acad Sci USA. 89:3070–3074. 1992.
|
|
11
|
Hammond JR, Johnstone RM and Gros P:
Enhanced efflux of [3H]vinblastine from Chinese hamster ovary cells
transfected with a full-length complementary DNA clone for the mdr1
gene. Cancer Res. 49:3867–3871. 1989.
|
|
12
|
Goldie JH and Coldman AJ: The Genetic
Origin of Drug Resistance in Neoplasms: Implications for Systemic
Therapy. Cancer Res. 44:3643–3653. 1984.
|
|
13
|
Goldie JH: Drug Resistance in Cancer: A
Perspective. Cancer Metastasis Rev. 20:63–68. 2001.
|
|
14
|
Shen X, Zheng JY, Shi H, et al: Survivin
knockdown enhances gastric cancer cell sensitivity to radiation and
chemotherapy in vitro and in nude mice. Am J Med Sci. 344:52–58.
2012.
|
|
15
|
Zheng WE, Chen H, Yuan SF, et al:
Overexpression of βIII-tubulin and survivin associated with drug
resistance to docetaxel-based chemotherapy in advanced gastric
cancer. J BUON. 17:284–290. 2012.
|
|
16
|
Wenying Z, Zhaoning J, Zhimin Y, et al:
Survivin siRNA inhibits gastric cancer in nude mice. Cell Biochem
Biophys. 62:337–341. 2012.
|
|
17
|
Ikeguchi M, Nakamura S and Kaibara N:
Quantitative analysis of expression levels of bax, bcl-2, and
survivin in cancer cells during cisplatin treatment. Oncol Rep.
9:1121–1126. 2002.
|
|
18
|
Cregan IL, Dharmarajan AM and Fox SA:
Mechanisms of cisplatin-induced cell death in malignant
mesothelioma cells: role of inhibitor of apoptosis proteins (IAPs)
and caspases. Int J Oncol. 42:444–452. 2013.
|
|
19
|
Bertazza L, Mocellin S, Marchet A, et al:
Survivin gene levels in the peripheral blood of patients with
gastric cancer independently predict survival. J Transl Med.
7:1112009.
|
|
20
|
Krieg A, Baseras B, Tomczak M, et al: Role
of survivin as prognostic and clinicopathological marker in gastric
cancer: a meta-analysis. Mol Biol Rep. 40:5501–5511. 2013.
|