Introduction
Laryngeal cancer is the most common malignant
neoplasm of the head and neck region. In Poland, laryngeal cancer
is the seventh most common neoplasm in men (1). Despite the advances in diagnostics and
therapeutic procedures, the majority of laryngeal cancer cases are
diagnosed at an advanced clinical stage. Between 1991 and 2001, a
multicenter, nationwide study was conducted, which included a total
of ~13,000 patients diagnosed with cancer of the larynx and
hypopharynx. More than half of all the cases had a stage T3 or T4
primary tumor. Cervical lymph node metastases were identified in
47.7% of the cases (2). The 5-year
survival rate for laryngeal cancer is currently ~50% (1).
Epigenetics was defined by Wu and Morris (3) as changes in gene function that are
mitotically and/or meiotically heritable and that do not entail a
change in the DNA sequence. DNA methylation along with histone
modifications and non-coding RNA dysregulation are the leading
epigenetic alterations in human cancer (4).
During DNA methylation, a methyl group is added to
an aromatic ring of cytosine in CpG dinucleotides. Approximately
70% of total CpG dinucleotides in the human genome are methylated.
However, certain CpG islands located in the promoter region of
multiple genes remain physiologically unmethylated (5). The methylation of promoter sites results
in the suppression of gene expression by several mechanisms.
5-methylcytosine may spontaneously become deaminated to thymine,
leading to the formation of point mutations (5). DNA methylation directly prevents the
binding of transcriptional factors to the promoter (6). The methyl-CpG-binding domain proteins
recognize methylation sites. These proteins bind the methylated
sequence and block the interaction between RNA polymerase and the
promoter (6,7). Promoter site methylation of the
suppressor gene may lead to carcinogenesis.
Environmental factors may lead to epigenetic changes
in DNA. Chang et al (8) proved
the effect of smoking combined with alcohol intake on methylation
of p15 in the epithelium of the upper airways. Van Engeland et
al (9) documented the correlation
between alcohol consumption and hypermethylation of the following
genes: Ras association domain-containing protein 1 (RASSF1A),
O-6-methylguanine-DNA methyltransferase (O-MGMT), adenomatous
polyposis coli, p16, p14 and human MutL homolog. Since alcohol
abuse and smoking are the main risk factors for the development of
laryngeal cancer, it may be hypothesized that abnormal DNA
methylation is common among patients suffering from this
neoplasm.
Hypermethylated in cancer 1 (HIC1) is a potential
suppressor gene located on chromosome 17 (17p13.3). HIC1 is known
for cooperating with TP53 in the regulation of apoptosis. The
promoter region of HIC1 abounds in CpG dinucleotides and
methylation of these CpG islands may lead to gene silencing
(10). Hypermethylation of HIC1 was
documented in the pathogenesis of several neoplasms. The aim of
this study was the assessment of HIC1 expression in patients with
laryngeal cancer.
Materials and methods
Study population and tissue
samples
The protocol of this study was approved by the
Ethics Committee of the Medical University of Silesia (Katowice,
Poland). The population study consisted of 21 men with
histologically confirmed laryngeal squamous cell carcinoma, treated
in the Laryngology Clinic of the Medical University of Silesia
between 2005 and 2009. Written informed consent was obtained from
all patients regarding the use of their tissue samples for the
purpose of this study. All the patients were clinically at stage
T3-T4, N0-N3 and M0 and underwent total laryngectomy.
Paraffin-embedded 20-µm tissue sections were collected from tumor
and corresponding healthy tissues.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
assay
Total RNA from the formalin-fixed paraffin-embedded
(FFPE) tissue samples was isolated using the AllPrep DNA/RNA FFPE
kit, including DNase treatment (Qiagen, Venlo, The Netherlands).
The integrity of RNA was assessed using an Agilent RNA 6000 Nano
kit on Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, USA).
The quantity and quality of RNA were measured spectrometrically on
the NanoDrop 2000 (NanoDrop products, Wilmington, DE, USA). The
total RNA concentration in each sample was 50 ng, with RNA
integrity number values of ~2. RNA was amplified and detected in
one-step RT-PCR on the LightCycler 480 (Roche, Basel, Switzerland)
using a QuantiFast Probe assay (Qiagen) for the human HIC1 (Entrez
target gene ID: 3090) duplexed with GAPDH_2_HS (as a reference
gene) in combination with the QuantiFast Probe (amplicon size, 79
bp). All the samples were tested in triplicate. The results were
normalized and analyzed using the ΔΔcycle threshold (Ct)
method (11).
Statistical analysis
All data are presented as the arithmetical mean ±
standard error of the mean (SEM) and the unpaired Student's t-test
was used for statistical analysis. Fold change (FC) was evaluated
in the target gene, or log ratio. FC was defined as the difference
per gene between the averages of the group: Target gene/control.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Relative expression of HIC1 in tumor
and healthy tissue samples
A competitive RT-PCR approach with specific RNA
competitive molecules for HIC1 and GADPH was used to evaluate the
expression levels of HIC1 in samples obtained from 21 patients with
laryngeal carcinoma (healthy and tumor tissues). According to the
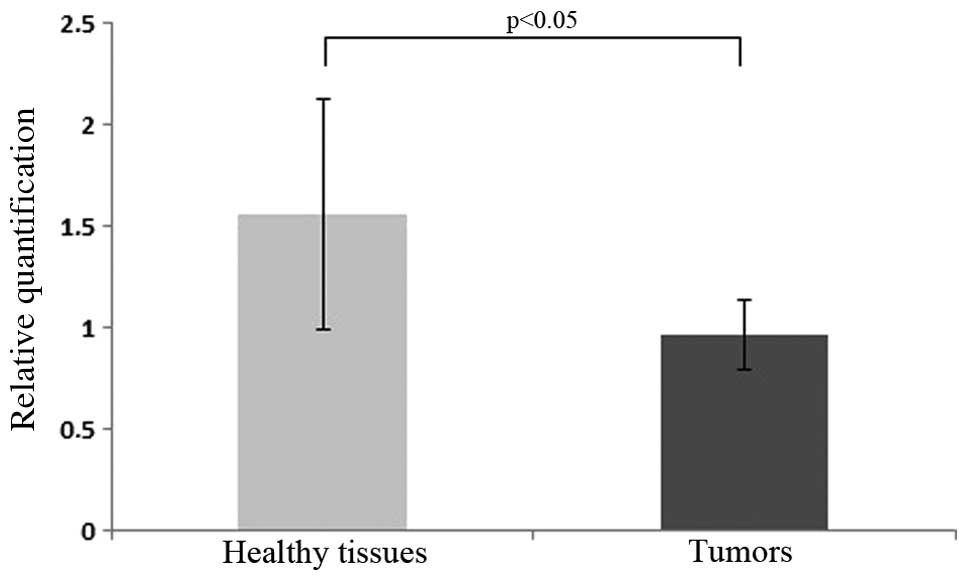
advanced relative quantification (second derivative), the mean Ct
detected in tumor samples reached 0.96 (SEM = 0.57), whereas the
mean Ct of healthy tissues amounted to 1.56 (SEM = 0.17). The
median tumor/normal tissue ratio for HIC1 expression was 0.615. The
evaluation revealed that the expression of HIC1 in tumors was 38.5%
lower compared to that in healthy tissues (Fig. 1) (P<0.05).
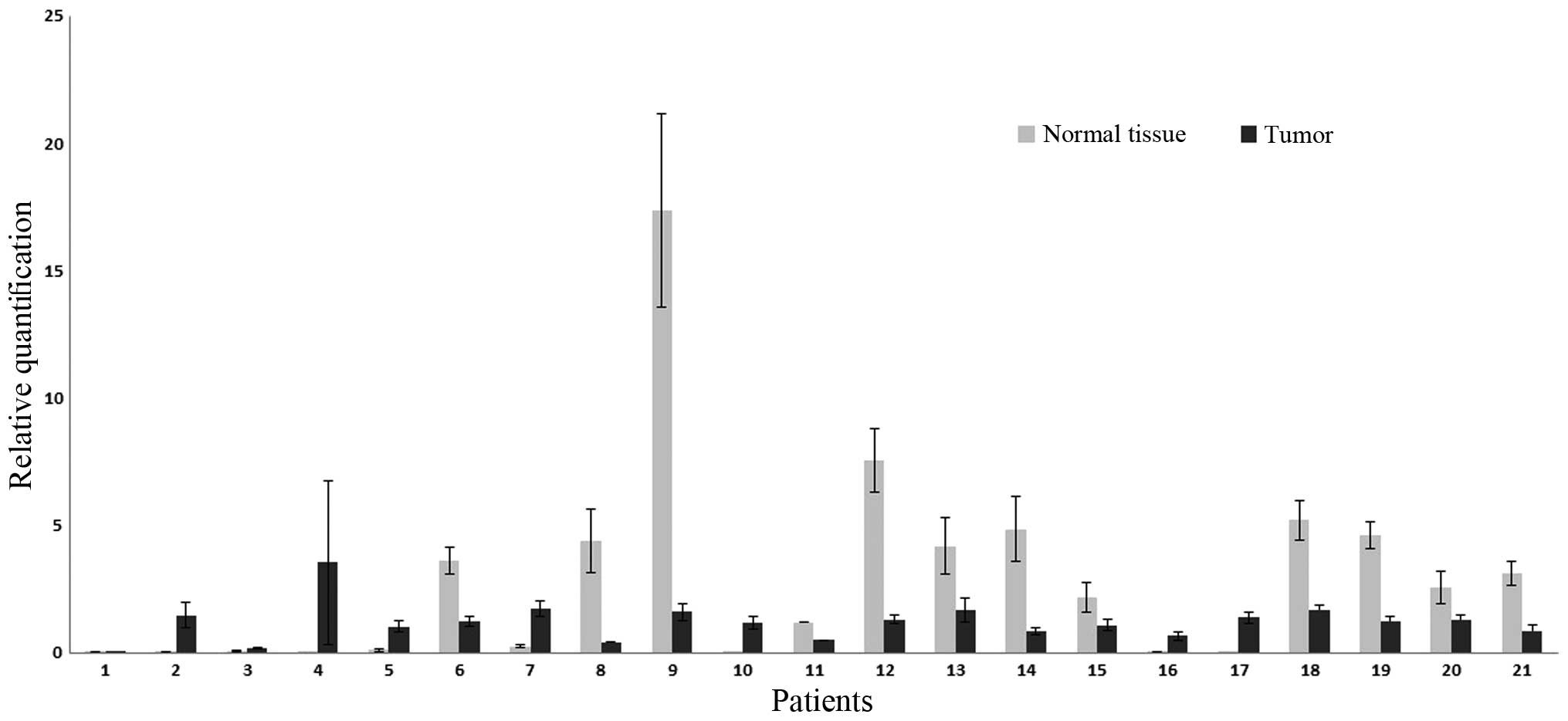
The inspection of the expression profiles revealed
that the differences in expression between samples and controls
(comparisons between pairs of cancerous and healthy tissues in each
of the 21 patients) were prominent in samples 6, 8, 9, 11, 12, 13,
14, 15, 18, 19, 20 and 21. The expression profiles of the samples
are shown in Fig. 2. The FC in HIC1
was 2.54.
Discussion
Little is known regarding the epigenetic background
of carcinogenesis in the larynx. improper methylation status of the
following genes has been identified thus far in laryngeal cancer:
Retinoic acid receptor β (RARβ), death-associated protein kinase
(DAPK), cyclin-dependent kinase inhibitor 2A (CDKN2A), MGMT,
RASSF1A, fragile histidine triad, chromodomain-helicase-DNA-binding
protein 5, cellular retinol binding protein 1 and HIC1 (12–19). the
majority of the studies reported certain correlations between the
clinical course of the disease and the molecular changes
identified. Kong et al (14)
suggested that the hypermethylation of DAPK was associated with
lymph node involvement. In addition, nodal metastases appear to
correlate with the methylation profile of MGMT (15). According to Smigiel et al
(17), CDKN2A hypermethylation is
reflected by a high grade of histological differentiation of the
tumor (G3) (17). Olasz et al
(18) reported that RARβ2
hypermethylation is observed in patients with well-differentiated
lesions.
HIC1 encodes the transcriptional factor responsible
for the propagation of the biological function of p53. HIC1
contains the C-domain of five zinc-finger motifs responsible for
binding with DNA at the site called the HIC1 responsive element
(HiRE). The N-terminal domain BTB/POZ mediates protein-protein
interactions (13). The main task of
HIC1 is to suppress the expression of class III NAD-dependent
histone deacetylase called sirtuin 1 (SIRT1), which causes
deacetylation of the p53 protein; this process attenuates the
proapoptotic action of p53 (20).
SIRT1 also affects apoptosis in pathways other than p53.
Deacetylation of the Ku70 protein leads to the sequestration of
B-cell lymphoma 2-associated X proteins in the cytoplasm (21). In addition, SIRT1 is involved in gene
silencing during the formation of facultative heterochromatin
(22).
The transcriptional activity of HIC1 is due to two
structural domains: BTB/POZ enables the formation of complexes
between HIC1 and SIRT1, whereas zinc finger motifs recognize and
bind HiRE sequences in DNA, resulting in the suppression of HIC1
gene expression (10,23).
Under physiological conditions, there is a feedback
loop between HIC1 and p53. HIC1 protects p53 from deacetylation,
whereas p53 activates the transcription of HIC1 (22,23).
Chronic silencing of HIC1 causes upregulation of SIRT1, persistent
deacetylation of p53 and loss of the proapoptotic function of this
protein. Thus, loss of expression of HIC1 may be involved in
carcinogenesis (20).
The association between HIC1 and neoplastic
transformation is not limited to the p53-dependent apoptosis
pathway. Briones et al (24)
recently reported that HIC1 may participate in the regulation of
neoplastic angiogenesis. The presence of the HiRE sequence within
the gene encoding the fibroblast growth factor-binding protein 1
(FGF-BP1) was detected. FGF-BP1 stimulates the differentiation of
endothelial cells and smooth myocytes during the formation of
embryonic blood vessels. FGF-BP1 also participates in neoplastic
vascularisation. Increased FGF-BP1 expression was detected in the
tissues of several tumors, including cancer of the colon (25).
Silencing of HIC1 may have an epigenetic background,
since the promoter site of this gene contains CpG dinucleotides
susceptible to methylation. Hypermethylation of HIC1 was detected
in the tissues of a number of solid tumors and appears to
predispose tissues to neoplastic transformation. Eguchi et
al (26) documented the
association between smoking and improper methylation at locus
17p13.3 in non-small-cell lung cancer. The alteration in
methylation status occurred more frequently among smokers compared
to that in non-smokers.
The incidence of hypermethylation of HIC1 may
increase along with the histological progression of the tumor.
Kanai et al (27) proved that
the frequency of this epigenetic process increased from normal
liver tissue, to precancerous state, to hepatocellular carcinoma.
Similar conclusions may be drawn on the basis of the study
conducted by Kanai et al (28), investigating the role of HIC1
methylation in gastric carcinogenesis.
The level of HIC1 hypermethylation may correspond to
the aggressiveness of the tumor and poor prognosis. Nicoll et
al (29) proved that the
undisturbed expression of HIC1 is reflected in the promising
outcome and lack of nodal involvement in breast cancer (29). Hayashi et al (30) suggested that low HIC1 expression may
be involved in the malignant progression of non-small-cell lung
cancer. Brieger et al (31)
analyzed the demethylation of HIC1 in head and neck squamous
carcinoma cell lines in vitro and confirmed that the
demethylation of the tumor suppressor gene HIC1 increases the
radiosensitivity of head and neck squamous carcinoma cells.
HIC1 hypermethylation is the process of binding
methyl groups to DNA. The present study did not analyze the
hypermethylation of HIC1 itself, but rather the expression of this
gene, i.e. the level of the mRNA transcript. Zheng et al
(32) analyzed the hypermethylation
of HIC1 and concluded that the hypermethylation did not
consistently correlate with the HIC1 expression level in breast and
non-small-cell lung cancer.
The study of Stephen et al (33) is the only published study suggesting
the effect of HIC1 methylation on laryngeal cancer. In that study,
the methylation status of 38 genes was assessed in patients with
laryngeal carcinoma. HIC1 was found to be methylated in 5 of a
total of 79 cases. Stephen et al (33) revealed that the hypermethylation of
HIC1 may be an independent predictor of poor survival in laryngeal
carcinoma. Patients with laryngeal cancer and HIC1 methylation had
a median survival time of 1.02 years, as compared to the median
survival time of 4.40 years of patients without HIC1 methylation.
To the best of our knowledge, the expression of HIC1 in laryngeal
cancer has not been evaluated to date.
In conclusion, the present study proved that the
relative expression of the HIC1 putative suppressor gene is
diminished in laryngeal cancer. However, as this difference was not
distinct in all the cases of laryngeal cancer, this finding
requires further investigation.
Acknowledgements
The equipment for molecular analysis was purchased
using the Silesian Bio-Farma Center for Biotechnology,
Bioengineering and Bioinformatics project (grant no.
POIG.02.01.00-00-166/08), the operational programme innovative
economy for 2007–2013, Priority Axis 2.
References
|
1
|
The national cancer registry. http://onkologia.org.pl/[(In Polish)].
Accessed. March 15–2014.
|
|
2
|
Bień S, Kamiński B, Żyłka S, Meżyk R and
Piasta Z: Evolution of the epidemiology and clinical
characteristics of larynx and hypopharynx carcinoma in poland from
1991 to 2001. Eur Arch Otorhinolaryngol. 1:S39–S46. 2008.
View Article : Google Scholar
|
|
3
|
Wu CT and Morris JR: Genes, genetics and
epigenetics: a correspondence. Sciences. 293:1103–1105. 2001.
View Article : Google Scholar
|
|
4
|
Wong TS, Gao W, Li ZH, Chan JY and Ho WK:
Epigenetic dysregulation in laryngeal squamous cell carcinoma. J
Oncol. 2012:7394612012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jabłońska J and Jesionek-Kupnicka D:
Zmiany epigenetyczne w nowotworach. Onkol Pol. 7:181–185. 2004.[(In
Polish)].
|
|
6
|
Severin PM, Zou X, Gaub HE and Schulten K:
Cytosine methylation alters dna mechanical properties. Nucleic
Acids Res. 39:8740–8751. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Baylin SB and Herman JG: Dna
hypermethylation in tumorigenesis: epigenetics joins genetics.
Trends Genet. 16:168–174. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang HW, Ling GS, Wei WI and Yuen AP:
Smoking and drinking can induce p15 methylation in the upper
aerodigestive tract of healthy individuals and patients with head
and neck squamous cell carcinoma. Cancer. 101:125–132. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
van Engeland M, Weijenberg MP, Roemen GM,
Brink M, de Bruïne AP, Goldbohm RA, van den Brandt PA, Baylin SB,
de Goeij AF and Herman JG: Effects of dietary folate and alcohol
intake on promoter methylation in sporadic colorectal cancer: the
netherlands cohort study on diet and cancer. Cancer Res.
63:3133–3137. 2003.PubMed/NCBI
|
|
10
|
Fleuriel C, Touka M, Boulay G, Guérardel
C, Rood BR and Leprince D: Hic1 (hypermethylated in cancer 1)
epigenetic silencing in tumors. Int J Biochem Cell Biol. 41:26–33.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Peralta R, Valdivia A, Alvarado-Cabrero I,
Gallegos F, Apresa T, Hernández D, Mendoza M, Romero P, Paniagua L,
Ibáñez M, Cabrera L and Salcedo M: Correlation between expression
of cellular retinol-binding protein 1 and its methylation status in
larynx cancer. J Clin Pathol. 65:46–50. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bazan V, Zanna I, Migliavacca M,
Sanz-Casla MT, Maestro ML, Corsale S, Macaluso M, Dardanoni G,
Restivo S, Quintela PL, Bernaldez R, Salerno S, Morello V, Tomasino
RM, Gebbia N and Russo A: Prognostic significance of p16ink4a
alterations and 9p21 loss of heterozygosity in locally advanced
laryngeal squamous cell carcinoma. J Cell Physiol. 192:286–293.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kong WJ, Zhang S, Guo C, Zhang S, Wang Y
and Zhang D: Methylation-associated silencing of death-associated
protein kinase gene in laryngeal squamous cell cancer.
Laryngoscope. 115:1395–1401. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Paluszczak J, Misiak P, Wierzbicka M,
Woźniak A and Baer-Dubowska W: Frequent hypermethylation of DAPK,
RARbeta, MGMT, RASSF1A and FHIT in laryngeal squamous cell
carcinomas and adjacent normal mucosa. Oral Oncol. 47:104–107.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang J, Chen H, Fu S, Xu ZM, Sun KL and Fu
WN: The involvement of chd5 hypermethylation in laryngeal squamous
cell carcinoma. Oral Oncol. 47:601–608. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Smigiel R, Sasiadek M, Krecicki T, Ramsey
D, Jagielski J and Blin N: Inactivation of the cyclin-dependent
kinase inhibitor 2a (cdkn2a) gene in squamous cell carcinoma of the
larynx. Mol Carcinog. 39:147–154. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Olasz J, Juhász A, Remenár E, Engi H, Bak
M, Csuka O and Kásler M: Rar beta2 suppression in head and neck
squamous cell carcinoma correlates with site, histology and age.
Oncol Rep. 18:105–112. 2007.PubMed/NCBI
|
|
19
|
Temam S, Bénard J, Dugas C, Trassard M,
Gormally E, Soria JC, Faivre S, Luboinski B, Marandas P, Hainaut P,
Lenoir G, Mao L and Janot F: Molecular detection of early-stage
laryngopharyngeal squamous cell carcinomas. Clin Cancer Res.
11:2547–2551. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen WY, Wang DH, Yen RC, Luo J, Gu W and
Baylin SB: Tumor suppressor hic1 directly regulates sirt1 to
modulate p53-dependent dna-damage responses. Cell. 123:437–448.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cohen HY, Miller C, Bitterman KJ, Wall NR,
Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R and Sinclair
DA: Calorie restriction promotes mammalian cell survival by
inducing the sirt1 deacetylase. Science. 305:390–392. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vaquero A, Scher M, Erdjument-Bromage H,
Tempst P, Serrano L and Reinberg D: Sirt1 regulates the histone
methyl-transferase suv39h1 during heterochromatin formation.
Nature. 450:440–444. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kelly KF and Daniel JM: Poz for
effect-poz-zf transcription factors in cancer and development.
Trends Cell Biol. 16:578–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Briones VR, Chen S, Riegel AT and
Lechleider RJ: Mechanism of fibroblast growth factor-binding
protein 1 repression by tgf-beta. Biochem Biophys Res Commun.
345:595–601. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tassi E, Henke RT, Bowden ET, Swift MR,
Kodack DP, Kuo AH, Maitra A and Wellstein A: Expression of a
fibroblast growth factor-binding protein during the development of
adenocarcinoma of the pancreas and colon. Cancer Res. 66:1191–1198.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Eguchi K, Kanai Y, Kobayashi K and
Hirohashi S: Dna hypermethylation at the d17s5 locus in non-small
cell lung cancers: its association with smoking history. Cancer
Res. 57:4913–4915. 1997.PubMed/NCBI
|
|
27
|
Kanai Y, Hui AM, Sun L, Ushijima S,
Sakamoto M, Tsuda H and Hirohashi S: Dna hypermethylation at the
d17s5 locus and reduced hic-1 mrna expression are associated with
hepatocarcinogenesis. Hepatology. 29:703–709. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kanai YI, Ushijima S, Ochiai A, Eguchi K,
Hui A and Hirohashi S: Dna hypermethylation at the d17s5 locus is
associated with gastric carcinogenesis. Cancer Lett. 122:135–141.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nicoll G, Crichton DN, McDowell HE,
Kernohan N, Hupp TR and Thompson AM: Expression of the
hypermethylated in cancer gene (hic-1) is associated with good
outcome in human breast cancer. Br J Cancer. 85:1878–1882. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hayashi M, Tokuchi Y, Hashimoto T, Hayashi
S, Nishida K, Ishikawa Y, Nakagawa K, Tsuchiya S, Okumura S and
Tsuchiya E: reduced hic-1 gene expression in non-small cell lung
cancer and its clinical significance. Anticancer Res. 21:535–540.
2001.PubMed/NCBI
|
|
31
|
Brieger J, Mann SA, Pongsapich W,
Koutsimpelas D, Fruth K and Mann WJ: Pharmacological genome
demethylation increases radiosensitivity of head and neck squamous
carcinoma cells. Int J Mol Med. 29:505–509. 2012.PubMed/NCBI
|
|
32
|
Zheng J, Wang J, Sun X, et al: HIC1
modulates prostate cancer progression by epigenetic modification.
Clin Cancer Res. 19:1400–1410. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stephen JK, Chen KM, Shah V, Havard S,
Kapke A, Lu M, Benninger MS and Worsham MJ: Dna hypermethylation
markers of poor outcome in laryngeal cancer. Clin Epigenetics.
1:61–69. 2010. View Article : Google Scholar : PubMed/NCBI
|