Introduction
Melanoma is a neoplastic lesion arising from
epidermal melanocytes. It is estimated that 132,000 new cases of
melanoma occur worldwide every year (1). According to the recent data from the
International Agency for Research on Cancer (IARC), the highest
reported national incidence rates for melanoma occurred in the
populations of Australia (39 cases per 100,000 individuals per
year) and New Zealand (34 cases per 100,000 individuals per year)
(2). A previous study indicated that
approximately 65–90% of all melanomas are attributable to
ultraviolet radiation exposure (3).
Ultraviolet radiation is subdivided into ultraviolet A, ultraviolet
B and ultraviolet C. Of these, ultraviolet A wavelengths (320–400
nm) cause single-stranded breaks and DNA protein crosslinking, and
generate free radicals that cause oxidative damage and DNA
double-strand breaks (DSBs) (4).
Ultraviolet B radiations (290–320 nm) induce damage in the form of
cyclobutane pyrimidine dimers and pyrimidine photoproducts, which
may lead to the formation of DSBs, chromosomal aberrations and
recombination during the course of replication arrest (5).
To date, mammalian cells utilize four major DNA
repair mechanisms to protect against genetic instability: base
excision repair, mismatch repair, nucleotide excision repair and
double-strand break repair (DSBR). DSBR is the most common form of
radiation-induced DNA damage and includes both homologous
recombination (HRR) and non-homologous end-joining (6). The X-ray repair cross-complementing
group 3 (XRCC3) protein is involved in the HRR pathway to repair
DNA damage and maintain genomic stability (7). The XRCC3 gene is localized on human
chromosomes 14q32.3. The most frequent polymorphism in XRCC3 is a
C/T transition resulting in an amino acid substitution from Thr to
Met at codon 241 (T241M). In addition, variants of the T241M
polymorphism may affect the function of the encoded protein and
consequently alter the DNA repair capacity (8).
In recent years, several studies have been performed
to evaluate the association between the T241M polymorphism and
melanoma risk. However, the published results have been
inconsistent. Meta-analysis is a useful tool in detecting an
association that could otherwise remain masked in the sample size
studies, particularly in those evaluating rare allele frequency
polymorphisms (9). The aim of this
meta-analysis was to investigate the association between T241M
polymorphism and susceptibility to melanoma using all eligible
case-control studies published to date.
Materials and methods
Literature search
We searched for studies in the PubMed and Embase
electronic databases using the terms ‘melanoma’, ‘skin cancer’,
‘T241M’, ‘XRCC3’, ‘excision repair cross-complementing group 3’ and
‘polymorphism’. The search was performed without any restrictions
on language and was focused on studies conducted in humans. Further
studies were identified by a hand search of references of original
or review articles on this topic. If data or data subsets were
published in more than one article, only the publication with the
largest sample size was included.
Inclusion and exclusion criteria
Eligible studies included in the present analysis
met the following criteria: i) studies that evaluated the
association between the XRCC3 T241M polymorphism and melanoma, ii)
a case-control study design, and iii) had detailed genotype
frequency of cases and controls or could be calculated from the
article text. The main exclusion criteria were: i) case reports,
letters, reviews, meta-analyses and editorial articles, ii)
non-case-control studies that evaluated the association between
XRCC3 T241M polymorphism and melanoma risk, iii) studies in which
the number of null and wild genotypes could not be ascertained, and
iv) duplicate data were included in the studies.
Data extraction
Two investigators independently extracted data
according to the inclusion criteria. Disagreement was resolved by
discussion between them. If no consensus was reached, an expert was
consulted to resolve the dispute and a final majority decision was
made. For each study, the following data was collected: the first
authors name, year of publication, country of origin, ethnicity,
area, number of patients and controls, distributions of genotypes
and alleles, and evidence of Hardy-Weinberg equilibrium (HWE).
These are listed in Table I.
 | Table I.Characteristics of studies included in
meta-analysis. |
Table I.
Characteristics of studies included in
meta-analysis.
|
|
|
|
|
| Genotypes of
cases | Genotypes of
controls |
|
|---|
|
|
|
|
|
|
|
|
|
|---|
| Study included
(ref.) | Year | Area | Ethnicity | Cases/controls | TT | MT | MM | TT | MT | MM | HWE test |
|---|
| Winsey et al
(11) | 2000 | England | Caucasian | 125/211 | 39 | 65 | 21 | 110 | 78 | 23 | 0.11 |
| Duan et al
(12) | 2002 | USA | Mixed | 305/319 | 119 | 148 | 38 | 116 | 158 | 45 | 0.45 |
| Figl et al
(13) | 2010 | Germany | Caucasian | 1184/1274 | 451 | 541 | 192 | 436 | 645 | 193 | 0.07 |
| Gonçalves et
al (14) | 2011 | Brazil | Mixed | 192/192 | 78 | 89 | 25 | 95 | 79 | 18 | 0.79 |
| Bertram et al
(15) | 2004 | England | Caucasian | 140/335 | 50 | 68 | 22 | 135 | 160 | 40 | 0.89 |
| Han et al
(16) | 2004 | USA | Mixed | 187/810 | 75 | 84 | 28 | 300 | 396 | 114 | 0.36 |
Data analysis
We tested whether genotype frequencies of controls
were in HWE using the χ2 test. The odds ratio (OR) and
corresponding 95% confidence interval (CI) were calculated to
evaluate the association between the XRCC3 T241M polymorphism and
melanoma risk under a homozygote comparison (TT vs. MM), a
heterozygote comparison (TT vs. MT), a dominant model (MM+MT vs.
TT) and a recessive mode (TT+MT vs. MM) between groups.
Between-study heterogeneities were estimated using the
I2 test. I2 values of 25, 50 and 75% were
defined as low, moderate and high estimates, respectively (10). If heterogeneity was observed among the
studies, the pooled OR was estimated using the fixed-effects model
(P>0.10 or I2<50%). Otherwise, the random-effects
model was used to estimate the pooled OR. Subgroup analyses by
ethnicity and sample size in cases were performed. Sensitivity
analysis was performed by comparing the random-effect model values
with the fixed-effect model values. Publication bias was
investigated using funnel plot and Begg's funnel plot (P<0.05
was considered to indicate a statistically significant difference).
Analyses were performed with Stata software (version 12.0; Stata
Corporation, College Station, TX, USA), using two-sided
P-values.
Results
Identification of eligible
studies
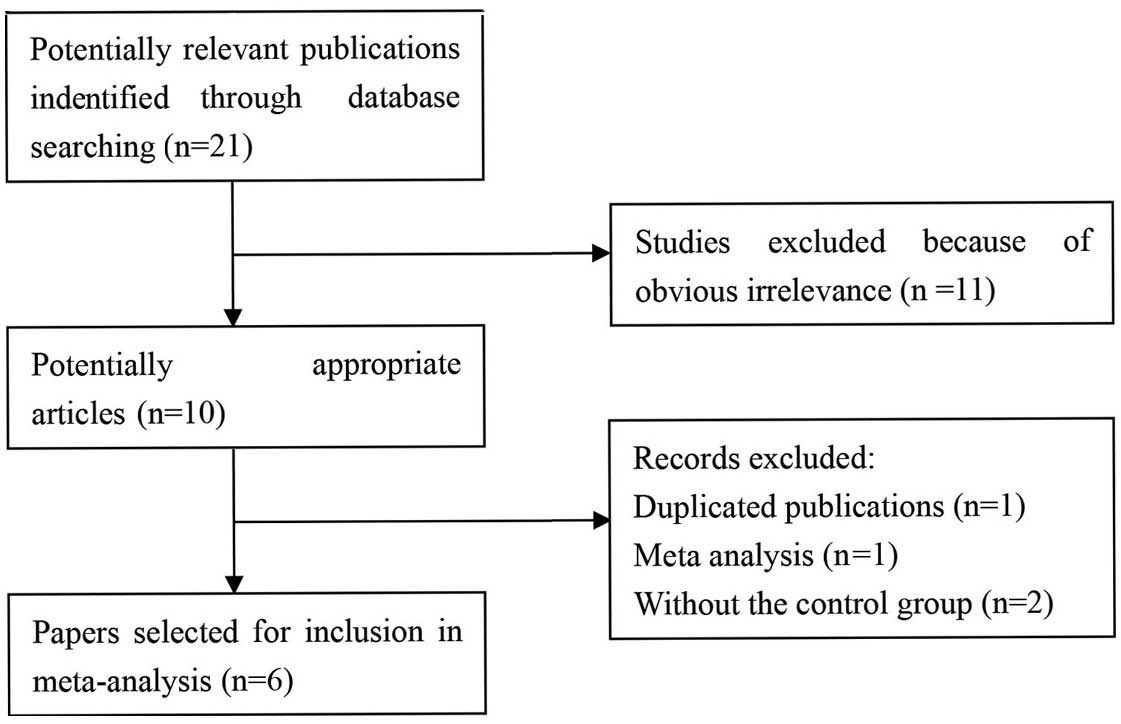
The search strategy retrieved 21 potentially
relevant studies. Based on the inclusion criteria, six case-control
studies with full text were included in this meta-analysis
(11–16) and 15 studies were excluded. The flow
chart for the study selection is summarized in Fig. 1. The six case-control studies selected
included a total of 2,133 cases and 3,141 healthy controls. The HWE
test was performed on genotype distribution of the controls; all of
them were in HWE (P>0.05). The studies had been carried out in
England, USA, Germany and Brazil. Among them, there were three
studies of Europeans (11,13,15). The
publishing year of the included studies ranged from 2000 to 2014.
All of the articles were written in English. The source of controls
was mainly based on healthy populations. Furthermore, the studies
were conducted with >500 subjects included in the subgroup
meta-analysis (12,13,16).
General characteristics and the allele and genotype distributions
in the published articles included in this meta-analysis are shown
in Table I.
Meta-analysis
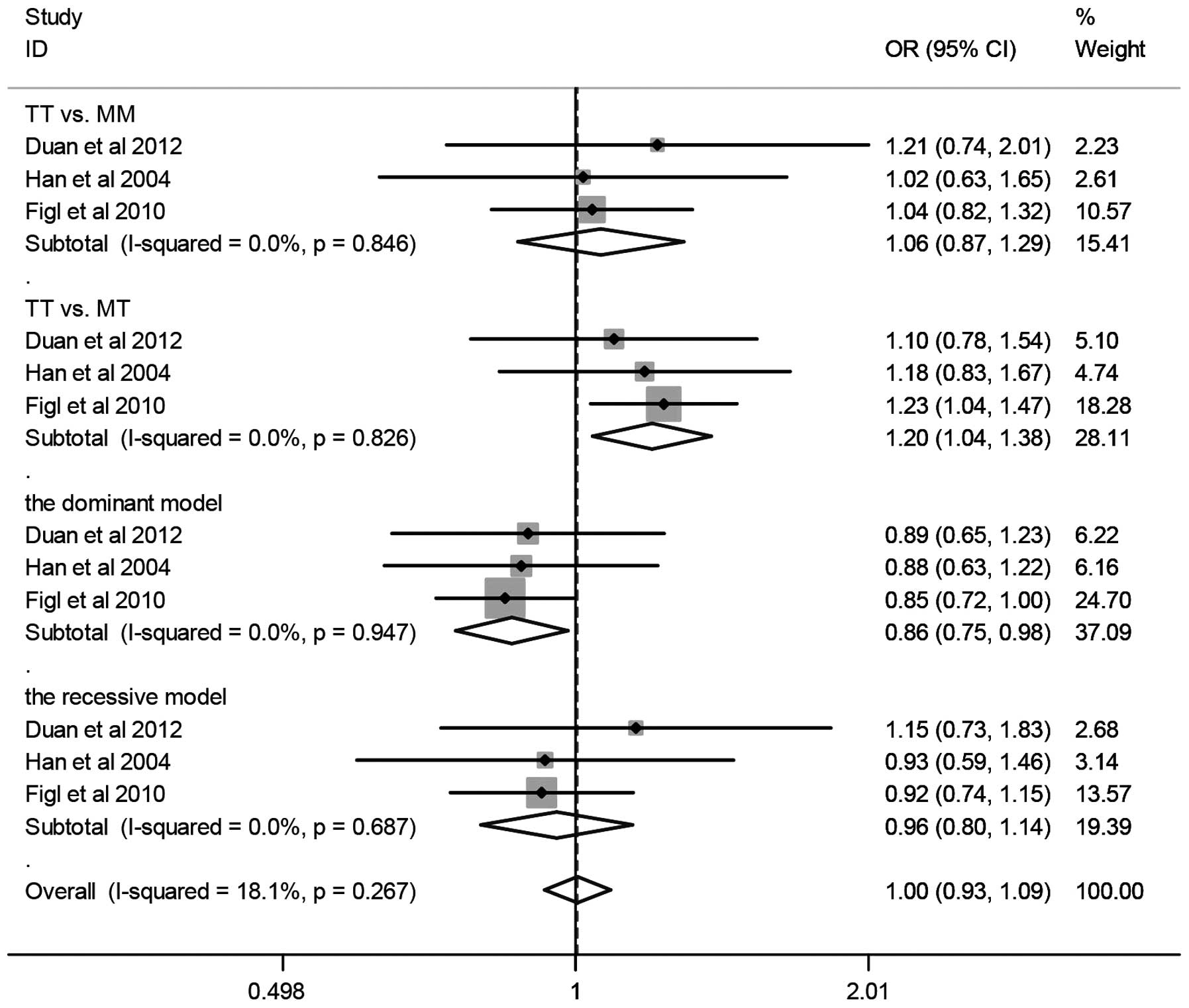
A summary of the meta-analysis findings of the
association between the XRCC3 T241M polymorphism and melanoma risk
is shown in Fig. 2 and Table II. The combined results based on all
studies revealed that variant genotypes are not associated with
increased melanoma risk in different genetic models (TT vs. MM:
OR=0.92, 95% CI=0.68–1.26; TT vs. MT: OR=0.95, 95% CI=0.72–1.25;
dominant model: OR=1.07, 95% CI=0.81–1.41; recessive model: OR
=0.94, 95% CI=0.80–1.10). When stratified according to ethnicity,
no significant association was detected in Caucasians (TT vs. MM:
OR=0.70, 95% CI=0.39–1.24; TT vs. MT: OR=0.80, 95% CI=0.44–1.46;
dominant model: OR=1.31, 95% CI=0.72–2.38; recessive model:
OR=0.86, 95% CI=0.71–1.05). In the stratified analysis by sample
size (>500 subjects), we detected a significant association
between T241M and melanoma (TT vs. MM: OR=1.06, 95% CI=0.87–1.29;
TT vs. MT: OR=1.20, 95% CI=1.04–1.38; dominant model: OR=0.86, 95%
CI=0.75–0.98; recessive model: OR =0.96, 95% CI=0.80–1.15).
Sensitivity analyses were conducted by altering the statistic
models. No material alteration was detected, indicating that our
results were statistically robust.
 | Table II.Summary of odds ratios and 95%
confidence interval of T241M polymorphism with melanoma risk. |
Table II.
Summary of odds ratios and 95%
confidence interval of T241M polymorphism with melanoma risk.
|
|
| Sample size |
| Test of
heterogeneity | Test of
association | Test of publication
bias |
|---|
|
|
|
|
|
|
|
|
|---|
| Subgroup | Genetic model | Cases | Controls | Type of model | I2 | P-value | OR | 95% CI | Z | P-value |
|---|
| Overall | TT vs. MM | 2133 | 3141 | Random | 54.1% | 0.05 | 0.83 | 1.62–1.12 | 0.24 | 0.81 |
|
| TT vs. MT |
|
| Random | 75.3% | 0.00 | 0.91 | 0.69–1.20 | 0.24 | 0.81 |
|
| Dominant model |
|
| Random | 77.8% | 0.00 | 1.14 | 0.86–1.51 | 0.24 | 0.81 |
|
| Recessive model |
|
| Fixed | 0.0% | 0.57 | 0.89 | 0.76–1.05 | 0.24 | 0.81 |
| Caucasians | TT vs. MM | 1449 | 1820 | Random | 74.6% | 0.02 | 0.70 | 0.39–1.24 | 0.00 | 1.00 |
|
| TT vs. MT |
|
| Random | 88.2% | 0.00 | 0.80 | 0.44–1.46 | 0.00 | 1.00 |
|
| Dominant model |
|
| Random | 89.1% | 0.00 | 1.31 | 0.72–2.38 | 0.00 | 1.00 |
|
| Recessive
model |
|
| Fixed | 0.0% | 0.39 | 0.86 | 0.71–1.05 | 0.00 | 1.00 |
| Sample size | TT vs. MM | 1676 | 2403 | Fixed | 0.0% | 0.85 | 1.06 | 0.87–1.29 | 1.04 | 0.30 |
| >500 | TT vs. MT |
|
| Fixed | 0.0% | 0.83 | 1.20 | 1.04–1.38 | 1.04 | 0.30 |
|
| Dominant model |
|
| Fixed | 0.0% | 0.95 | 0.86 | 0.75–0.98 | 1.04 | 0.30 |
|
| Recessive
model |
|
| Fixed | 0.0% | 0.69 | 0.96 | 0.80–1.15 | 1.04 | 0.30 |
Publication bias
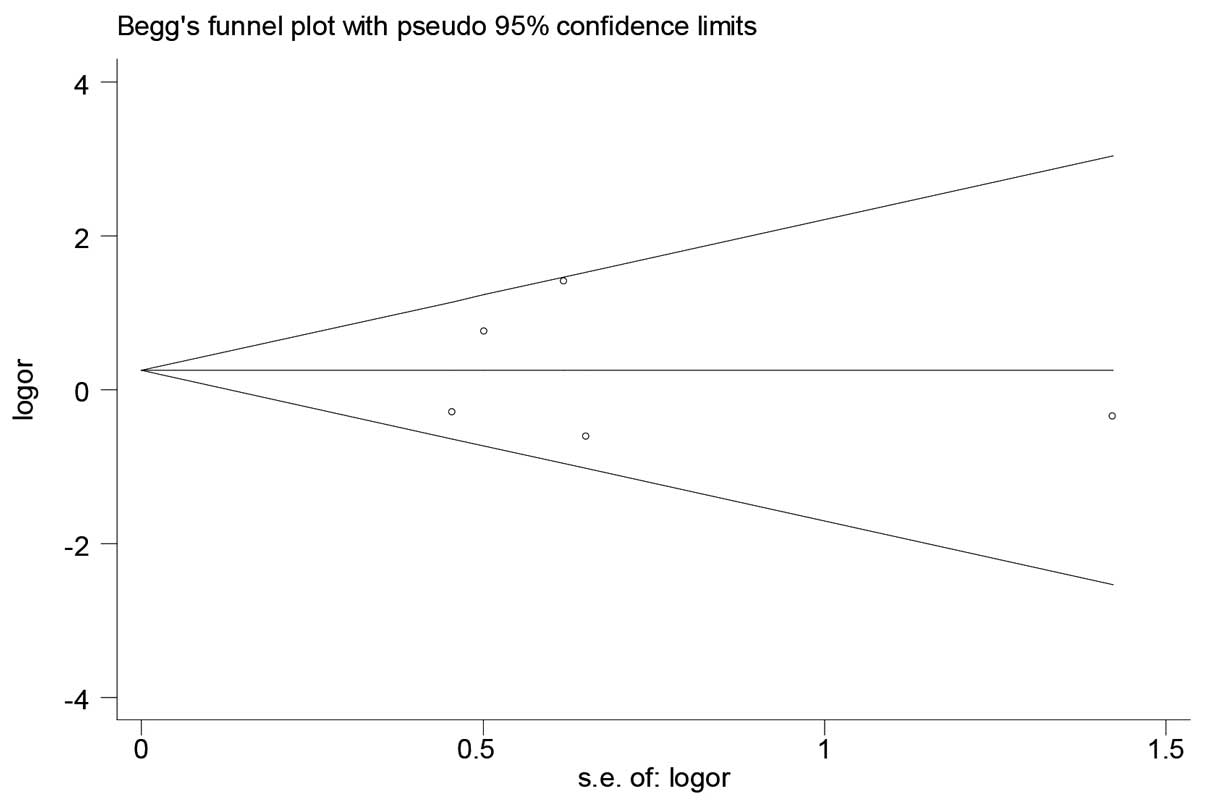
The funnel plot and Begg's test were used to assess
the publication bias of the literature. There was no evidence of
publication bias in our study (Fig.
3). The results implied that the publication bias was low in
the present meta-analysis (all P>0.05). Information concerning
the Begg's funnel plot is given in Table
II.
Discussion
The incidence rate of cutaneous melanoma, which is
etiologically linked to sun exposure, has rapidly grown over the
years (17). Evidence suggests that
cancer may be initiated by DNA damage, and that most DNA damage may
be removed by DNA repair enzymes, including XRCC3. Numerous studies
have evaluated the association of the XRCC3 T241M polymorphism with
the risk of various cancer types, including colorectal, bladder,
lung, breast and pancreatic cancer (18–20). A
variety of studies have focused on the association between the
XRCC3 T241M polymorphism and melanoma. However, the observed
associations of these studies were inconclusive. The most likely
reason for the inconsistencies among these studies is that they are
single case-control studies with small sample sizes. To resolve
these conflicting results, we conducted this meta-analysis to
combine the same type of studies to increase the sample size and
statistical power, and hence obtain a more reliable result.
The present meta-analysis, including 2,133 cases and
3,141 controls from six case-control studies, explored the
association between the XRCC3 T241M polymorphism and melanoma risk.
The results of the meta-analysis revealed that the T241M
polymorphism is not associated with increased or decreased risk of
melanoma in the overall population. Considering that the result may
be affected by ethnicity, an ethnicity-related subgroup analysis
was performed, and no significant association was identified in
Caucasians. Our meta-analysis involved several studies with small
samples. There may be a selective bias for the correlation between
the XRCC3 T241M polymorphism and melanoma development, so the
association should be re-evaluated in studies with large sample
sizes. When stratifying by sample size (>500), this
meta-analysis detected a significant association, suggesting the
possibility of publication bias by smaller studies. Nevertheless,
caution should be exercised when considering this conclusion.
Sensitivity analysis was performed by comparing random-effect model
values with fixed-effect values, and the results revealed that this
meta-analysis was realistic and accurate. There was no evidence of
publication bias in this meta-analysis (all P>0.05).
The mechanism of how XRCC3 T241M polymorphism is
associated with melanoma risk remains unclear. XRCC3 codes for a
protein participating in HRR of DSB. It is a member of an emerging
family of Rad-51-related proteins that may take part in homologous
recombination to repair DSB and maintain chromosome stability
(7), and the XRCC3 T241M polymorphism
affects the DNA repair capacity of its encoded protein, and thus
contributes to the development of melanoma (21). In addition, the potential influence of
the XRCC3 T241M polymorphism may be affected by gene-gene and
gene-environment interactions.
The present study has certain limitations. Firstly,
there are only six studies included in our meta-analysis. More
well-designed studies with large sample sizes are needed to further
identify this association more comprehensively. Secondly, studies
included in the present meta-analysis mainly provided data with
regard to Caucasians, and other ethnicities including Asians and
Africans should be investigated in future studies. Thirdly,
subgroup analyses according to age, radiation exposure,
histological types and other factors have not been performed due to
insufficient relevant data available in the primary studies.
Finally, only published English studies were included in this
study, so publication and potential language biases may occur.
In conclusion, our meta-analysis indicates that
XRCC3 T241M polymorphism is associated with risk of melanoma.
Large-scale case-control and population-based association studies
are warranted to validate the risk identified in the current
meta-analysis and investigate the potential gene-gene and
gene-environment interactions on melanoma risk.
References
|
1
|
Foster PJ, Dunn EA, Karl KE, et al:
Cellular magnetic resonance imaging: in vivo imaging of melanoma
cells in lymph nodes of mice. Neoplasia. 10:207–216. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Matullo G, Palli D, Peluso M, et al:
XRCC1, XRCC3, XPD gene polymorphisms, smoking and (32)P. DNA
adducts in a sample of healthy subjects. Carcinogenesis.
22:1437–1445. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Glanz K, Buller DB and Saraiya M: Reducing
ultraviolet radiation exposure among outdoor workers: state of the
evidence and recommendations. Environ Health. 6:222007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Elmets C and Mukhtar H: Ultraviolet
radiation and skin cancer: progress in pathophysiological
mechanisms. Dermatology Foundation. Prog Dermatol. 30:1–16.
1996.
|
|
5
|
Lehmann A: Dual functions of DNA repair
genes: molecular, cellular and clinical implications. Bioessays.
20:146–155. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Altieri F, Grillo C, Maceroni M, et al:
DNA damage and repair: from molecular mechanisms to health
implications. Antioxid Redox Signal. 10:891–937. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brenneman MA, Weiss AE, Nickoloff JA, et
al: XRCC3 is required for efficient repair of chromosome breaks by
homologous recombination. Mutat Res. 459:89–97. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Matullo G, Guarrera S and Carturan S: DNA
repair gene polymorphisms, bulky DNA adducts in white blood cells
and bladder cancer in a case. control study. Int J Cancer.
92:562–567. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Attia J, Thakkinstian A and D'Este C:
Meta-analyses of molecular association studies: methodologic
lessons for genetic epidemiology. J Clin Epidemiol. 56:297–303.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Higgins JP and Thompson SG: Quantifying
heterogeneity in a meta-analysis. Stat Med. 21:1539–1558. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Winsey SL, Haldar NA, Marsh HP, et al: A
variant within the DNA repair gene XRCC3 is associated with the
development of melanoma skin cancer. Cancer Res. 60:5612–5616.
2000.PubMed/NCBI
|
|
12
|
Duan Z, Shen H, Lee JE, et al: DNA repair
gene XRCC3 241Met variant is not associated with risk of cutaneous
malignant melanoma. Cancer Epidemiol Biomarkers Prev. 11:1142–1143.
2002.PubMed/NCBI
|
|
13
|
Figl A, Scherer D, Nagore E, et al:
Single-nucleotide polymorphisms in DNA. repair genes and cutaneous
melanoma. Mutat Res. 702:8–16. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Goncalves FT, Francisco G, de Souza SP, et
al: European ancestry and polymorphisms in DNA repair genes modify
the risk of melanoma: a case. control study in a high UV index
region in Brazil. J Dermatol Sci. 64:59–66. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bertram CG, Gaut RM, Barrett JH, et al: An
assessment of a variant of the DNA repair gene XRCC3 as a possible
nevus or melanoma susceptibility genotype. J Invest Dermatol.
122:429–432. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Han J, Colditz GA, Samson LD, et al:
Polymorphisms in DNA double-strand break repair genes and skin
cancer risk. Cancer Res. 64:3009–3013. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Leiter U and Garbe C: Epidemiology of
melanoma and nonmelanoma skin cancer. the role of sunlight. Adv Exp
Med Biol. 624:89–103. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Z, Li C, Xu Y, et al: A
meta-analysis on XRCC1 and XRCC3 polymorphisms and colorectal
cancer risk. Int J Colorectal Dis. 25:169–180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jiao L, Hassan MM, Bondy ML, et al: XRCC2
and XRCC3 gene polymorphism and risk of pancreatic cancer. Am J
Gastroenterol. 103:360–367. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jin MJ, Chen K, Song L, et al: The
association of the DNA repair gene XRCC3 Thr241Met polymorphism
with susceptibility to colorectal cancer in a Chinese population.
Cancer Genet Cytogenet. 163:38–43. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Araujo FD, Pierce AJ, Stark JM, et al:
Variant XRCC3 implicated in cancer is functional in
homology-directed repair of double-strand breaks. Oncogene.
21:4176–4180. 2002. View Article : Google Scholar : PubMed/NCBI
|