Introduction
AC3-33 (GenBank name: C3orf33, accession no.
FLJ31139), also known as chromosome 3 open reading frame 33,
encodes a classical secretory protein with a predicted molecular
mass of 29.3 kDa (1). Transcription
factor activator protein-1 (AP-1) is crucial in the regulation of
cellular proliferation, transformation and death (2). Using a dual-luciferase reporter assay
system, our previous study found that AC3-33 significantly
inhibited AP-1 transcriptional activity. Further investigation
indicated that AC3-33 significantly inhibited the transcriptional
activity of Elk1 and c-jun, but not of c-fos; additionally, AC3-33
significantly inhibits Elk1 transcriptional activity via the
extracellular-signal-regulated kinases 1/2/mitogen-activated
protein kinases pathway. This occurs via disruption of ERK1/2 MAPK
pathway (3). AC3-33 is highly
expressed in a number of tissues, including the adrenal glands and
cervix, and expression is comparatively significantly reduced in
the human leukemia cell lines, K562 and KG1a (4). However, the expression of AC3-33
in multiple organ tumors and cancer-adjacent normal tissue remains
to be elucidated.
In the present study, RNA in situ
hybridization was used to detect the AC3-33 gene expression
in multiple organ tumors and cancer-adjacent normal tissue. An
improved understanding of the expression of AC3-33 may offer
more information as to the role of AC3-33 in the
pathological process of tumorigenesis, which may subsequently
provide a new insight into AC3-33 and its potential
applications in the treatment and diagnosis of human disease.
Materials and methods
Tissue microarray
Tissue microarray was purchased from Chaoying
Biotechnology (Xian, China; MCN602). Specimens for microarray were
obtained from a total of 56 cases of multiple organ tumors and
adjacent normal tissue. This included 10 organ types (esophagus,
stomach, colon, rectum, liver, lung, kidney, breast, uterine
cervix, ovary), three tissue cores for cancerous tissue, three
adjacent normal tissue cores for each organ and a single specimen
per case. For all specimens, details of age, gender, organ,
pathological diagnosis, clinical grade, TNM classification,
clinical stage, specimen type and results were recorded. This study
was approved by the ethics committee of Hebei United University
(Tangshan, China).
Preparation of digoxigenin-labeled
probes for RNA in situ hybridization
Sense and anti-sense probes that matched the AC3-33
corresponding sequence were: Anti-sense,
TATAA*GTTCTCTGAACTTCAGTATTAAGGAGCAGTTGTTCATGTTGTCTTTC-DIG; and
sense, GAAATG*TTAAACTACGTGGACGATTACGCCGAATAACTGAGAATGGTTTA-DIG. The
asterisk indicates that the 3′ terminal was labeled with
digoxigenin. All probes were synthesized by Sangon Biotech
(Shanghai, China).
RNA in situ hybridization
Hybridization procedures were performed in this
study as described. Hybridization conditions were as follows:
Anti-sense or sense probe concentration, 20 ng/ul; anti-digoxigenin
antibody (catalog no. ab76907; Abcam, Cambridge, UK) dilution,
1:500; was hing temperature, room temperature; dyeing temperature,
37°C; dyeing time, 2 h. Deparaffinized sections were mounted on
Denhardt-coated glass slides (D2532; Sigma Aldrich, St. Louis, MO,
USA) and treated with pepsin (0.25 mg/ml in diethylpyrocarbonate
H2O-HCl) for 30 min in a 37°C water bath. The treated
sections were then processed for in situ hybridization at
42–47°C for 24 h. The hybridization mixture contained the labeled
oligonucleotide probe, 50% formamide, 10 mmol/l Tris-HCl, 1 mmol/l
vanadyl-ribonucleoside complex (Sigma-Aldrich; catalog no. 94740),
1 mmol/l cetrimonium bromide (Sigma-Aldrich; catalog no. 855820, pH
7.0), 0.15 mol/l NaCl, 1 mmol/l EDTA (pH 7.0), 1×Denhardt's mixture
and 10% dextran sulfate. Following hybridization, the slides were
was hed three times, 30 min each time, in 0.1 mol/l Tris buffered
saline (TBS) at 47°C, and subsequently treated with TBS (100 mmol/l
Tris, pH 7.5, 150 mmol/l NaCl) containing 1% blocking reagent
(Roche Diagnostics, Shanghai, China) and 0.03% Triton X-100 for 30
min at room temperature and incubated for 30 min with
antidioxigenin alkaline phosphatase-conjugated antibodies (Roche
Diagnostics) diluted at 1:4000 in TBS containing 0.03% Triton X-100
and a 1% blocking reagent. After was hing three times in TBS and
0.05% Tween, 15 min each time, the slides were rinsed in a
diammonimum phosphate (DAP) buffer (100 mmol/l Tris, pH 9.5, 100
mmol/l NaCl, 50 mmol/l MgCl2) and subsequently
hybridization signals were visualized using nitroblue tetrazolium
and 5-brom-4-chlor-3-indolyl phosphate as substrates [DAP in 10%
polyvinyl alcohol (Sigma-Aldrich; catalog no. 341584)]. Positive
expression was determined to be 1+, 2+ and 3+ staining, and
negative expression was observed as no staining.
Results
The association between AC3-33
expression and multiple pathological cell types
The AC3-33 gene expression in multiple organ
and cancer-adjacent normal tissue was detected by RNA in
situ hybridization. As shown in Table
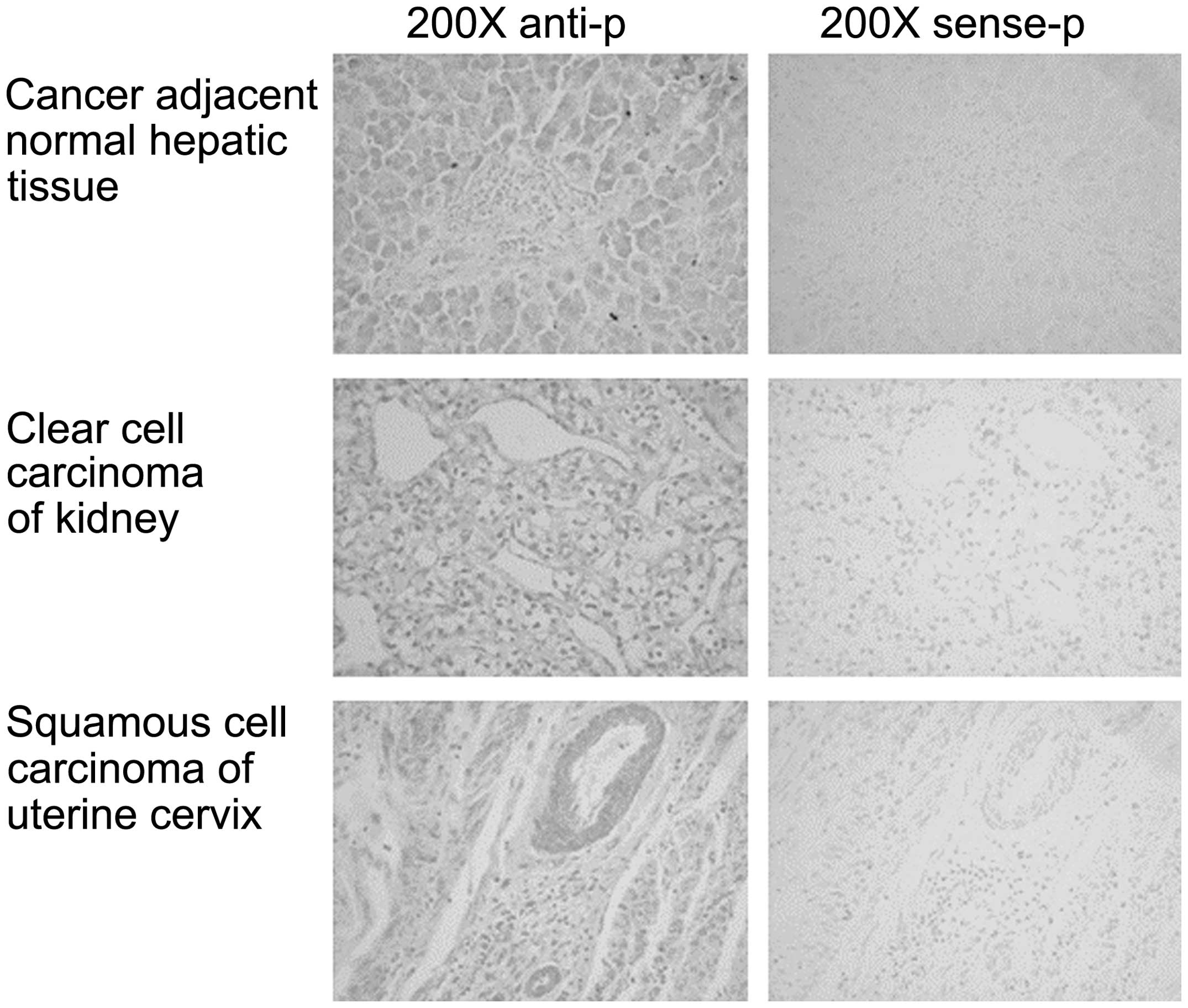
I and Fig. 1, the expression
level of AC3-33 varies between the different tissues. The
expression of AC3-33 is positive in squamous cell carcinoma
(SCC) of the esophagus and adenocarcinoma of the rectum, but is
negative in cancer-adjacent normal esophageal tissue and
cancer-adjacent normal rectal tissue. AC3-33 exhibits
positive expression in hepatocellular carcinoma and cancer-adjacent
normal hepatic tissue, clear cell carcinoma of the kidney and
cancer-adjacent normal kidney tissue. Negative AC3-33
expression was observed for adenocarcinoma of the stomach and
cancer-adjacent normal gastric tissue, adenocarcinoma of the colon
and cancer-adjacent normal colon tissue, serous adenocarcinoma of
the ovary and cancer-adjacent normal ovarian tissue. AC3-33
exhibited positive expression in squamous cell carcinoma of the
lung, invasive ductal carcinoma of the breast and squamous cell
carcinoma of the uterine cervix. However, the expression of AC3-33
in cancer-adjacent normal breast tissue is partially positive.
 | Table I.The association between AC3-33
expression and multiple organ tumors and cancer adjacent normal
tissue. |
Table I.
The association between AC3-33
expression and multiple organ tumors and cancer adjacent normal
tissue.
| Organ | Pathological
diagnosis | Tumors, n | AC3-33 mRNA-positive
tumors (+/+/++) | AC3-33 mRNA-negative
tumors |
|---|
| Esophagus | Squamous cell
carcinoma | 3 | 3 (1/2/0) | 0 |
|
| Cancer adjacent
normal esophageal tissue | 3 | 0 | 3 |
| Stomach | Adenocarcinoma | 3 | 0 | 3 |
|
| Cancer adjacent
normal gastric tissue | 3 | 0 | 3 |
| Colon | Adenocarcinoma | 3 | 0 | 3 |
|
| Cancer adjacent
normal colon tissue | 3 | 0 | 3 |
| Rectum | Adenocarcinoma | 3 | 3 (1/2/0) | 0 |
|
| Cancer adjacent
normal rectal tissue | 3 | 0 | 3 |
| Liver | Hepatocellular
carcinoma | 3 | 3 (0/3/0) | 0 |
|
| Cancer adjacent
normal hepatic tissue | 3 | 3 (0/0/3) | 0 |
| Lung | Squamous cell
carcinoma | 3 | 3 (1/2/0) | 0 |
|
| Cancer adjacent
normal lung tissue | 1 | 1 (0/1/0) | 0 |
| Kidney | Clear cell
carcinoma | 3 | 3 (0/2/1) | 0 |
|
| Cancer adjacent
normal kidney tissue | 3 | 3 (0/3/1) | 0 |
| Breast | Invasive ductal
carcinoma | 3 | 3 (0/2/1) | 0 |
|
| Cancer adjacent
normal breast tissue | 4 | 3 (1/2/0) | 1 |
| Uterine cervix | Squamous cell
carcinoma | 3 | 3 (0/2/1) | 0 |
| Ovary | Serous
adenocarcinoma | 3 | 0 | 3 |
|
| Cancer adjacent
normal ovarian tissue | 3 | 0 | 3 |
Discussion
With >300,000 new cases per year, cancer of the
esophagus, predominantly SCC, is one of the 10 most frequently
diagnosed tumor types (5). Esophageal
cancer often occurs in developing countries, and the incidence is
greatly different between different regions (5). The development of molecular oncology in
the last decade has provided much information with regard to
genetic abnormalities in cancer, and the clinical characteristics
of cancer patients can now be predicted on the basis of these
genetic abnormalities. Expression of N-myc (6), int-2 (7),
cyclin D1 (8) and p53 (9) may be useful markers for predicting the
outcome and distant organ metastasis in patients with SCC of the
esophagus. The current study also found that AC3-33 exhibits
positive expression in esophagal SCC, but negative expression in
cancer-adjacent normal esophageal tissue. These results indicate
that AC3-33 may be a novel prognostic factor.
Colorectal cancer (also known as colon cancer,
rectal cancer, bowel cancer or colorectal adenocarcinoma) is a
cancer due to uncontrolled cell growth in the colon or rectum, or
in the appendix (10). Genetic
analysis has shown that colon and rectal tumors are genetically the
same cancer (11). Although the
prognosis of rectal adenocarcinoma is associated with
histopathological features, including invasion of the rectal wall
or perirectal fat and lymph node involvement, a number of patients
experience recurrence despite undergoing potentially curative
procedures and early pathological staging (12–13). It
has been proposed that genetic alterations acquired during tumor
development may predict prognosis (14). For example, the expression of the p53
protein has been found to predict a worse prognosis in rectal
adenocarcinoma (14). Similarly to
SCC of the esophagus, the current study identified positive
AC3-33 expression in rectal adenocarcinoma, but negative
expression in cancer-adjacent normal rectal tissue.
In conclusion, the differential expression of
AC3-33 may be significant in the development and progression
of rectal adenocarcinoma and esophagal SCC, and may be used as a
prognostic indicator. However, the mechanism of AC3-33 function
appears to be complex and further investigations are required to
elucidate the role and molecular mechanisms of AC3-33 in the
development and progression of rectal adenocarcinoma and esophagal
SCC.
Acknowledgements
This study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81072093; 30671092;
81302323), the Natural Science Foundation of Hebei Province (grant
nos. C2009001260; C2013209024; C2014209140), the General Higher
Education Young Talents Program of Hebei Province (BJ2014027), and
the Science and Technology Support Program of Tangshan City (grant
no. 14120208a).
References
|
1
|
Zhang X, Ma X, Xue Y, et al: Prokaryotic
expression and characterization of human AC3-33 protein. Front
Biosci (Elite Ed). 2:1134–1142. 2010. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Angel P and Karin M: The role of Jun, Fos
and the AP-1 complex in cell-proliferation and transformation.
Biochim Biophys Acta. 1072:129–157. 1991.PubMed/NCBI
|
|
3
|
Hao D, Gao P, Liu P, et al: AC3-33, a
novel secretory protein, inhibits Elk1 transcriptional activity via
ERK pathway. Mol Biol Rep. 38:1375–1382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu P, Deng WW, Gao P, et al: Molecular
cloning and preliminary function study of a novel human gene AC3-33
related to suppress AP-1 activity. Yi Chuan. 30:575–582. 2008.(In
Chinese). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bollschweiler E and Holscher AH: Carcinoma
of the esophagus-actual epidemiology in Germany. Onkologie.
24:180–184. 2001.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Suita S, Zaizen Y, Kaneko M, et al: What
is the benefit of aggressive chemotherapy for advanced
neuroblastoma with N-myc amplification? A report from the Japanese
study group for the treatment of advanced neuroblastoma. J Pediatr
Surg. 29:746–750. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kitagawa Y, Ueda M, Ando N, Shinozawa Y,
Shimizu N and Abe O: Significance of int-2/hst-1 coamplification as
a prognostic factor in patients with esophageal squamous carcinoma.
Cancer Res. 51:1504–1508. 1991.PubMed/NCBI
|
|
8
|
Shinozaki H, Ozawa S, Ando N, et al:
Cyclin D1 amplification as a new predictive classification for
squamous cell carcinoma of the esophagus, adding gene information.
Clin Cancer Res. 2:1155–1161. 1996.PubMed/NCBI
|
|
9
|
Yamasaki M, Miyata H, Fujiwara Y, et al:
p53 genotype predicts response to chemotherapy in patients with
squamous cell carcinoma of the esophagus. Ann Surg Oncol.
17:634–642. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
National Cancer Institute, . Colon Cancer
Treatment (PDQ®). http://www.cancer.gov/cancertopics/pdq/treatment/colon/Patient/page1/AllPagesAccessed.
29–June. 2014
|
|
11
|
Comprehensive molecular characterization
of human colon and rectal cancer. Nature. 487:330–337. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Olson RM, Perencevich NP, Malcolm AW,
Chaffey JT and Wilson RE: Patterns of recurrence following curative
resection of adenocarcinoma of the colon and rectum. Cancer.
45:2969–2974. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ratto C, Sofo L, Ippoliti M, Merico M,
Doglietto GB and Crucitti F: Prognostic factors in colorectal
cancer. Literature review for clinical application. Dis Colon
Rectum. 41:1033–1049. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jurach MT, Meurer L and Moreira LF:
Expression of the p53 protein and clinical and pathologic
correlation in adenocarcinoma of the rectum. Arq Gastroenterol.
43:9–14. 2016.
|