Introduction
Renal cell carcinoma (RCC) is the most common
neoplasm in the kidney, with an estimated 5-year survival rate of
50–60%. In the United States, RCC incidence has been increasing
with an estimated 65,150 new cases and 13,680 deaths in 2013
(1). In the Republic of Korea, RCC
accounts for ~1% of all primary malignancies and is the 10th most
common cancer in males (2). RCC has
several subtypes, each derived from various parts of the nephron,
with each presenting different genetic characteristics,
histological features and clinical phenotypes. The most common
subtype is the clear cell type, accounting for >75% of cases
(3). Recent research in genetics
has enabled the discovery of alterations in different pathways and
the benefits of molecular profiling research have already been
incorporated into clinical oncology, such as targeted therapy
(4). Development of targeted
therapies has changed the treatment of metastatic RCC, and further
information will become available from the genomic approach to
tumor classification, prognostic markers and predictive indicators
of response to treatment, along with personal susceptibility to
developing cancer when exposed to risk factors (5).
DNA hypermethylation plays a critical role in the
regulation of gene expression in differentiation, development and
disease (6). Changes in DNA
methylation are recognized as one of the most common forms of
molecular alteration in cancer development. Hypermethylation of CpG
islands located in the promoter regions of tumor suppressor genes
is established as the most frequent mechanism for gene
inactivation. Previously, we reported the hypermethylation status
of the Mut-S-Homologon-2 (MSH2) gene in clear cell RCC (7). A high-throughput genotyping assay
(GoldenGate Methylation Cancer Panel I microarray; Illumina, San
Diego, CA, USA) (6) was adapted to
determine the methylation status of 1,505 specific CpG sites in 807
cancer-related genes. Tissue specimens consisted of 62 cancer
tissues and 62 matched adjacent normal tissues obtained from clear
cell RCC patients of the Kyung Hee University Hospital (Seoul,
Korea). The results revealed that the mean β-value difference
between cancer and normal tissues was 0.30±0.28 for MSH2. We
examined the methylation status of CpG sites by bisulfite
sequencing. The results showed that the methylation rate of the
MSH2 gene was 54.8% in cancer tissue and 26.1% in normal tissue.
The MSH2 gene was hypermethylated in cancer tissue compared with
normal tissue.
The DNA mismatch repair (MMR) system is essential to
maintain the stability of the genome during repeated duplication
(8). Main functions of the MMR
system include the correction of biosynthetic errors, DNA damage
surveillance and prevention of recombination. The MMR system is
composed of a few well-conserved proteins, including Mut-S-Homolog
proteins and Mut-L-Homolog proteins. Genetic studies have revealed
that MSH2, which is one of the Mut-S-Homolog proteins, is required
for all mismatch corrections in nuclear DNA during replication;
whereas Mut-S-Homolog-3 and Mut-S-Homolog-6, which are also
Mut-S-Homolog proteins, are required for the repair of certain
overlapping types of mismatched DNA (9).
In present study aimed to investigate tumoral MSH2
immunohistochemical expression in clear cell RCC, as well as the
associations between tumoral MSH2 immunohistochemical expression
and clinicopathological parameters.
Material and methods
Patients and tissue specimens
Tissue samples from 129 clear cell RCC cases, 88
males (median age, 61.0±12.31 years) and 41 females (median age,
60.0±10.97 years), were used. All neoplasms were surgically
resected at the Kyung Hee University Hospital and the Kyung Hee
University Hospital at Gangdong (Seoul, Korea) from January 2000 to
December 2012. Tumors were graded according to criteria of the
American Joint Committee on Cancer (10). The clinical parameters, including
tumor grade, recurrence, progression and overall survival, were
analyzed along with the immunohistochemical results. The
institutional review board of the Kyung Hee University Hospital at
Gangdong approved this study (KHNMC IRB 2013-040). Written informed
consent was obtained from all patients.
Immunohistochemical staining
The tissue microarrays were assembled using a
commercially available manual tissue microarrayer (Quick-Ray;
Unitma Co., Ltd., Seoul, Korea) (11). Three representative tumor cores with
diameters of 2.0 mm were punched from each tumor tissue block. Each
of the tissue microarray blocks contained three normal kidney
tissue cores. Immunohistochemistry was performed on 4-μm tissue
sections from each tissue microarray block using the Bond Polymer
Intense Detection system (Vision BioSystems, Mount Waverley,
Victoria, Australia). Sections were incubated for 15 min at ambient
temperature with primary mouse anti-human MSH2 monoclonal antibody
(1:3,000, clone G219-1129; BD Biosciences, San Jose, CA, USA),
using a biotin-free polymeric horseradish peroxidase-linker
antibody conjugate system in a Bond-max automatic slide stainer
(Vision BioSystems). Nuclei were counterstained with hematoxylin.
The negative control was treated in an identical manner using mouse
IgG instead of the primary antibody. The degree of expression by
immunohistochemistry was classified by three pathologists blinded
to the data. Semiquantitative analysis of immunoreactivity was
performed according to intensity and proportion. The intensity
score was determined as 0, no staining; 1, weak but detectable
staining; 2, distinct staining; and 3, strong staining. The
proportion score was determined as 1, 0–10%; 2, 11–50%; 3, 51–80%;
and 4, 81–100%. The total score was the sum of the intensity score
and the proportion score. Total scores were as follows: 1 and 2,
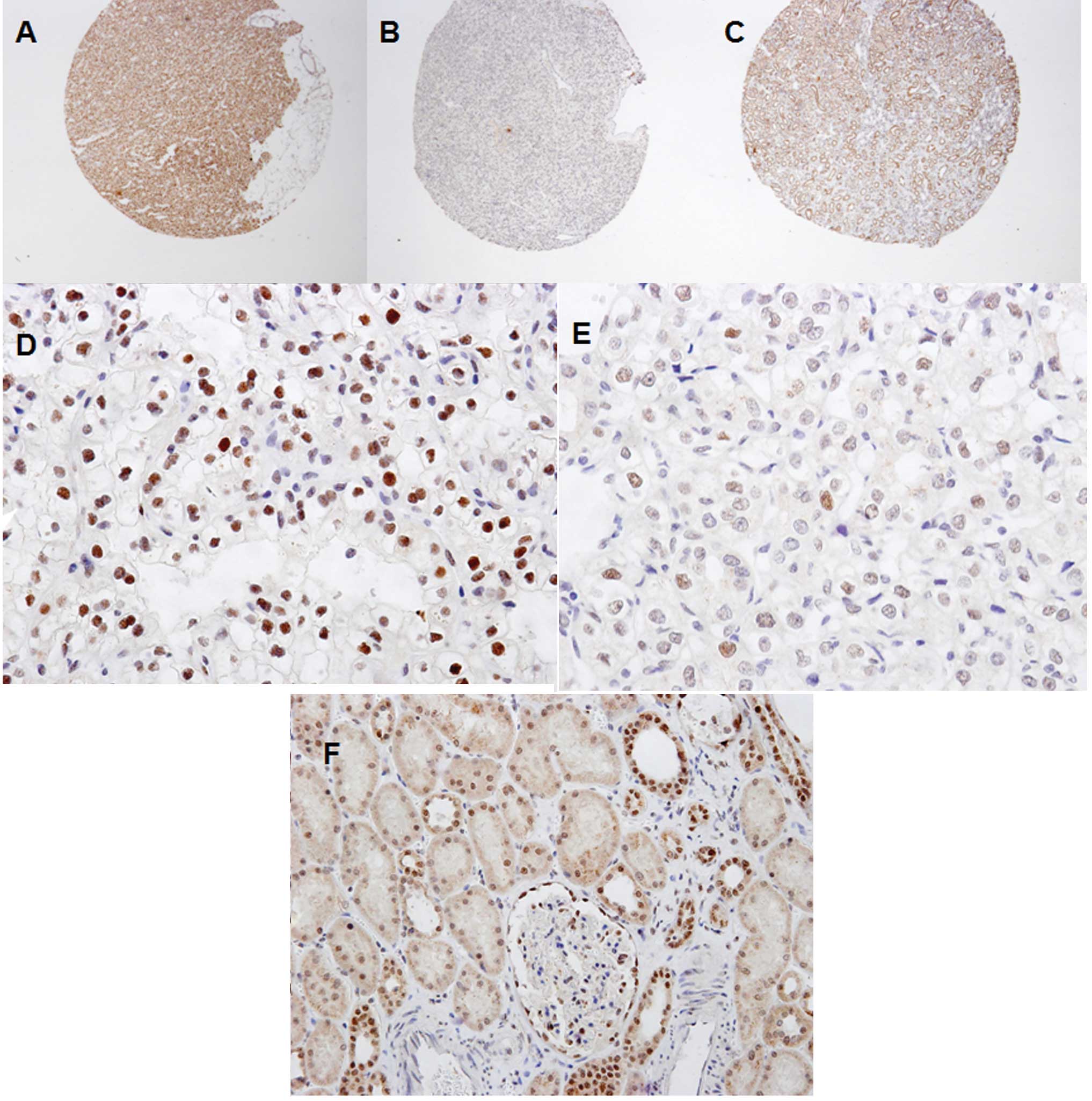
negative staining; and 3–7, positive staining (Fig. 1A–C) (12–14).
Statistical analysis
The χ2 test and linear-by-linear
association were used to evaluate the association between the
degree of expression determined by immunohistochemistry (negative
staining group, MSH2-negative group; positive staining group,
MSH2-positive group) with clinicopathological variables. Survival
was estimated using the Kaplan-Meier method, and comparisons among
survival curves were made using the log-rank test. P<0.05 was
considered to indicate a statistically significant difference.
Statistical analyses were performed using SPSS software, version 16
(SPSS, Inc., Chicago, IL, USA).
Results
Immunohistochemical expression pattern of
clear cell renal cell carcinoma
In the cancer tissue, nuclear MSH2 protein
expression was diffusely strong (Fig.
1D) or focally weak (Fig. 1E).
Negative and positive MSH2 expression was identified in 53 (41.1%)
and 76 (58.9%) out of 129 cases, respectively. In the normal
tissue, MSH2 protein was expressed in the nuclei of tubular
epithelial cells and parietal cells of Bowman’s capsule (Fig. 1F).
Associations between MSH2 protein
expression and clinical variables
Overall, 129 clear cell RCC cases (median age, 61
years) were included in the study. The comparison between
MSH2-negative patients (n=53) and MSH2-positive patients (n=76) is
shown in Table I. There were 35
males and 18 females in MSH2-negative group, and 53 males and 23
females in MSH2-positive group (P=0.657). The median patient age
(±SD) was 61.00±11.06 years in the MSH2-negative group and
60.00±12.50 years in the MSH2-positive group (P=0.823).
 | Table IAnalysis of MSH2 immunohistochemistry
results and clinicopathological parameters of 129 clear cell renal
cell carcinoma patients. |
Table I
Analysis of MSH2 immunohistochemistry
results and clinicopathological parameters of 129 clear cell renal
cell carcinoma patients.
| Parameter | MSH2-negative group
(n=53, %) | MSH2-positive group
(n=76, %) | P-value |
|---|
| Males/females, n | 35/18 | 53/23 | 0.703a |
| Median age, years
(±SD) | 61.00±11.06 | 60.00±12.50 | 0.823b |
| Clinical T stage,
n | | | 0.021c |
| T1a | 17 (32) | 43 (57) | |
| T1b | 14 (26) | 16 (21) | |
| T2a | 9 (17) | 6 (8) | |
| T2b | 1 (2) | 2 (3) | |
| T3a | 9 (17) | 7 (9) | |
| T3b | 1 (2) | 2 (3) | |
| T3c | 0 (0) | 0 (0) | |
| T4 | 1 (2) | 0 (0) | |
| Clinical N stage,
n | | | 0.072a |
| N0 | 49 (92) | 75 (99) | |
| N1 | 4 (8) | 1 (1) | |
| Clinical M stage,
n | | | 0.759a |
| M0 | 48 (91) | 70 (92) | |
| M1 | 5 (9) | 6 (8) | |
| Fuhrman’s nuclear
grade, n | | | 0.118c |
| Grade I | 3 (6) | 4 (5) | |
| Grade II | 20 (38) | 40 (53) | |
| Grade III | 22 (42) | 26 (34) | |
| Grade IV | 8 (15) | 6 (8) | |
T stage was significantly higher in the
MSH2-negative group than in the MSH2-positive group (P=0.021).
There were no significant differences in terms of N stage, M stage
and Fuhrman’s grade between the two groups (N stage; P=0.072, M
stage; P=0759, Fuhrman grade; P=0118).
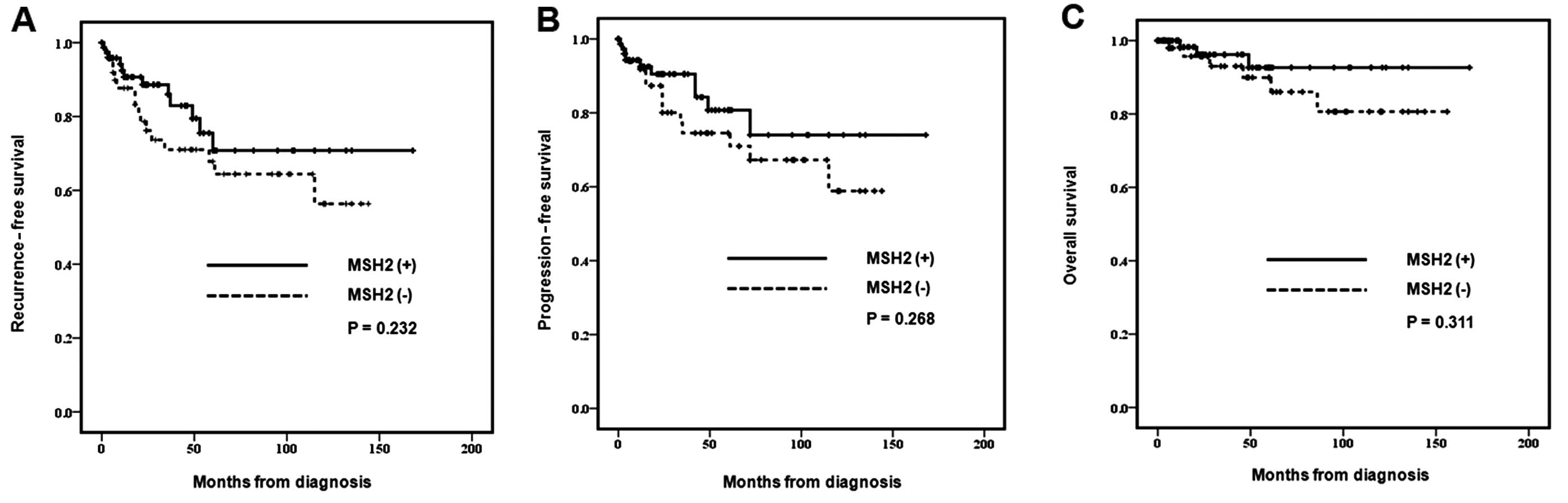
Kaplan-Meier survival analysis
The MSH2-negative group showed decreased rates of
recurrence-free survival, progression-free survival and overall
survival, without statistically significant results (P=0.232,
P=0.268 and P=0.311, respectively) (Fig. 2).
Discussion
DNA methylation in cancer has been recognized for
over 30 years (15–16). Two mechanisms of losses of
methylation for carcinogenesis have been proposed. A weakening of
transcriptional repression in normally silent regions could cause
the harmful expression of inserted viral genes and repeat elements,
and of normally silenced genes (17–18).
Losses of methylation may affect the functional stability of
chromosomes in cancer (18).
Pericentromeric regions of chromosomes depend on correct levels of
DNA methylation for the stability of DNA. As hypermethylation in
gene promoters occurs early on in tumor progression, inhibiting or
reversing these changes has a potential benefit for cancer
prevention (20). Reactivation of
the types of genes that are epigenetically silenced in cancer may
have a profound antitumor effect. Early detection of cancer is
essential to survival of cancer patients. The use of
hypermethylation of CpG islands as tumor markers has benefited from
extremely sensitive polymerase chain reaction (PCR) methods to
detect methylated DNA sequences (16).
Several studies have reported associations between
MSH2 gene methylation and gastrointestinal and urothelial tumor
carcinogenesis (15,21,22).
Heritable germline epimutations in MSH2 have been identified in a
small number of Lynch syndrome families, which did not exhibit
germline mutations in the MSH2 gene (23). Nagasaka et al reported the
methylation status of the MSH2 gene in 268 colorectal cancers
tissues, which comprised 222 sporadic colorectal cancers and 46
Lynch syndrome tumors that did not express MSH2 (24). The results showed that there was
frequent MSH2 hypermethylation in Lynch syndrome tumors with MSH2
deficiency. The authors concluded that high levels of aberrant
methylation at CpG island methylator phenotype-related markers in
MSH2-methylated tumors highlights the possibility that MSH2 is a
target which is susceptible to aberrant methylation in Lynch
syndrome. Tumor-specific alterations of MSH2 hypermethylation in
circulating DNA have been associated with esophageal squamous cell
carcinoma (25). Promoter
hypermethylation of MSH2 was analyzed using real-time
methylation-specific PCR in paired tumor and plasma samples of 209
patients with esophageal squamous cell carcinoma. Aberrant MSH2
methylation was found in 101 of 209 esophageal squamous cell
carcinoma patients. Follow-up analysis indicated significantly
lower disease-free survival rates for patients with high MSH2
methylation compared with those with MSH2 unmethylation after
surgery.
Recently, it was reported that patients with Lynch
syndrome and MSH2 mutation are at an increased risk of developing
urothelial carcinoma (26). Using
cancer data of the Familial Gastrointestinal Cancer Registry in
Toronto, Lynch syndrome patients with MSH2 mutations are at an
increased risk of developing not only upper tract urothelial cancer
but also bladder cancer, and could be offered appropriate screening
(26). Stoehr et al reported
that MMR proteins hMLH1 and hMSH2 are differentially expressed in
the three main subtypes of sporadic RCC (clear cell, papillary and
chromophobe) (27). Expression of
hMLH1 and hMSH2 was investigated in 166 RCC patients of the main
subtypes (101, clear cell; 30, papillary; 32, chromophobe; and 3,
mixed RCC) by immunohistochemistry. Expression of hMLH1 and hMSH2
was reduced in 83.7% (118/141) and 51.2% (65/127) of clear cell RCC
and papillary RCC cases, respectively. None of the clear cell RCC
tumors showed high hMLH1 expression, while papillary and
chromophobe RCC demonstrated preserved high hMLH1 expression in
25.0 and 33.3% of cases, respectively. Subtype specificity was
present in the hMSH2 staining; chromophobe RCC showed high
expression in 41.7% of cases, while clear cell and papillary tumors
did not retain high expression. Therefore, diminished MMR protein
expression was linked to tumor entity and may contribute to the
different biological behavior of the RCC subtypes.
Previously, we utilized a high-throughput method for
analyzing the methylation status of 807 preselected genes; the
GoldenGate Methylation Cancer Panel I microarray (6,7). The
results revealed that the mean β-value difference between cancer
and normal tissues was 0.30±0.28 for MSH2, and the methylation rate
of the MSH2 gene was 54.8% in cancer tissue and 26.1% in normal
tissue. The MSH2 gene was hypermethylated in cancer tissue compared
with normal tissue. In the present study, using immunohistochemical
staining, T stage was found to be significantly higher in
MSH2-negative clear cell RCC patients, compared with those that
were MSH2-positive. There was no significant difference in terms of
N stage, M stage and Fuhrman’s grade between the MSH2-negative and
MSH2-positive group. The MSH2-negative group showed decreased rates
of recurrence-free survival, progression-free survival and overall
survival, without statistically significant results. The product of
the MSH2 gene corrects any mismatched nucleotide errors in the DNA
strand using the mismatch-repair mechanism and maintains the
precise fidelity of DNA replication. Chen et al (28) reported a new primary RCC cell line
and revealed a deletion of 1476 bp encoding 492 amino acids of MSH2
cDNA, truncated forms of MSH2, and microsatellite instability using
a clear cell RCC cell line. Chen et al concluded that
genomic instability caused by a variety of mutational events
appears to be the important cornerstone of carcinogenesis. However,
Leach et al reported that complete inactivation of mismatch
repair is uncommon in RCC (29).
Leach et al demonstrated that genetic alterations affecting
expression were limited to MLH1 since MSH2, MSH6, PMS2 were
detectable in their RCC cell lines. Chen et al and Leach
et al reported that a damaged mismatch repair system is
related to RCC (28,29). In order to demonstrate this outcome,
one of the genetic methods utilized were microsatellite mutations.
In our study, we observed similar results using DNA methylation. We
believe that this indicates that the results are not only due to
genetic factors, such as mutations, but also epigenetic factors,
such as DNA methylation.
In conclusion, the current study identified that
MSH2-negative staining was associated with a higher T stage in
clear cell RCC patients. MSH2 protein expression may be a useful
marker for predicting TNM stage and prognosis and, thus, MSH2 may
be a prognostic factor in clear cell RCC. The results of the
present study may lead to further large-scale studies regarding the
identification of tumor markers for clear cell RCC.
Acknowledgements
This study was supported by a grant from the Kyung
Hee University in 2013 (KHU-20130367).
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013.
|
|
2
|
Kim H, Cho NH, Kim DS, et al: Renal cell
carcinoma in South Korea: a multicenter study. Hum Pathol.
35:1556–1563. 2004.
|
|
3
|
Rini BI, Campbell SC and Escudier B: Renal
cell carcinoma. Lancet. 373:1119–1132. 2009.
|
|
4
|
Figlin R, Sternberg C and Wood CG: Novel
agents and approaches for advanced renal cell carcinoma. J Urol.
188:707–715. 2012.
|
|
5
|
Audenet F, Yates DR, Cancel-Tassin G,
Cussenot O and Rouprêt M: Genetic pathways involved in
carcinogenesis of clear cell renal cell carcinoma: genomics towards
personalized medicine. BJU Int. 109:1864–1870. 2012.
|
|
6
|
Bibikova M, Lin Z, Zhou L, et al:
High-throughput DNA methylation profiling using universal bead
arrays. Genome Res. 16:383–393. 2006.
|
|
7
|
Yoo KH, Park YK, Kim HS, Jung WW and Chang
SG: Epigenetic inactivation of HOXA5 and MSH2 gene in clear cell
renal cell carcinoma. Pathol Int. 60:661–666. 2010.
|
|
8
|
Peltomäki P: Role of DNA mismatch repair
defects in the pathogenesis of human cancer. J Clin Oncol.
21:1174–1179. 2003.
|
|
9
|
Marsischky GT, Filosi N, Kane MF and
Kolodner R: Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in
MSH2-dependent mismatch repair. Genes Dev. 10:407–420. 1996.
|
|
10
|
Frank I, Blute ML, Leibovich BC, Cheville
JC, Lohse CM and Zincke H: Independent validation of the 2002
American Joint Committee on cancer primary tumor classification for
renal cell carcinoma using a large, single institution cohort. J
Urol. 173:1889–1892. 2005.
|
|
11
|
Won KY, Kim HS, Sung JY, et al: Tumoral
FOXP3 has potential oncogenic function in conjunction with the p53
tumor suppressor protein and infiltrated Tregs in human breast
carcinomas. Pathol Res Pract. 209:767–773. 2013.
|
|
12
|
Kouso H, Yoshino I, Miura N, et al:
Expression of mismatch repair proteins, hMLH1/hMSH2, in non-small
cell lung cancer tissues and its clinical significance. J Surg
Oncol. 98:377–383. 2008.
|
|
13
|
Uehara H, Miyamoto M, Kato K, et al:
Deficiency of hMLH1 and hMSH2 expression is a poor prognostic
factor in esophageal squamous cell carcinoma. J Surg Oncol.
92:109–115. 2005.
|
|
14
|
Olasz J, Mándoky L, Géczi L, Bodrogi I,
Csuka O and Bak M: Influence of hMLH1 methylation, mismatch repair
deficiency and microsatellite instability on chemoresistance of
testicular germ-cell tumors. Anticancer Res. 25:4319–4324.
2005.
|
|
15
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003.
|
|
16
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996.
|
|
17
|
Walsh CP, Chaillet JR and Bestor TH:
Transcription of IAP endogenous retroviruses is constrained by
cytosine methylation. Nat Genet. 20:116–117. 1998.
|
|
18
|
Cui H, Cruz-Correa M, Giardiello FM, et
al: Loss of IGF2 imprinting: a potential marker of colorectal
cancer risk. Science. 299:1753–1755. 2003.
|
|
19
|
Xu GL, Bestor TH, Bourc’his D, et al:
Chromosome instability and immunodeficiency syndrome caused by
mutations in a DNA methyltransferase gene. Nature. 402:187–191.
1999.
|
|
20
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.
|
|
21
|
Martin-Subero JI, Ammerpohl O, Bibikova M,
et al: A comprehensive microarray-based DNA methylation study of
367 hematological neoplasms. PLoS One. 4:e69862009.
|
|
22
|
Vlaykova T, Mitkova A, Stancheva G, et al:
Microsatellite instability and promoter hypermethylation of MLH1
and MSH2 in patients with sporadic colorectal cancer. J BUON.
16:265–273. 2011.
|
|
23
|
Lynch HT and de la Chapelle A: Hereditary
colorectal cancer. N Engl J Med. 348:919–932. 2003.
|
|
24
|
Nagasaka T, Rhees J, Kloor M, et al:
Somatic hypermethylation of MSH2 is a frequent event in Lynch
Syndrome colorectal cancers. Cancer Res. 70:3098–3108. 2010.
|
|
25
|
Ling ZQ, Zhao Q, Zhou SL and Mao WM: MSH2
promoter hypermethylation in circulating tumor DNA is a valuable
predictor of disease-free survival for patients with esophageal
squamous cell carcinoma. Eur J Surg Oncol. 38:326–332. 2012.
|
|
26
|
Skeldon SC, Semotiuk K, Aronson M, et al:
Patients with Lynch syndrome mismatch repair gene mutations are at
higher risk for not only upper tract urothelial cancer but also
bladder cancer. Eur Urol. 63:379–385. 2013.
|
|
27
|
Stoehr C, Burger M, Stoehr R, et al:
Mismatch repair proteins hMLH1 and hMSH2 are differently expressed
in the three main subtypes of sporadic renal cell carcinoma.
Pathobiology. 79:162–168. 2012.
|
|
28
|
Chen HC, Bhattacharyya N, Wang L, et al:
Defective DNA repair genes in a primary culture of human renal cell
carcinoma. J Cancer Res Clin Oncol. 126:185–190. 2000.
|
|
29
|
Leach FS, Koh M, Sharma K, et al: Mismatch
repair gene mutations in renal cell carcinoma. Cancer Biol Ther.
1:530–536. 2002.
|