Introduction
Significant progress has been made in the
development of prognostic factors for chronic lymphocytic leukemia
(CLL), which is difficult to predict using clinical parameters, as
some patients exhibit indolent disease for decades while others
suffer from rapid progression of the disease and require early
treatment (1). Furthermore, the
development of CLL is associated with immune suppression in the
host, which contributes to the failure to mount an effective immune
response against the cancer cells (2); however, no prognostic factors for
indicating the host immune status of CLL patients exist at
present.
Myeloid-derived suppressor cells (MDSCs), a subset
of T cells, are a heterogeneous population of immature myeloid
cells with immunosuppressive function. Therefore, the presence of
MDSCs in the tumor microenvironment has been widely investigated
(3). Previously, a novel population
of MDSCs, termed cluster of differentiation 14 (CD14)+
human leukocyte antigen (HLA)-DRlow/− MDSCs, were
identified in melanoma, hepatocarcinoma and B-cell non-Hodgkin
lymphoma patients (4–6), indicating that low or lacking HLA-DR
expression is associated with a CD14+ cell subset, which
is highly suppressive of lymphocyte function.
Therefore, the present study analyzed
CD14+HLA-DRlow/− MDSCs from 49 CLL patients,
monoclonal B-cell lymphocytosis (MBL) patients and healthy
volunteers. Furthermore, the correlation between CLL patient
survival and CD14+HLA-DRlow/− expression was
also investigated.
Patients and methods
Patients
Peripheral blood samples were collected from 49
untreated CLL patients (males, 34; females, 15; mean age, 63 years;
age range, 50–85 years) and 23 MBL patients (males, 18; females, 5;
mean age, 67 years; age range, 54–86 years) who met the World
Health Organization diagnostic criteria for CLL and MBL (7), as well as 21 age-matched healthy
controls (males, 15; females, 6; mean age, 57 years; age range,
48–65 years). The MBL patients were screened from a cohort of
healthy adults from the general population with normal peripheral
blood lymphocyte counts using the sensitive technique of multicolor
flow cytometry. Individuals with abnormal peripheral blood
lymphocyte counts were denoted as MBL cases and the remainder were
classified as controls. Any individuals with immune or chronic
infectious diseases were excluded from the present study. Of the 49
CLL patients, 12, 18, 13 and six were classified as stages I, II,
III and IV, respectively, based on the Rai staging criteria
(8). All patients provided informed
consent in accordance with the Declaration of Helsinki and the
study was approved by the Zhejiang Province People’s Hospital
Review Board (Hanzhou, China).
Flow cytometry
For flow cytometry diagnosis and prognosis of the
CLL patients, the following mouse anti-human monoclonal antibodies
(MoAbs), labeled with fluorescein isothiocyanate (FITC),
phycoerythrin (PE), allophycocyanin (APC) and peridinin
chlorophyll-a protein (PerCP) were used: CD5 (cat. no. A07710),
CD10 (cat. no. A07708), CD19 (cat. no. A07708), CD20 (cat. no.
A07708), CD22 (cat. no. IM0779u), CD23 (cat. no. A07710), CD38
(cat. no. A07778), CD79a (cat. no. A07705), Flinders Medical Centre
(FMC-7; cat. no. A07791), κ (cat. no. A07706), λ (cat. no. A07706)
and ζ-chain-associated protein kinase-70 (ZAP-70; cat. no. 731902).
For the lymphocyte subset analyzed, the following mouse anti-human
MoAbs labeled with FITC, PE, APC and PerCP were used: CD3 (cat. no.
6607013), CD4 (cat. no. A07751), CD8 (cat. no. 6607013), CD14 (cat.
no. 6603262), CD16 (cat. no. 6607073), CD19 (cat. no. 6607073),
CD25 (cat. no. IM0478u), CD56 (cat. no. 6607073), CD127 (cat. no.
IM1980u) and HLA-DR (cat. no. IM0463u). All of the abovementioned
products were purchased from Immunotech (Marseille, France). Cells
stained with separate antibodies were defined as CD3+ T
cells (CD3+), CD4+ T cells
(CD3+CD4+), CD8+ T cells
(CD3+CD8+), regulatory T cells
(Treg cells;
CD4+CD25highCD127low T helper), B
cells (CD3−CD19+), natural killer (NK;
CD3−CD16+CD56+), MDSCs
(CD14+HLA-DRlow/−) and
CD5+CD19+ cells. CD38 and cytoplasmic ZAP-70
expression were analyzed within the CD19+CD5+
lymphocytes. Additionally, the expression the signal transduction
molecule ZAP-70 was provided as the percentage of mean fluorescence
intensity (MFI) of the gated events, and regarded as high when
>20%. The expression of CD38 was also provided as a percentage
of the MFI of the gated events, and was regarded as high when
>30%.
Immunoglobulin heavy chain (IGHV) gene
mutation
The IGHV gene mutation status of the
CD19+CD5+ cells of all patients was assessed
as previously described (9).
Briefly, IgVH gene mutations were detected by polymerase chain
reaction and specific primers (cat. no. 5-101-0010), using the IGH
Somatic Hypermutation Assay for Gel Detection kit (Invivoscribe
Technologies, Inc., San Diego, CA, USA), according to the
manufacturer’s instructions. Sequences exhibiting <98% homology
with the corresponding germline IGHV genes were considered to be
mutated.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism software (version 5.01; GraphPad Software, Inc., La Jolla,
CA, USA) and the following statistical tests were performed: The
log-rank test, the Mann Whitney U test and Spearman’s rank
correlation. In addition, quantitative data are presented as the
mean ± standard deviation. *P<0.05,
**P<0.01 and ***P<0.001 were considered
to indicate a statistically significant difference.
Results
Correlation between
CD14+HLA-DRlow/− MDSC upregulation and CLL
tumor progression
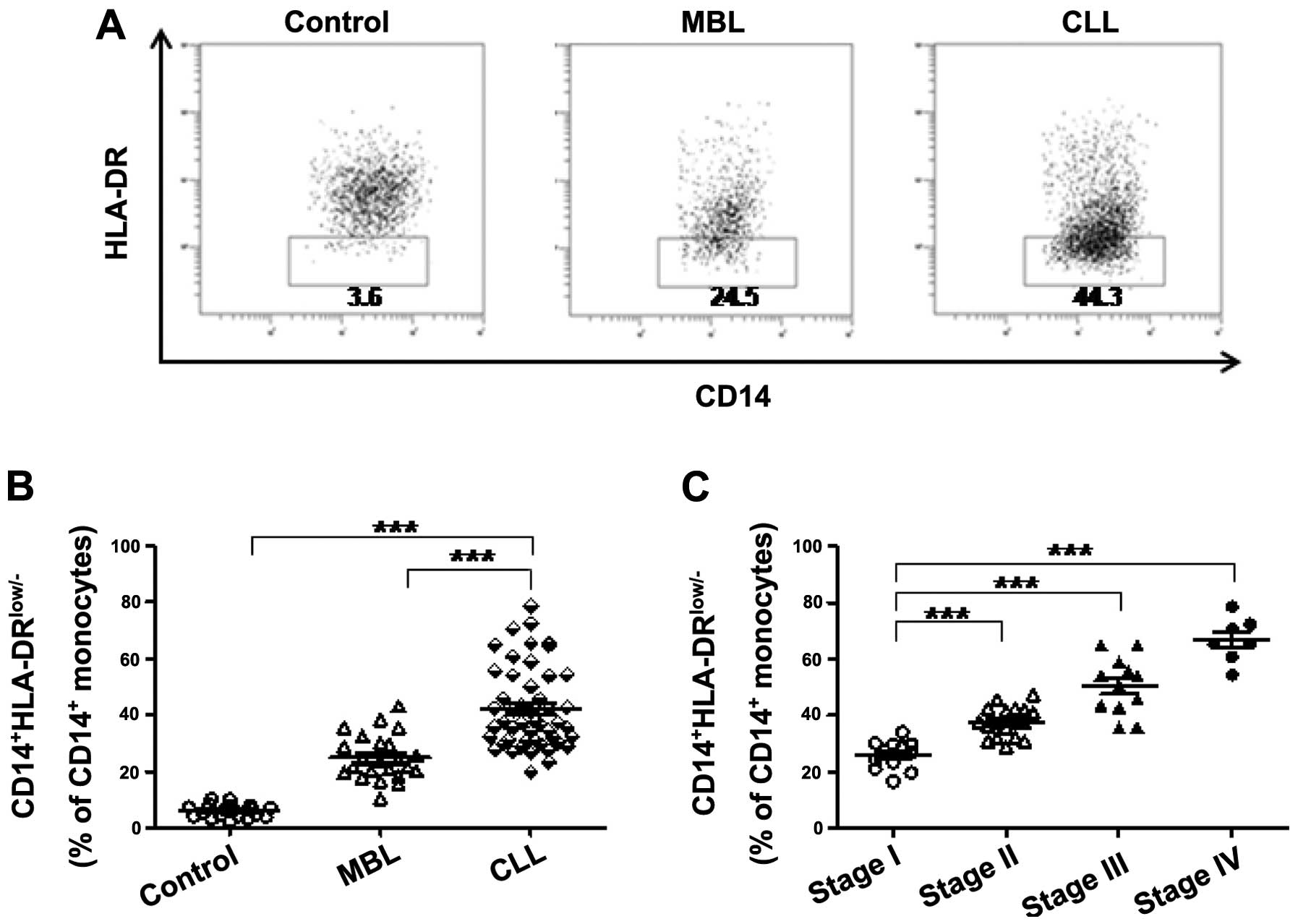
Analysis of the frequency of
CD14+HLA-DRlow/− MDSC cells in 49 CLL, 23 MBL
and 21 control cases revealed that the frequency of
CD14+HLA-DRlow/− cells was significantly
elevated in CLL patients compared with MBL and control cases
(Fig. 1A and B), where MBL is a
condition that resembles CLL but does not meet all of the criteria
for CLL (10). The
clinicopathological data of the studied patients and control cases,
including the number of subjects, median age, gender, Rai stage,
IGHV mutation status, CD38 status and ZAP-70 status, were recorded
(Table I). Furthermore, statistical
analyses revealed that CD14+HLA-DRlow/− cells
were significantly associated with the clinical stage of disease in
CLL patients (Fig. 1C), indicating
that these cells may participate in the progression of CLL.
 | Table IClinicopathologic characteristics of
the different groups of patients. |
Table I
Clinicopathologic characteristics of
the different groups of patients.
| Chronic lymphocytic
lymphocytosis | Monoclonal B cell
leukemia | Control |
|---|
| Subjects, n | 49 | 23 | 21 |
| Median age (range),
years | 63 (50–85) | 67 (54–86) | 57 (48–65) |
| Gender, n |
| Male | 34 | 18 | 15 |
| Female | 15 | 5 | 6 |
| Rai stage, n |
| 0 – II | 30 | NA | NA |
| III – IV | 19 | NA | NA |
| Immunoglobulin
heavy chain mutation status, n |
| Mutated (≥2%) | 31 | NA | NA |
| Unmutated
(<2%) | 18 | NA | NA |
| Cluster of
differentiation 38 status, n |
| Low (<30%) | 32 | NA | NA |
| High (≥30%) | 17 | NA | NA |
| ζ-chain-associated
protein kinase-70 status, n |
| Low (<20%) | 30 | NA | NA |
| High (≥20%) | 19 | NA | NA |
CD14+HLA-DRlow/−
MDSC and poor prognosis for CLL patients
Most notably, the present study identified that the
overall survival time in CLL patients whose samples were obtained
prior to the administration of any therapy (n=49) was significantly
different between those exhibiting low and high
CD14+HLA-DRlow/− expression levels, and that
CD14+HLA-DRlow/− expression was inversely
correlated with survival time (P<0.0001; Fig. 2A). Furthermore, 15 patients in the
high CD14+HLA-DRlow/− expression group (n=24;
>40% CD14+ monocytes) and three patients in the low
expression group (n=25; <40% CD14+ monocytes)
succumbed to CLL within four years, indicating that
CD14+HLA-DRlow/− MDSCs may be a prognostic
factor in CLL patients. CD38, ZAP-70 and the IGHV gene are typical
immunophenotypic and genetic prognostic markers (11,12),
which have previously been used to predict the survival of CLL
patients in the clinic. Firstly, when comparing CLL patients
grouped according to CD38 expression,
CD14+HLA-DRlow/− MDSC levels in
CD38low and CD38high CLL patients were
significantly higher compared with the control group (both
P<0.0001). In addition, the
CD14+HLA-DRlow/− MDSC count was significantly
higher in the CD38high subgroup compared with the
CD38low subgroup (P=0.0335; Fig. 2B), the
CD14+HLA-DRlow/− MDSC frequency was
significantly elevated in the ZAP-70low and
ZAP-70high CLL patients when compared with the control
group (both P<0.0001), and was significantly higher in the
ZAP-70high subgroup compared with ZAP-70low
subgroup (P=0.0003) (Fig. 2C).
Following data analysis based on the mutational status of IGHV, a
similar frequency pattern was observed: The frequency of
CD14+HLA-DRlow/− MDSCs was significantly
higher in the IGHV-unmutated (U-CLL) and IGHV-mutated (M-CLL) CLL
patients when compared with the control (both P<0.0001), and
these MDSCs were significantly more elevated in the U-CLL patients
compared with the M-CLL patients (P=0.0019) (Fig. 2D). Furthermore, when patients were
divided based on combinations of good (M-CLL/CD38low) or
poor (U-CLL/CD38high) prognosis markers, the
M-CLL/CD38low and U-CLL/CD38high subgroups of
patients exhibited significantly higher
CD14+HLA-DRlow/− MDSC frequency compared with
the controls (both P<0.0001), and the
CD14+HLA-DRlow/− frequency in the
U-CLL/CD38high subgroup was higher than in the
M-CLL/CD38low subgroup (P=0.0052) (Fig. 2E). In the present study, it was
identified that CD14+HLA-DRlow/− MDSCs are
associated with CD38 and ZAP-70 expression, as well as IGHV
mutations. Thus, CD14+HLA-DRlow/− MDSCs may
be prognostic factors in CLL patients.
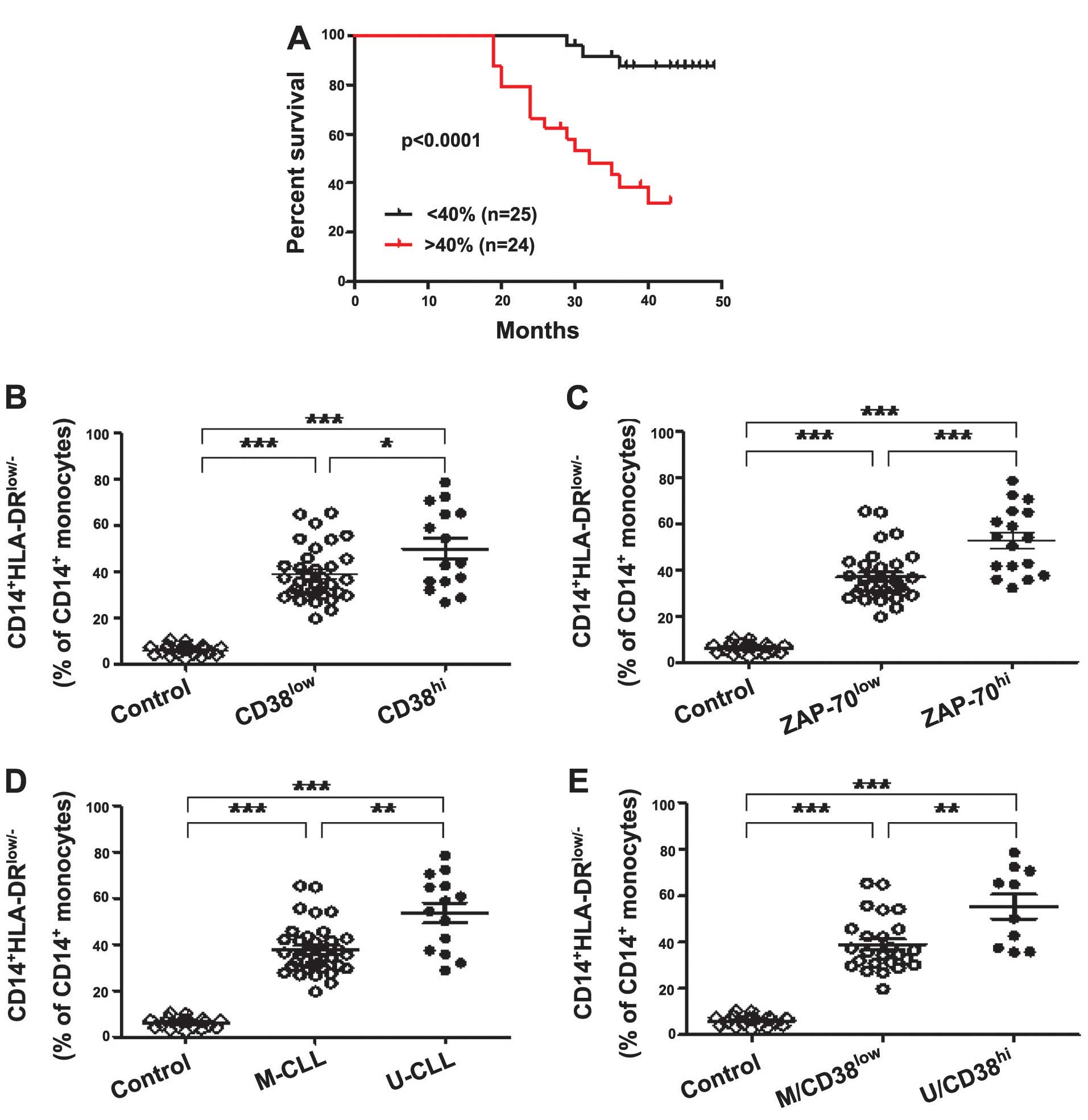 | Figure 2Upregulation of
CD14+HLA-DRlow/− MDSCs predicts poor survival
in CLL patients. (A) Kaplan-Meier curves of CLL patients with
<40% (n=25) versus >40% (n=24)
CD14+HLA-DRlow/− MDSCs in the
CD14+ monocytes (log-rank test, P<0.0001). (B)
Frequency of CD14+HLA-DRlow/− MDSCs in
CD14+ monocytes of CLL patients stratified by CD38
expression levels compared with the control. A significantly higher
frequency of CD14+HLA-DRlow/− MDSCs were
identified in CD38high (n=15) and CD38low
patients (n=34) compared with the healthy controls (n=21) (both
P<0.0001). The CD38high subgroup tended to have
higher frequency of CD14+HLA-DRlow/− MDSCs
compared with the CD38low subgroup (P=0.0335). (C) A
significantly higher frequency of
CD14+HLA-DRlow/− MDSCs in CD14+
monocytes was identified in ZAP-70high (n=17) and
ZAP-70low patients (n=32) compared with the healthy
controls (n=21) (both P<0.0001), and the ZAP-70high
subgroup exhibited a higher frequency than the ZAP-70low
subgroup (P=0.0003). (D) A significantly higher frequency of
CD14+HLA-DRlow/− MDSCs in CD14+
monocytes was identified in U-IGHV (n=14) and M-IGHV patients
(n=35) compared with in the healthy controls (n=21) (both
P<0.0001), and the ZAP-70high subgroup exhibited a
higher frequency than the ZAP-70low subgroup (P=0.0019).
(E) A significantly higher frequency of
CD14+HLA-DRlow/− MDSCs in CD14+
monocytes was identified in U/CD38high (n=10) and
M/CD38low patients (n=35) compared with in the healthy
controls (n=21) (both P<0.0001), and the U/CD38high
subgroup exhibited a higher frequency than the M/CD38low
subgroup (P=0.0052). *P<0.05, **P<0.01
and ***P<0.001. CD14, cluster of differentiation 14;
HLA, human leukocyte antigen; MDSCs, myeloid-derived suppressor
cells; ZAP-70, ζ-chain-associated protein kinase-70; CLL, chronic
lymphocytic leukemia; M, mutated; U, unmutated. |
CD14+HLA-DRlow/−
MDSCs predominantly inhibit the CD4+ T-cell response
during CLL disease progression
As previously established, MDSCs suppress immunity
by disrupting the innate and adaptive immune responses (3). In the present study, the frequency of
various lymphocyte subsets were assessed, and it was identified
that the frequency of CD3+ T, CD4+ T,
CD8+ T and NK cells in CLL patients were significantly
reduced compared with in the MBL patients and the control group
(Table II). By contrast, B,
Treg and CD19+CD5+ cells were
significantly elevated in CLL patients compared with MBL patients
and the control group (Table II).
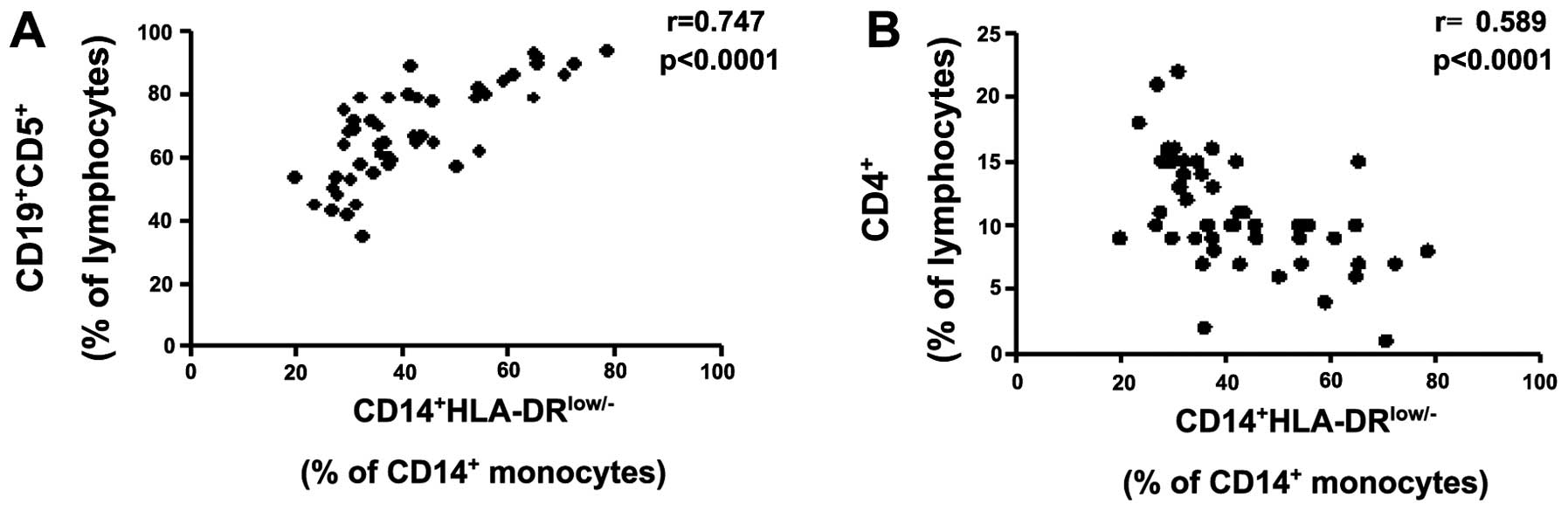
Furthermore, Spearman’s correlation analysis revealed that
CD14+HLA-DRlow/− MDSCs were significantly
positively correlated with CD19+CD5+ cells
(Fig. 3A) and negatively correlated
with the CD4+ T cells (Fig.
3B); however, no significant correlation was identified with
CD3+ T, B, Treg or NK cells (data not shown).
Data obtained in the present study was inconsistent with previous
reports, which described MDSCs as potent inducers of
Treg cells in liver cancer patients (4). However, in general,
CD14+HLA-DRlow/− MDSCs appear to disturb
systemic immunity during CLL disease progression, predominantly by
inhibiting the CD4+ T-cell response. Additionally, no
correlation was identified between
CD14+HLA-DRlow/− MDSCs, and age, serum
hemoglobin concentration, platelet and white blood cell count, Rai
stage and lymphocytosis (data not shown).
 | Table IILymphocyte subsets in the CLL, MBL
and control groups. |
Table II
Lymphocyte subsets in the CLL, MBL
and control groups.
| Mean ± standard
deviation, % |
|---|
|
|
|---|
| Lymphocyte | CLL | MBL | Control |
|---|
|
CD3+ | 14.61±3.72 | 35.68±9.35 | 69.42±7.39 |
|
CD4+ | 8.15±6.38 | 22.73±5.15 | 35.17±4.56 |
|
CD8+ | 6.57±2.62 | 21.61±7.33 | 23.46±4.78 |
|
CD3−CD16+CD56+NK | 3.92±2.40 | 6.85±1.77 | 8.63±2.77 |
|
CD4+CD25+/highCD127low/− | 10.09±3.21 | 5.84±1.76 | 4.38±1.42 |
|
CD3−CD19+ | 68.74±14.56 | 39.46±5.60 | 12.31±4.29 |
|
CD5+CD19+ | 51.68±9.43 | 35.16±6.18 | 2.85±1.36 |
Discussion
Currently, the use of clinical, biological and
genetic parameters allows CLL patients to be diagnosed and
characterized with a mild, intermediate or aggressive onset and
prognosis of this heterogeneous disease (13). However, excepting allogeneic stem
cell transplantation, there is no curative treatment available for
CLL patients, possibly due to CLL patients exhibiting a defective
immune antitumor response. Furthermore, in individuals with
established CLL, MDSCs are likely to be a major factor in reducing
the efficacy of therapeutic treatment strategies, which require an
immunocompetent host (14).
Therefore, improved understanding of the immune status of
individual CLL patients may be useful for determining which
patients may exhibit indolent disease for a number of decades, and
which patients may progress rapidly and require early treatment. In
addition, the elimination of MDSCs is a priority for CLL patients
who are candidates for active immunotherapy.
Presently, there is a requirement to monitor the
number of immunosuppressive CD14+HLA-DRlow/−
MDSCs (>40% CD14+ monocytes) in CLL patients, as the
survival time from diagnosis is currently <3 years. However,
additional multivariate statistical analysis with other prognostic
factors in long-term multicenter studies should initially be
performed to clarify the observations of the current study, as the
follow-up period of CLL patients in the present study was only
conducted for ~4 years. Furthermore, the specific immunosuppressive
function of CD14+HLA-DRlow/− MDSCs in CLL
patients requires further investigation.
In conclusion, to the best of our knowledge, the
present study reports for the first time a population of
immunosuppressive monocytes, characterized by the
CD14+HLA-DRlow/− phenotype, which were
significantly elevated in CLL patients and were poor predictors of
survival in CLL patients (n=49). In addition, these
CD14+HLA-DRlow/− MDSCs were associated with
the prognostic factors CD38, ZAP-70 and IGHV. Finally, Spearman’s
correlation analysis revealed that the poor survival of upregulated
CD14+HLA-DRlow/−MDSC CLL patients
predominantly occurred via the inhibition of the CD4+
T-cell response, resulting in a dysregulation of the balance of
T-cell subsets in vivo, including significant proliferation
of CD5+CD19+ cells in CLL patients. Thus,
CD14+HLA-DRlow/− MDSCs may be used as a novel
prognostic marker for the survival of CLL patients and eliminating
CD14+HLA-DRlow/− MDSCs may represent a
promising strategy for the treatment of CLL patients.
Acknowledgements
The authors would like to thank all of the members
of the Department of Clinical Laboratory (Zhejiang Provincial
People’s Hospital, Hangzhou, China) for their assistance and
encouragement. The present study was supported by the National
Natural Science Foundation (grant no. 30973568), the Zhejiang
Provincial Natural Science Fund (grant no. LH12H16019), the
Zhejiang Provincial Program for the Cultivation of High-level
Innovative Health Talents (grant no. 2012), and the Zhejiang
Provincial Health Bureau (grant no. 2010KYA015).
References
|
1
|
Rassenti LZ, Jain S, Keating MJ, et al:
Relative value of ZAP-70, CD38, and immunoglobulin mutation status
in predicting aggressive disease in chronic lymphocytic leukemia.
Blood. 112:1923–1930. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Scrivener S, Goddard RV, Kaminski ER and
Prentice AG: Abnormal T-cell function in B-cell chronic lymphocytic
leukaemia. Leuk Lymphoma. 44:383–389. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gabrilovich DI and Nagaraj S:
Myeloid-derived suppressor cells as regulators of the immune
system. Nat Rev Immunol. 9:162–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hoechst B, Ormandy LA, Ballmaier M, et al:
A new population of myeloid-derived suppressor cells in
hepatocellular carcinoma patients induces
CD4+CD25+Foxp3+ T cells.
Gastroenterology. 135:234–243. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Filipazzi P, Valenti R, Huber V, et al:
Identification of a new subset of myeloid suppressor cells in
peripheral blood of melanoma patients with modulation by a
granulocyte-macrophage colony-stimulation factor-based antitumor
vaccine. J Clin Oncol. 25:2546–2553. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lin Y, Gustafson MP, Bulur PA, Gastineau
DA, Witzig TE and Dietz AB: Immunosuppressive
CD14+HLA-DR(low)/− monocytes in B-cell
non-Hodgkin lymphoma. Blood. 117:872–881. 2011. View Article : Google Scholar :
|
|
7
|
Jaffe ES: The 2008 WHO classification of
lymphomas: implications for clinical practice and translational
research. Hematology Am Soc Hematol Educ Program. 523–531. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hallek M, Cheson BD, Catovsky D, et al:
International Workshop on Chronic Lymphocytic Leukemia: Guidelines
for the diagnosis and treatment of chronic lymphocytic leukemia: a
report from the International Workshop on Chronic Lymphocytic
Leukemia updating the National Cancer Institute - Working Group
1996 guidelines. Blood. 111:5446–5456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gharagozlou S, Kardar GA, Rabbani H and
Shokri F: Molecular analysis of the heavy chain variable region
genes of human hybridoma clones specific for coagulation factor
VIII. Thromb Haemost. 94:1131–1137. 2005.
|
|
10
|
Ghia P and Caligaris-Cappio F: Monoclonal
B-cell lymphocytosis: right track or red herring? Blood.
119:4358–4362. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Orchard JA, Ibbotson RE, Davis Z, et al:
ZAP-70 expression and prognosis in chronic lymphocytic leukaemia.
Lancet. 363:105–111. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Damle RN, Wasil T, Fais F, et al: Ig V
gene mutation status and CD38 expression as novel prognostic
indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847.
1999.PubMed/NCBI
|
|
13
|
Hallek M: Signaling the end of chronic
lymphocytic leukemia: new frontline treatment strategies. Blood.
122:3723–3734. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marx J: Cancer immunology. Cancer’s
bulwark against immune attack: MDS cells. Science. 319:154–156.
2008. View Article : Google Scholar : PubMed/NCBI
|