Introduction
Despite recent improvements in its diagnosis, lung
cancer remains a significant cause of mortality among malignant
diseases due to its high incidence rate, malignant behavior and a
lack of major advancements in treatment strategies (1). In Japan in 2011, the majority of
respiratory surgeries performed were a result of lung cancer
(48.9%) and >33,000 patients underwent surgery for lung cancer
(2). The clinical behavior of lung
cancer is predominantly associated with its stage; thus, the
treatment of lung cancer by surgery is only achieved in cases
presenting in an early stage (3).
In addition to epidermal growth factor receptor
(EGFR) and anaplastic lymphoma kinase gene alternations,
genomic studies in lung adenocarcinoma have identified other
potential therapeutic targets, including activating mutations in
Kras, BRAF, HER2 and PIK3CA, in
frequencies >1% (4–6). BRAF mutations in lung
adenocarcinoma would be of interest as these mutations may be
associated with increased sensitivity to agents directly targeting
BRAF or BRAF-mediated downstream signaling pathways (7,8). For
example, BRAF V600E is a driver mutation that can be
effectively targeted with selective BRAF and/or MEK inhibitors
(9–11). Previous reports identified
BRAF mutations in 1–4% of cases of lung adenocarcinoma
(12–15), and 40–50% of lung cancer cases have
been demonstrated to harbor non-V600E mutations distributed in
exons 11 and 15 (12–17). A number of these non-V600E mutations
exhibit only intermediate or low kinase activity, and the analysis
of preclinical data indicates that non-V600E-mutant BRAF kinases
may be resistant to BRAF-targeted therapy (17,18).
Although BRAF copy number gain has been
investigated in thyroid tumors (19), to the best of our knowledge, the
association between BRAF gene mutation and copy number gain
in Japanese lung adenocarcinoma patients has not previously been
reported. In the present study, the possibility that BRAF
copy number gain represents a novel mechanism for BRAF gene
mutation is investigated. To determine the BRAF copy number
status in Japanese lung adenocarcinoma patients, quantitative
polymerase chain reaction (qPCR) amplification was performed. The
findings were compared with the clinicopathological features of the
lung cancer patients and data from fluorescence in situ
hybridization (FISH) performed using BRAF-specific and
chromosome 7 centromeric probes. Typically, increases in
BRAF copy number are moderate; however, in V600E lung
adenocarcinomas, BRAF copy number increases occur with
significant prevalence.
Patients and methods
Patients
The study group included 29 lung adenocarcinoma
patients who had undergone surgery at the Department of Oncology,
Immunology and Surgery, Nagoya City University Hospital (Nagoya,
Japan) between 2002 and 2011. All tumor samples were immediately
frozen and stored at −80°C until assaying.
The clinical and pathological characteristics of the
29 lung adenocarcinoma patients were as follows: Stage I, 16 cases;
stage II, six cases; and stage III, seven cases. The mean age of
the patients was 67.5 years (range, 47–84 years). Among the 29 lung
adenocarcinoma patients, eight were female and 10 were non-smokers.
The samples from these patients had previously been analyzed for
EGFR or Kras gene status (20,21)
and were considered to be wild-type. This study was approved by the
ethics committee of Nagoya City University (Nagoya, Japan) and
written informed consent was obtained from all patients.
PCR assays for BRAF
Genomic DNA was extracted from the lung cancer
tissues using the Wizard® SV Genomic DNA Purification
system (Promega Corporation, Madison, WI, USA), according to the
manufacturer’s instruction. The DNA concentration was determined
using a NanoDrop spectrophotometer (ND-1000, version 3.0; Thermo
Fisher Scientific, Wilmington, DE, USA) and adjusted to a
concentration of 2.5 ng/ml. BRAF copy number was analyzed by
performing qPCR assays on a 7500 Real-Time PCR system (Applied
Biosystems Life Technologies, Foster City, CA, USA) using a
QuantiTect SYBR Green® PCR kit (Qiagen, Valencia, CA,
USA), with 5 μl DNA from each tumor sample (20,21).
The DNA of each tumor sample was quantified by comparing the target
locus (BRAF) to the reference long interspersed nucleotide
element (Line-1), a repetitive element for which the copy
number per haploid genome is similar in all healthy and neoplastic
human cells (22). The
quantification was based on a standard curve previously determined
from a serial dilution of healthy human genomic DNA (Roche
Diagnostics, Indianapolis, IN, USA) and the relative BRAF
copy number was normalized to the healthy human genomic DNA
(calibrator). Furthermore, the change in BRAF gene copy
number relative to Line-1 and the calibrator was determined
using the following formula: (T BRAF/T Line-1)/(C BRAF/C Line-1),
where T and C represent the quantity present in the tumor DNA and
the calibrator, respectively. BRAF copy number was
determined by assaying BRAF for each sample using the
following primers: Forward, 5′-TCATAATGCTTGCTCTGATAGGA-3′ and
reverse, 5′-GGCCAAAAATTTAATCAGTGGA-3′. In addition, the total DNA
content was estimated by assaying Line-1 elements for each
sample using the following primers: Forward,
5′-AAAGCCGCTCAACTACATGG-3′ and reverse,
5′-TGCTTTGAATGCGTCCCAGAG-3′. PCR was performed in triplicate for
each primer set and the cycling conditions were as follows: Initial
denaturation at 95°C for 15 min followed by 40 cycles at 94°C for
15 sec, 56°C for 30 sec and 72°C for 34 sec.
BRAF FISH analysis
Unstained 5-μm sections of formalin-fixed and
paraffin-embedded tumor tissue were submitted to dual-color FISH
analysis using four probe sets. The BRAF/CEN 7q probe sets
were developed at GSP Research, Inc. (Kawasaki, Japan) and were
labeled with Texas Red® (TexRed) and fluorescein
isothiocyanate (FITC). The probe sets were as follows: BRAF1 (390
kb; 140.3–140.7 MB) at chromosome 7p12-TexRed; and CEN 7q (820 kb;
64.2–65.1 MB)-FITC at chromosome 7q11.21. The lung adenocarcinoma
slides were deparaffinized and pre-incubated with Pretreatment
Solution (GSP Research, Inc.) at 95–99°C for 30 min, followed by
protease digestion buffer at 37°C for 10–20 min. The slides were
subsequently washed and dried. In addition, labeled probe sets (10
μl) were cohybridized at 37°C for 72 h following denaturation at
75°C for 5 min. A stringency wash was conducted at 72°C with 2X
saline-sodium citrate/0.3% Nonidet P-40 (Sigma-Aldrich, St. Louis,
MO, USA) for 1–2 min and the slides were counterstained with DAPI.
The slides were then visualized using the Leica MM AF imaging
system (Leica Microsystems, Wetzlar, Germany).
Statistical analysis
Statistical analyses of unpaired samples were
performed using the Mann-Whitney U test, and correlation
coefficients were determined by rank correlation using Spearman’s
rank correlation analysis and the χ2 test. All analyses
were performed using StatView software (Abacus Concepts, Inc.,
Berkeley, CA, USA) and P<0.05 was considered to indicate a
statistically significant difference.
Results
BRAF gene status in Japanese lung
adenocarcinoma patients
The clinicopathological data of the 29 lung cancer
patients is indicated in Table I.
Using primers sets for BRAF, 3/29 patients were identified
to express >3 copies of the BRAF gene. BRAF gene
copy status was not significantly correlated with gender (male,
9.5% vs. female, 12.5%; P>0.9999), tobacco-smoking (non-smoker,
0% vs. smoker, 15.8%; P=0.5320), pathological tumor (pT) status
(pT1, 18.2% vs. pT2–4, 5.6%; P=0.5394), tumor stage (stage I vs.
stage II–IV, P=0.9999) or age (<65 vs. ≥65, P=0.5320). No
non-V600E BRAF-mutant cases exhibited an increased BRAF copy
number; however, BRAF V600E status was correlated with an
BRAF increased copy number.
 | Table IClinicopathological data of 29 lung
cancer patients. |
Table I
Clinicopathological data of 29 lung
cancer patients.
| BRAF gene
status | |
|---|
|
| |
|---|
| Factor | Increased (n=3) | Normal (n=26) | P-value |
|---|
| Mean age,
yearsa (mean±SD) | 75.0±7.0 | 66.7±9.8 | 0.1670 |
| Age, years [n
(%)] |
| <65 | 0 (0.0) | 9 (36.6) | 0.5320 |
| ≥65 | 3 (100.0) | 17 (65.4) | |
| Gender, n (%) |
| Male | 2 (66.7) | 19 (73.1) | 0.9999 |
| Female | 1 (33.3) | 7 (26.9) | |
| Tumor stage, n
(%) |
| I | 2 (66.7) | 14 (53.8) | 0.9999 |
| II–IV | 1 (33.3) | 12 (46.2) | |
| Lymph node
metastasis, n (%) |
| N0 | 2 (66.7) | 17 (65.4) | 0.9999 |
| N+ | 1 (33.3) | 9 (36.6) | |
| Smoking status, n
(%) |
| Never-smoker | 0 (0.0) | 10 (38.5) | 0.5320 |
| Smoker | 3 (100.0) | 16 (61.5) | |
| BRAF mutation, n
(%) |
| V600E | 3 (100.0) | 2 (7.7) | 0.0027 |
| Non-V600E or
wild-type | 0 (0.0) | 24 (92.3) | |
| Pathological T
status, n (%) |
| T1 | 2 (66.7) | 9 (34.6) | 0.5394 |
| T2–4 | 1 (33.3) | 17 (65.4) | |
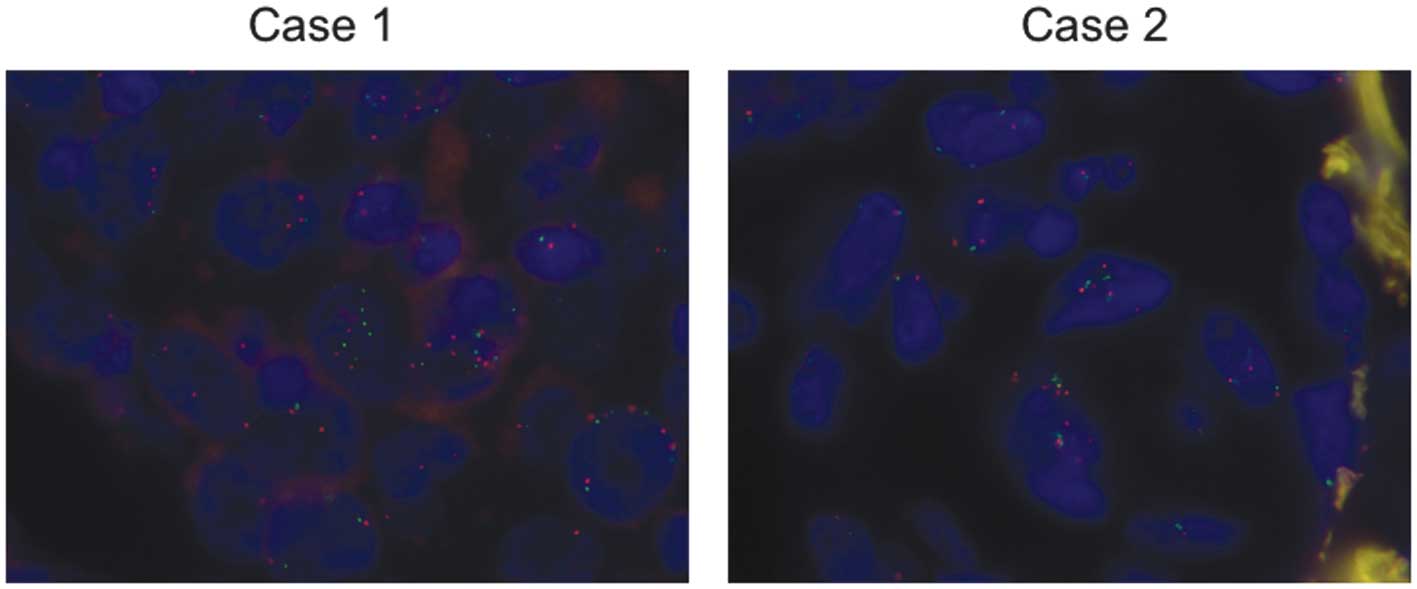
FISH
The screening of seven BRAF-mutant tumors by
FISH using a BRAF-specific probe revealed two cases (28.6%)
with BRAF gene amplification (Fig
1). The two cases were V600E mutants and demonstrated an
association between the BRAF copy number and chromosome 7
centromeric signals, indicating an association between numerical
changes of the BRAF locus and whole chromosome 7 amplification. The
BRAF copy number in the FISH-positive cases (whole
chromosome 7 amplification) was three, 4/5 stage I cases were
FISH-negative and 1/2 stage II cases were FISH-positive.
Discussion
In the present study, increased BRAF gene
copy number was identified in 10.3% of Japanese lung adenocarcinoma
patients without EGFR or Kras mutations. The
BRAF gene status was correlated with BRAF V600E
mutation and whole chromosome 7 amplification.
A previous report demonstrated that the clinical
outcomes of BRAF mutation-positive patients to
platinum-based combination chemotherapy resembled those of
wild-type lung cancer patients (23). Within the BRAF-mutant cohort,
patients with V600E mutations exhibited lower response rates to
platinum-based chemotherapy and shorter progression-free survival
compared with non-V600E mutation patients (23,24).
Previous studies have identified that V600E-mutated tumors are
frequently associated with a more aggressive histotype (24,25).
Furthermore, current second-generation BRAF inhibitors, such
as vemurafenib and dabrafenib, have potent, selective activity
against the V600-mutant BRAF kinases. One study in the literature
described a BRAF V600E-mutant lung cancer patient responding
to vemurafenib (7) and two studies
described a response to dabrafenib (8,26).
Polysomy of chromosome 7 has been identified in the
majority of solid tumors (27) and
it is well-established that clonal numerical changes of chromosome
7 are common in lung cancer (28,29).
Comparative genomic hybridization analysis demonstrated that 65% of
lung cancer cases exhibit overrepresentation of chromosome 7p
(28). This chromosome 7p gain has
been associated with lymph node metastasis in lung cancer (29) and a detailed analysis of chromosome
7 identified various regions of alteration (30), including EGFR. Although gains
of chromosome 7 result in an increase in the copy number of various
genes located on this chromosome, data from the present study
indicate that BRAF may also represent a target for its
selection and clonal progression (19). The present study supports this role
of BRAF due to the identification of chromosome 7 amplification in
the EGFR/Kras wild-type, BRAF V600E-mutant
cases screened. In a previous study, no overlap was identified
between BRAF copy number changes and RAS mutations
that are known to activate MAPK (19).
The numerical changes in BRAF determined in
the present study included gains of three copies of the gene, which
would be expected to result in its modest overexpression. However,
one of the lymph node-positive V600E cases demonstrated increased
copy number. Furthermore, one patient with an increased BRAF copy
number had experienced cancer recurrence. Thus, BRAF copy
number gain may serve as a marker of the more aggressive behavior
of V600E lung adenocarcinoma (19).
In conclusion, the present study determined
BRAF amplification in lung cancer for the first time and
demonstrated that BRAF copy number gain may be present in
BRAF V600E cases. BRAF copy number gain is rare in
lung adenocarcinomas, however, it does occur in the aggressive
V600E subtype.
Acknowledgements
The authors would like to thank Miss Ito Yamamoto
for her technical assistance. The present study was supported by
Grants-in-Aid for Scientific Research, Japan Society for the
Promotion of Science (grant nos. 23659674, 24592097 and
25293303).
References
|
1
|
Ginsberg RJ, Kris MK and Armstrong JG:
Cancer of the lung. Principles and Practice of Oncology. 4th
edition. JB Lippincott; Philadelphia, PA: pp. 673–682. 1993
|
|
2
|
Amano J, Kuwano H and Yokomise H: Thoracic
and cardiovascular surgery in Japan during 2011: Annual report by
the Japanese Association for Thoracic Surgery. Gen Thorac
Cardiovasc Surg. 61:578–607. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Postus PE: Chemotherapy for non-small cell
lung cancer: the experience of the Lung Cancer Cooperative Group of
the European Organization for Research and Treatment of Cancer.
Chest. 113(Suppl 1): 28S–31S. 1997. View Article : Google Scholar
|
|
4
|
Ding L, Getz G, Wheeler DA, et al: Somatic
mutations affect key pathways in lung adenocarcinoma. Nature.
455:1069–1075. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun Y, Ren Y, Fang Z, et al: Lung
adenocarcinoma from East Asian never-smokers is a disease largely
defined by targetable oncogenic mutant kinases. J Clin Oncol.
28:4616–4620. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weir BA, Woo MS, Getz G, et al:
Characterizing the cancer genome in lung adenocarcinoma. Nature.
450:893–898. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gautschi O, Pauli C, Srobel K, et al: A
patient with BRAF V600E lung adenocarcinoma responding to
vemurafenib. J Thorac Oncol. 7:e23–e24. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Falchook GS, Long GV, Kurzrock R, et al:
Dabrafenib in patients with melanoma, untreated brain metastases,
and other solid tumors: a phase 1 dose-escalation trial. Lancet.
379:1893–1901. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chapman PB, Hauschild A, Robert C, et al;
BRIM-3 Study Group. Improved survival with vemurafenib in melanoma
with BRAF V600E mutation. N Engl J Med. 364:2507–2516. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Flaherly KT, Infante JR, Daud A, et al:
Combined BRAF and MEK inhibition in melanoma with BRAF V600E
mutations. N Engl J Med. 367:1694–1703. 2012. View Article : Google Scholar
|
|
11
|
Flaherly KT, Robert C, Hersey P, et al;
METRIC Study Group. Improved survival with MEK inhibition in
BRAF-mutated melanoma. N Engl J Med. 367:107–114. 2012. View Article : Google Scholar
|
|
12
|
Marchetti A, Felicioni L, Malatesta S, et
al: Clinical features and outcome of patients with non-small-cell
lung cancer haboring BRAF mutations. J Clin Oncol. 29:3574–3579.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Cardarella S, Ogino A, Nishio M, et al:
Clinical, pathologic, and biologic features associated with BRAF
mutations in non-small cell lung cancer. Clin Cancer Res.
19:4532–4540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sasaki H, Shitara M, Yokota K, et al: Braf
and erbB2 mutations correlate with smoking status in lung cancer
patients. Exp Ther Med. 3:771–775. 2012.PubMed/NCBI
|
|
15
|
Paik PK, Arcila ME, Fara M, et al:
Clinical characteristics of patients with lung adenocarcinomas
haboring BRAF mutations. J Clin Oncol. 29:2046–2051. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sasaki H, Shimizu S, Tani Y, et al:
Usefulness of immunohistochemistry for the detection of the BRAF
V600E mutation in Japanese lung adenocarcinoma. Lung Cancer.
82:51–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pratilas CA, Hanrahan AJ, Halilovic E, et
al: Genetic predictors of MEK dependence in non-small cell lung
cancer. Cancer Res. 68:9375–9383. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wan PT, Garnett MJ, Roe SM, et al; Cancer
Genome Project. Mechanism of activation of the RAF-ERK signaling
pathway by oncogenic mutations of B-RAF. Cell. 116:855–867. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ciampi R, Zhu Z and Nikiforov YE: BRAF
copy number gains in thyroid tumors detected by fluorescence in
situ hybridization. Endocr Pathol. 16:99–105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Endo K, Sasaki H, Yano M, et al:
Evaluation of the epidermal growth factor receptor gene mutation
and copy number in non-small cell lung cancer with gefitinib
therapy. Oncol Rep. 16:533–541. 2006.PubMed/NCBI
|
|
21
|
Sasaki H, Okuda K, Kawano O, et al: Nras
and Kras mutation in Japanese lung cancer patients: Genotyping
analysis using LightCycler. Oncol Rep. 18:623–628. 2007.PubMed/NCBI
|
|
22
|
Wang TL, Maierhofer C, Speicher MR, et al:
Digital karyotyping. Proc Natl Acad Sci USA. 99:16156–16161. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cardarella S, Ogino A, Nishino M, et al:
Clinical, pathologic, and biologic features associated with BRAF
mutations in non-small cell lung cancer. Clin Cancer Res.
19:4532–4540. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marchetti A, Felicioni L, Malatesta S, et
al: Clinical features and outcome of patients with non-small-cell
lung cancer haboring BRAF mutations. J Clin Oncol. 29:3574–3579.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Oliveira Duarte Achcar R, Nikiforova MN
and Yousem SA: Micropappillary lung adenocarcinoma: EGFR, K-ras,
and BRAF mutational profile. Am J Clin Pathol. 131:694–700. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rudin CM, Hong K and Streit M: Molecular
characterization of acquired resistance to the BRAF inhibitor
dabrafenib in patient with BRAF-mutant non-small-cell lung cancer.
J Thorac Oncol. 8:e41–e42. 2013.PubMed/NCBI
|
|
27
|
El-Naggar AK, Dinh M, Tucker SL, et al:
Numerical chromosomal changes in DNA hypodiploid solid tumors;
restricted loss and gain of certain chromosomes. Cytometry.
37:107–112. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Balsara BR, Sonoda G, du Manoir S, et al:
Comparative genomic hybridization analysis detects frequent, often
high-level, overrepresentation of DNA sequences at 3q, 5p, 7p, and
8q in human non-small cell lung carcinomas. Cancer Res.
57:2116–2120. 1997.PubMed/NCBI
|
|
29
|
Ubagai T, Matsuura S, Tauchi H, et al:
Coparative genomic hybridization analysis suggests a gain of
chromosome 7p associated with lymph node metastasis in non-small
cell lung cancer. Oncol Rep. 8:83–88. 2001.
|
|
30
|
Garnis C, Lockwood WW, Vucic E, et al:
High resolution analysis of non-small cell lung cancer cell lines
by whole genome tiling path array CGH. Int J Cancer. 118:1556–1564.
2006. View Article : Google Scholar
|