Introduction
MicroRNAs (miRNAs) are a family of small endogenous
noncoding RNAs with a length of ~22 nucleotides. These molecules
are able to negatively regulate gene expression at the
post-transcriptional level by binding to the 3′-untranslated region
(3′-UTR) of target messenger RNA (mRNA), resulting in mRNA
degradation and/or translational repression (1,2). Notably,
one mRNA sequence can be targeted by multiple miRNAs, while one
miRNA has multiple mRNA targets (3).
Depending on specific target genes, including oncogenes and tumor
suppressor genes, miRNAs regulate numerous cellular functions,
including cell development, proliferation, differentiation,
apoptosis, signal transduction, tumorigenesis and cancer
progression (4,5). In addition, miRNAs are potential
prognostic and diagnostic biomarkers, as well as therapeutic
targets for the treatment of various neoplastic diseases (6,7).
Circulating miRNAs were initially described in 2008, and since
then, >79 miRNAs have been reported to be plasma or serum
biomarkers of several tumors, including prostate, lung, breast,
colon, ovarian, esophageal, melanoma and gastric cancer (8,9). As a
hematopoietic cell-specific miRNA, miRNA-150 (miR-150) plays an
important role in normal hematopoiesis and hematological
malignancies (10). A large number of
studies have indicated that the aberrant expression of miR-150 is
closely associated with tumorigenesis, cancer development,
malignant behavior and a curative effect by influencing oncogenes
and/or tumor suppressor genes (11–13). In
addition to hematological malignancies, miR-150 is involved in a
variety of solid tumors, including breast, lung and gastric
cancer.
The role of miR-150 in normal and malignant
hematopoiesis has been summarized in detail in a review article by
He et al (14). In the present
review, the functions and regulatory mechanism of miR-150 as an
oncogene or tumor suppressor gene in certain solid tumors were
discussed. In addition, its potential application as a tumor
biomarker, targeted therapeutic strategy and index of prognosis in
these cancer types was investigated.
miR-150 and cancer
Increasing evidence has indicated that miRNAs are
associated with the molecular mechanisms of various clinical
diseases and can potentially regulate numerous aspects of cellular
biological progress (4,5). In addition, different tissues exhibit
different expression patterns. Monticelli et al were the
first to investigate the systematic miRNA gene profiling in
hematopoietic cells, demonstrating that its profiling was different
from non-hematopoietic cells (15).
As an important hematopoietic cell-specific miRNA, miR-150 is
mainly expressed in B-cells, T-cells and natural killer cells, and
plays a critical role in the differentiation of numerous
hematopoietic cell lineages, particularly in lymphocyte development
and function (14,16). In addition, a recent study has
identified that serum circulating miR-150 is a sensor of general
lymphocyte activation and may serve as a biomarker of human
lymphocyte activation in healthy and disease conditions (17). miR-150 has been previously reported to
be differentially expressed in various hematopoietic cell lineages
of a specific developmental stage or characteristically up- or
downregulated in various types of hematopoietic malignancies,
including leukemia, lymphoma and myelodysplastic syndrome (14). In chronic myeloid leukemia (CML),
miR-150 has been demonstrated to be involved in the mechanism of
apoptosis induced by cisplatin in the human CML cell line, K562
(18). Xie et al (18)demonstrated a negative correlation
between the expression levels of miR-150 and p53 following
treatment of K562 cells with cisplatin, indicating that cisplatin
induced apoptosis in the K562 cells by inhibiting miR-150
expression, which then upregulated p53 expression. Therefore,
miR-150 may serve as a novel, clinically-useful biomarker in
myeloid leukemia diagnosis and may have a curative effect. In
addition, miR-150 is significantly downregulated in the majority of
acute myeloid leukemia (AML) cases, which is not associated with
the DNA copy number changes, methylation or mutations (11). Furthermore, the results of a recent
study revealed that the plasma expression of miR-150 was
significantly downregulated in AML patients at diagnosis when
compared with healthy controls; however, miR-150 plasma expression
in complete remission AML patients resembled that of the healthy
controls (19). Receiver operating
characteristic curve analyses revealed that plasma miR-150 may
serve as a valuable biomarker for the differentiation of AML
patients from control individuals, with a sensitivity and
specificity of 80 and 70%, respectively (19). The expression signature of miR-150 in
the plasma indicated that it may serve as a valuable diagnostic and
potentially prognostic biomarker for human AML (19). Furthermore, in cutaneous T-cell
lymphoma, upregulation of miR-150 inhibited tumor invasion and
metastasis by targeting the chemokine CCL20 receptor, CCR6
(20). These results have provided a
novel insight into the function of miR-150 as a tumor suppressor in
the pathogenesis of hematological malignancies, as well as a basis
for novel therapies targeting miR-150 for the treatment of these
hematological malignancies discussed above.
In hematological malignancies, miR-150 dysregulation
has been also demonstrated to be involved in the tumorigenesis and
development of a number of solid tumors, as well as function as a
biomarker of clinical diagnosis, outcome and prognosis of these
solid cancer types(Table I and
Fig. 1).
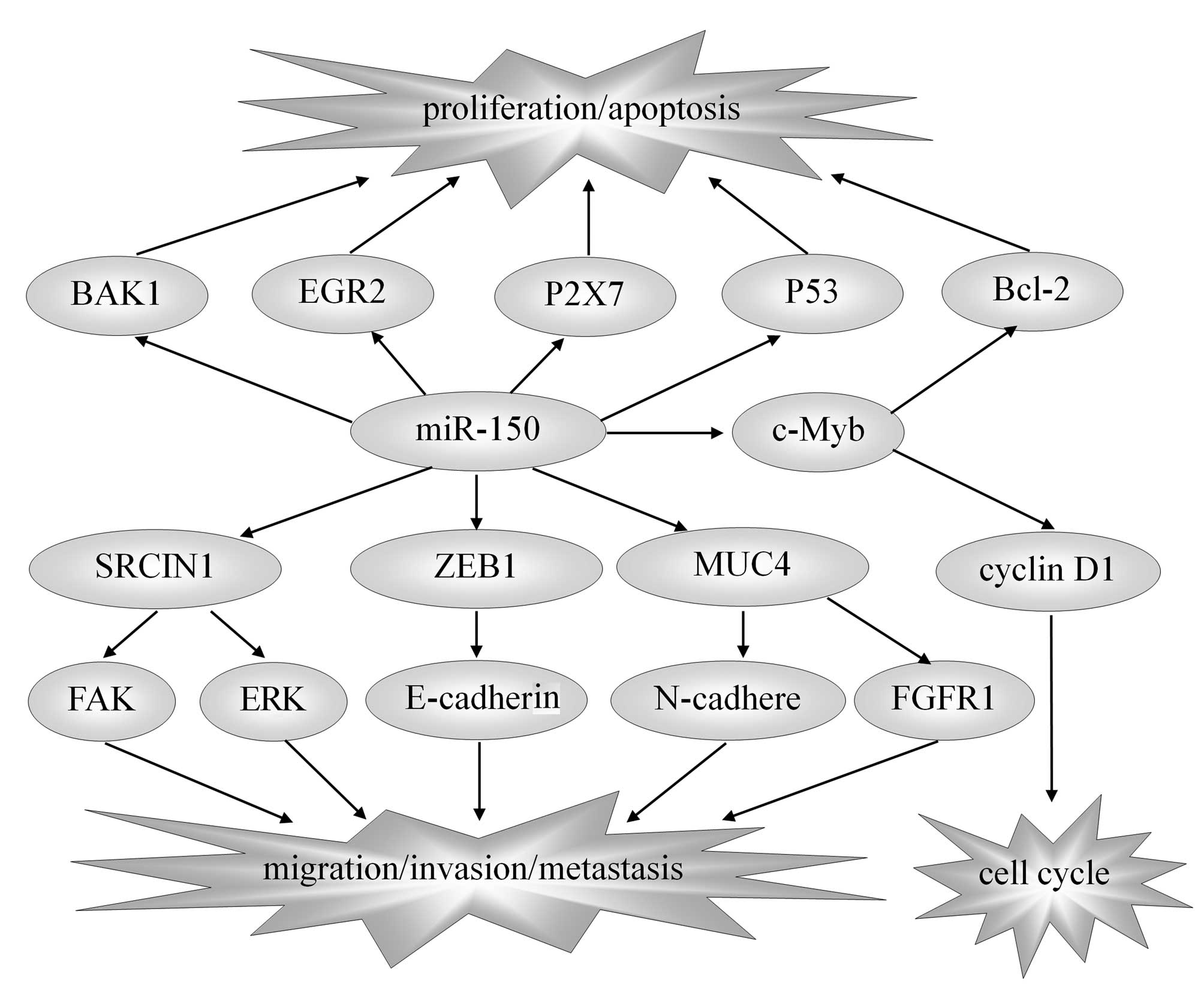 | Figure 1.miR-150 and target genes in solid
tumors. The reported direct targets of miR-150 include EGR2, P2X7,
P53, c-Myb, SRCIN1, ZEB1, MUC4 and BAK1. Though regulating these
target genes, miR-150 influences the proliferation, apoptosis,
metastasis and prognosis of solid tumors, thus playing the role of
anti-tumor or carcinogenesis. miR-150, microRNA-150; EGR2, early
growth response factor 2; SRCIN1, sarcoma gene kinase signalling
inhibitor 1; ZEB1, zinc-finger E-box binding homeobox 1; MUC4,
mucin 4; BAK1, BRI1-associated receptor kinase 1; Bcl-2, B-cell
lymphoma 2; FAK, focal adhesion kinase; ERK, extracellular
signal-regulated kinase. |
 | Table I.Level, targets and functions of
miR-150 in solid tumors. |
Table I.
Level, targets and functions of
miR-150 in solid tumors.
| Type of cancer | Level | Targets | Functions |
|---|
| Pancreatic
cancer | ↓ | MUC4 | Overexpression of
miR-150 inhibits growth, clonogenicity and invasion, as well as
enhances intercellular adhesion in pancreatic cancer cells by
downregulating MUC4. |
| Esophageal squamous
cell carcinoma | ↓ | ZEB1 | Through
targeted-degradation of ZEB1, miR-150 induces MET-like changes and
evidently inhibits tumorigenicity and tumor proliferation. |
| Colorectal
cancer | ↓ | – | Exosomal miR-150
expression appears to mirror pathological changes in colorectal
cancer patients and is a promising biomarker for non-invasive
diagnosis of the disease. |
| Gastric cancer | ↑ | EGR2 | In gastric cancer,
forced miR-150 specifically represses the expression of
pro-apoptotic gene, EGR2, at the translational level, as a result
of promoting proliferation and growth of gastric cancer. |
| Breast cancer | ↑ | P2X7 | The anti-apoptosis
and maintain-growth role of miR-150 for the development of breast
cancer was induced directly by downregulating P2X7. |
| Non-small cell lung
cancer | ↑ | SRCIN1, P53,
BAK1 | miR-150 promotes
the proliferation and migration of lung cancer cells through
specifically targeting the 3′-UTR of p53, SRCIN1 and BAK1. |
| Liver cancer | ↓ | c-Myb | miR-150 may be
involved in maintenance of the CD133+ liver cancer stem
cell phenotype through the negative regulation of the downstream
target, c-Myb. |
miR-150 in pancreatic cancer
Mucins (MUCs) are a family of high molecular weight
glycoproteins, which are widely expressed in epithelial cells.
Under normal physiological conditions, MUCs have a protective role
on the adjoining epithelial tissues, whereas carcinomas and
neoplastic lesions are often associated with an altered expression
of MUCs (21). MUC4 is a specifically
and restrictedly upregulated membrane-bound glycoprotein in
pancreatic tumors and its potential role as a marker for pancreatic
adenocarcinoma has been identified (22). In addition, a number of studies have
indicated that the silencing of MUC4 expression resulted in altered
tumor cell phenotypic characteristics (adhesion, aggregation and
motility), decreased growth and a marked reduction in metastatic
incidences in an orthotopic mouse model of pancreatic cancer
(21). Furthermore, MUC4
overexpression potentiated pancreatic tumor cell proliferation,
survival and invasive properties by stabilizing fibroblast growth
factor receptor 1 through N-cadherin upregulation (Fig. 1). This supports the aforementioned
observations and indicates the important role of the MUC4 in
pancreatic adenocarcinoma development and progression (21,23,24). A
recent study has demonstrated the presence of a highly conserved
miR-150 binding site at the 3′-UTR of MUC4 and that its direct
interaction with miR-150 downregulated the endogenous MUC4 protein
expression levels, as shown in Table
I (25). In addition, miR-150
overexpression inhibited the malignant behavior and enhanced the
homotypic interactions of pancreatic cancer (25). Therefore, as a novel tumor suppressor
miRNA, restoring the miR-150 expression levels may have a
therapeutic effect in pancreatic cancer.
miR-150 in esophageal cancer
miR-150 expression has also been demonstrated to be
significantly lower in esophageal squamous cell carcinoma compared
with the normal esophageal mucosa levels (26). In addition, its deregulation
contributed to a number of malignant characteristics, including
cancer progression, higher clinical staging and poor prognosis
(26). Zinc-finger E-box binding
homeobox 1 (ZEB1), a crucial epithelial-mesenchymal transition
(EMT)-inducer, may promote tumor invasion and migration through
E-cadherin gene silencing in cancer (27–29). A
recent study has indicated that, through the targeted-degradation
of ZEB1, miR-150 induced mesenchymal-epithelial transition-like
changes and evidently inhibited tumorigenicity and tumor
proliferation (Fig. 1) (26). These results clarified the
EMT-regulatory ability and clinicopathological significance of
miR-150, and provided new insights into the prevention of
metastasis and a promising novel candidate for targeted therapeutic
strategies in esophageal cancer.
miR-150 in colorectal cancer
A recent study identified that the expression levels
of miR-150 were downregulated in primary colorectal cancer and
metastasis compared with the normal mucosa levels, while the
expression was almost stably maintained in the subsequent primary
tumor-to-metastasis transition (30).
In addition, its expression gradually decreased during the tumor
development, and patients with lower miR-150 expression levels in
the tissues exhibited lower survival rates and reduced response to
adjuvant chemotherapy, which was independent of other clinical risk
factors associated with the clinical outcome (31,32). These
observations suggested that miR-150 should be considered as a
potential biomarker associated with the prognosis and therapeutic
outcome of colorectal cancer. However, the serum exosomal
expression of miR-150 was significantly higher in primary
colorectal cancer patients compared with healthy controls and
significantly downregulated following surgical resection of the
tumors (33). miRNA was also found to
be secreted at significantly higher levels in colon cancer cell
lines compared with the levels in a normal colon-derived cell line
(33). The true positive rate of
miR-150 for identification of colorectal cancer was 55.7%, while
low false positive rates were observed for identification of the
healthy controls (33). By contrast,
the sensitivities of the carcinoembryonic antigen and carbohydrate
antigen 19–9, which are known as biomarkers of colorectal cancer,
were 30.7 and 16.0%, respectively (33). The exosomal miR-150 expression
appeared to mirror pathological changes of colorectal cancer
patients; therefore, it may be a promising biomarker for
non-invasive diagnosis of the disease (Table I).
miR-150 in gastric cancer
In contrast to the low miR-150 expression in
esophageal squamous cell carcinoma, high expression levels have
been identified in gastric, breast and endometrial cancer tissues
(34–37). In gastric cancer, miR-150
overexpression specifically repressed the expression of the
pro-apoptotic gene, early growth response factor 2, at the
translational level, as a result of promoting proliferation and
growth of gastric cancer (38). The
higher expression level of miR-150 in undifferentiated gastric
cancer was associated with shorter postoperative patient survival;
however, it was not a significantly independent prognostic factor
in undifferentiated gastric cancer patients (35).
miR-150 in breast cancer
Huang et al revealed that blocking the action
of miR-150 with inhibitors in breast cancer cell lines resulted in
cell death, while ectopic expression of miR-150 promoted growth and
clonogenicity, and reduced apoptosis (37). In addition, these authors identified
that low levels of P2X7 receptor, an adenosine triphosphate-gated
cation channel inducing apoptosis by leading Ca2+
release, were linked to the development of breast cancer (37). Furthermore, as shown in Table I, the 3′-UTR of P2X7 receptor contains
a highly conserved miR-150-binding motif and directly interacts
with miR-150, downregulating endogenous P2X7 protein levels and
promoting breast cancer growth and malignant behaviors (37), which is consistent with the results of
a previous study (36). These
observations provided further understanding of the anti-apoptosis
and growth-regulation role of miR-150 in the development of
malignancies; therefore, targeting miR-150 may provide a potential
therapeutic strategy for blocking proliferation in certain solid
cancer types.
miR-150 in lung cancer
In previous studies, researchers identified that
inhibition of miR-150 expression effectively delayed cell
proliferation and promoted apoptosis in the lung carcinoma cells,
A549, and was accompanied by increased p53 protein expression,
which has a specific miR-150 binding site (39–41).
Antisense oligonucleotide specific to miR-150 increased the
chemotherapeutic sensitivity of A549 cells to anticancer drugs,
which was promising for lung cancer therapy (40,41). In
addition, miR-150 was aberrantly upregulated in non-small cell lung
cancer (NSCLC) and promoted the growth of cancer cells through
specifically targeting the 3′-UTR of p53 (42). These results indicated that miR-150
may promote lung cancer tumorigenesis by targeting p53.
Overexpression of p53 promoted the expression of miRNAs, including
miR-34a, miR-184, miR-181a and miR-148, which affected cell cycle
progression in NSCLC tumorigenesis (43). These findings indicated that miR-150,
p53 protein and relevant miRNAs comprised a complicated regulatory
network in NSCLC tumorigenesis. In addition, miR-150 was found to
be significantly upregulated in lung cancer clinical specimens,
while sarcoma gene (SRC) kinase signalling inhibitor 1 (SRCIN1),
which is an important regulator of SRC activity, was identified as
a direct target of miR-150 (44).
Therefore, the repression of SRCIN1 by miR-150 triggered the
activation of the Src/focal adhesion kinase and
Src/Ras/extracellular signal-regulated kinase signaling pathways,
which eventually promoted the proliferation and migration of lung
cancer cells (Fig. 1); this promotion
by miR-150 cannot be reversed by the overexpression of SRCIN1
(44). Furthermore, miR-150
functioned as an oncogene by directly targeting human
BRI1-associated receptor kinase 1 (BAK1) in NSCLC cells (45). These observations highlighted a novel
molecular interaction between miR-150 and BAK1, and provided a
novel strategy for NSCLC therapy through the downregulation of
miR-150 expression. However, the underlying regulatory mechanism of
miR-150 in NSCLC was controversial (46). A recent study demonstrated that
miR-150 expression in peripheral blood mononuclear cells was
significantly higher in lung adenocarcinoma patients compared with
lung squamous cell carcinoma patients (47). This finding indicated that miR-150 may
be a potential noninvasive molecular biomarker for the
identification of histological subtypes of NSCLC and may assist the
selection of effective therapeutic strategies to improve the
treatment outcome.
However, in small cell lung cancer (SCLC), the
expression levels of miR-150 were much lower in the tumor samples
compared with normal lung samples (48). The miR-150/miR-886-3p signature, which
may be used as an independent predictor of survival, was
significantly correlated with the overall survival and
progression-free survival of SCLC patients (48). Therefore, miR-150 may predict cancer
progression and survival in early-stage SCLC patients and may be a
promising prognostic biomarker and potential therapeutic targets in
SCLC patients. However, the precise target of the miR-150 and the
mechanisms underlying its involvement in tumor formation and
prognosis in small cell lung cancer remain to be elucidated.
miR-150 in liver cancer
miR-150 was found to be significantly downregulated
in hepatocellular carcinomas and may be a suitable candidate in the
differentiation between tumoral and normal human primary
hepatocytes (49). Zhang et al
compared the miRNA profiles of CD133+ and
CD133− primary hepatocellular carcinoma subpopulations
and identified upregulation of miR-150 expression in
CD133− subpopulations (50). In addition, overexpression of miR-150
resulted in a significant reduction of CD133+ cells,
along with significant inhibition of cell growth and tumorsphere
formation (50). The levels of the
cell cycle regulator, cyclin D1, and cell survival regulator,
B-cell lymphoma-2, decreased in cells transfected with miR-150,
which was consistent with the outcome of cell cycle arrest and cell
apoptosis, as shown in Fig. 1
(50). Furthermore, these authors
demonstrated that miR-150 may be involved in the maintenance of the
CD133+ liver cancer stem cell phenotype through the
direct negative regulation of the downstream target, c-Myb, and its
potential function in liver cancer stem cells may provide a novel
therapeutic approach for hepatocellular carcinomas (50).
Conclusions
miRNAs have received increasing attention since
their discovery, and one study has indicated that miRNAs regulate
various cellular biological processes, as well as participate in
the pathogenesis of diseases, particularly cancer (51). In addition, the circulating miRNA
levels are useful in the diagnosis or evaluation of activity in
human diseases (51).
In the present review, the critical role of miR-150
as an oncogene or tumor suppressor gene in relevant solid tumors
was investigated, as well as its potential as a tumor biomarker,
targeted therapeutic strategy and index of prognosis in different
cancer types. In the aforementioned cancer types, certain
relatively definite conclusions have been reached on the mechanisms
underlying the role of miR-150 as an oncogene or cancer suppressor
gene in the pathogenesis of tumors. However, in other cancer types,
including osteosarcoma, Ewing sarcoma, hepatoblastoma and
adrenocorticotropic hormone-secreting pituitary tumor, the
regulatory mechanisms of miR-150 are unclear (52–56). In
addition, it is unclear why miR-150 functions both as an
onco-microRNA and a tumor suppressor microRNA in solid tumors. This
may depend on factors including the pathological type, histological
origin, cellular microenvironment and localization of the
respective neoplasm.
The miR-150 expression regulation provides a
promising novel candidate used as a tumor biomarker, targeted
therapeutic strategy and index of prognosis in cancer. However, the
use of circulating miRNAs as clinical biomarkers may face certain
technical challenges. For instance, dilution effects in blood may
limit the amount of RNA per volume of starting material, while
cellular detritus and hemolysis may potentially impact
reproducibility and sensitivity (17). Several studies have recently attempted
to use miRNAs in the serum or plasma as a highly sensitive and
non-invasive diagnostic or prognostic biomarker of various cancer
types (17,33). However, currently, no collective view
exists on which miRNA should be selected as a marker. At present,
the role and mechanisms of miR-150 are not fully understood;
however, future studies will aim to elucidate the existing
controversial findings.
Acknowledgements
The present study was supported by grants from the
National Basic Research Program of China (973 Program, No.
2012CB9333004) and the Tianjin Natural Science Foundation (No.
14JCYBJC27100).
References
|
1
|
Lee S and Vasudevan S:
Post-transcriptional stimulation of gene expression by microRNAs.
Adv Exp Med Biol. 768:97–126. 2013.PubMed/NCBI
|
|
2
|
Bartel DP: MicroRNAs: target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhou P, Xu W, Peng X, et al: Large-scale
screens of miRNA-mRNA interactions unveiled that the 3′UTR of a
gene is targeted by multiple miRNAs. PLoS One. 8:e682042013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Huang Y, Shen XJ, Zou Q, Wang SP, Tang SM
and Zhang GZ: Biological functions of microRNAs: a review. J
Physiol Biochem. 67:129–139. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ebert MS and Sharp PA: Roles for microRNAs
in conferring robustness to biological processes. Cell.
149:515–524. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Markou A, Liang Y and Lianidou E:
Prognostic, therapeutic and diagnostic potential of microRNAs in
non-small cell lung cancer. Clin Chem Lab Med. 49:1591–1603.
2011.PubMed/NCBI
|
|
7
|
Mulrane L, Klinger R, McGee SF, et al:
MicroRNAs: a new class of breast cancer biomarkers. Expert Rev Mol
Diagn. 14:347–363. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Pritchard CC, Kroh E, Wood B, et al: Blood
cell origin of circulating microRNAs: a cautionary note for cancer
biomarker studies. Cancer Prev Res (Phila). 5:492–497. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schwarzenbach H, Nishida N, Calin GA and
Pantel K: Clinical relevance of circulating cell-free microRNAs in
cancer. Nat Rev Clin Oncol. 11:145–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vasilatou D, Papageorgiou S, Pappa V,
Papageorgiou E and Dervenoulas J: The role of microRNAs in normal
and malignant hematopoiesis. Eur J Haematol. 84:1–16. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jiang X, Huang H, Li Z, et al: Blockade of
miR-150 maturation by MLL-fusion/MYC/LIN-28 is required for
MLL-associated leukemia. Cancer Cell. 22:524–535. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Watanabe A, Tagawa H, Yamashita J, et al:
The role of microRNA-150 as a tumor suppressor in malignant
lymphoma. Leukemia. 25:1324–1334. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhao JJ, Lin J, Lwin T, et al: MicroRNA
expression profile and identification of miR-29 as a prognostic
marker and pathogenetic factor by targeting CDK6 in mantle cell
lymphoma. Blood. 115:2630–2639. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He Y, Jiang X and Chen J: The role of
mir-150 in normal and malignant hematopoiesis. Oncogene.
33:3887–3893. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Monticelli S, Ansel KM, Xiao C, et al:
MicroRNA profiling of the murine hematopoietic system. Genome Biol.
6:R712005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Allantaz F, Cheng DT, Bergauer T, et al:
Expression profiling of human immune cell subsets identifies
miRNA-mRNA regulatory relationships correlated with cell type
specific expression. PLoS One. 7:e299792012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
de Candia P, Torri A, Pagani M and
Abrignani S: Serum microRNAs as biomarkers of human lymphocyte
activation in health and disease. Front Immunol.
5:432014.PubMed/NCBI
|
|
18
|
Xie SY, Li YJ, Wang PY, Jiao F, Zhang S
and Zhang WJ: MiRNA-regulated expression of oncogenes and tumor
suppressor genes in the cisplatin-inhibited growth of K562 cells.
Oncol Rep. 23:1693–1700. 2010.PubMed/NCBI
|
|
19
|
Fayyad-Kazan H, Bitar N, Najar M, et al:
Circulating miR-150 and miR-342 in plasma are novel potential
biomarkers for acute myeloid leukemia. J Transl Med. 11:312013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ito M, Teshima K, Ikeda S, et al:
MicroRNA-150 inhibits tumor invasion and metastasis by targeting
the chemokine receptor CCR6 in advanced cutaneous T-cell lymphoma.
Blood. 123:1499–1511. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Singh AP, Moniaux N, Chauhan SC, Meza JL
and Batra SK: Inhibition of MUC4 expression suppresses pancreatic
tumor cell growth and metastasis. Cancer Res. 64:622–630. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andrianifahanana M, Moniaux N, Schmied BM,
et al: Mucin (MUC) gene expression in human pancreatic
adenocarcinoma and chronic pancreatitis: a potential role of MUC4
as a tumor marker of diagnostic significance. Clin Cancer Res.
7:4033–4040. 2001.PubMed/NCBI
|
|
23
|
Chaturvedi P, Singh AP, Moniaux N, et al:
MUC4 mucin potentiates pancreatic tumor cell proliferation,
survival and invasive properties and interferes with its
interaction to extracellular matrix proteins. Mol Cancer Res.
5:309–320. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Rachagani S, Macha MA, Ponnusamy MP, et
al: MUC4 potentiates invasion and metastasis of pancreatic cancer
cells through stabilization of fibroblast growth factor receptor 1.
Carcinogenesis. 33:1953–1964. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Srivastava SK, Bhardwaj A, Singh S, et al:
MicroRNA-150 directly targets MUC4 and suppresses growth and
malignant behavior of pancreatic cancer cells. Carcinogenesis.
32:1832–1839. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yokobori T, Suzuki S, Tanaka N, et al:
MiR-150 is associated with poor prognosis in esophageal squamous
cell carcinoma via targeting the EMT inducer ZEB1. Cancer Sci.
104:48–54. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Korpal M, Lee ES, Hu G and Kang Y: The
miR-200 family inhibits epithelial-mesenchymal transition and
cancer cell migration by direct targeting of E-cadherin
transcriptional repressors ZEB1 and ZEB2. J Biol Chem.
283:14910–14914. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gregory PA, Bert AG, Paterson EL, et al:
The miR-200 family and miR-205 regulate epithelial to mesenchymal
transition by targeting ZEB1 and SIP1. Nat Cell Biol. 10:593–601.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Thompson EW and Haviv I: The social
aspects of EMT-MET plasticity. Nat Med. 17:1048–1049. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pizzini S, Bisognin A, Mandruzzato S, et
al: Impact of microRNAs on regulatory networks and pathways in
human colorectal carcinogenesis and development of metastasis. BMC
Genomics. 14:5892013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y, Zhang P, Wang F, et al: MiR-150 as a
potential biomarker associated with prognosis and therapeutic
outcome in colorectal cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yong FL, Law CW and Wang CW: Potentiality
of a triple microRNA classifier: miR-193a-3p, miR-23a and
miR-338-5p for early detection of colorectal cancer. BMC Cancer.
13:2802013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ogata-Kawata H, Izumiya M, Kurioka D, et
al: Circulating exosomal microRNAs as biomarkers of colon cancer.
PLoS One. 9:e929212014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Inoue T, Iinuma H, Ogawa E, Inaba T and
Fukushima R: Clinicopathological and prognostic significance of
microRNA-107 and its relationship to DICER1 mRNA expression in
gastric cancer. Oncol Rep. 27:1759–1764. 2012.PubMed/NCBI
|
|
35
|
Katada T, Ishiguro H, Kuwabara Y, et al:
MicroRNA expression profile in undifferentiated gastric cancer. Int
J Oncol. 34:537–542. 2009.PubMed/NCBI
|
|
36
|
Zhou L, Qi X, Potashkin JA, Abdul-Karim FW
and Gorodeski GI: MicroRNAs miR-186 and miR-150 downregulate
expression of the pro-apoptotic purinergic P2×7 receptor by
activation of instability sites at the 3′-untranslated region of
the gene that decrease steady-state levels of the transcript. J
Biol Chem. 283:28274–28286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang S, Chen Y, Wu W, et al: miR-150
promotes human breast cancer growth and malignant behavior by
targeting the pro-apoptotic purinergic P2×7 receptor. PLoS One.
8:P12013.
|
|
38
|
Wu Q, Jin H, Yang Z, et al: MiR-150
promotes gastric cancer proliferation by negatively regulating the
pro-apoptotic gene EGR2. Biochem Biophys Res Commun. 392:340–345.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cheng AM, Byrom MW, Shelton J and Ford LP:
Antisense inhibition of human miRNAs and indications for an
involvement of miRNA in cell growth and apoptosis. Nucleic Acids
Res. 33:1290–1297. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang PY, Li YJ, Zhang S, et al: Regulating
A549 cells growth by ASO inhibiting miRNA expression. Mol Cell
Biochem. 339:163–171. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li YJ, Zhang YX, Wang PY, et al:
Regression of A549 lung cancer tumors by anti-miR-150 vector. Oncol
Rep. 27:129–134. 2012.PubMed/NCBI
|
|
42
|
Zhang N, Wei X and Xu L: MiR-150 promotes
the proliferation of lung cancer cells by targeting P53. FEBS Lett.
587:2346–2351. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wang DT, Ma ZL, Li YL, et al: MiR-150, p53
protein and relevant miRNAs consist of a regulatory network in
NSCLC tumorigenesis. Oncol Rep. 30:492–498. 2013.PubMed/NCBI
|
|
44
|
Cao M, Hou D, Liang H, et al: MiR-150
promotes the proliferation and migration of lung cancer cells by
targeting SRC kinase signalling inhibitor 1. Eur J Cancer.
50:1013–1024. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gu XY, Wang J, Luo YZ, et al:
Down-regulation of miR-150 induces cell proliferation inhibition
and apoptosis in non-small-cell lung cancer by targeting BAK1 in
vitro. Tumour Biol. 35:5287–5293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sun Y, Su B, Zhang P, et al: Expression of
miR-150 and miR-3940-5p is reduced in non-small cell lung carcinoma
and correlates with clinicopathological features. Oncol Rep.
29:704–712. 2013.PubMed/NCBI
|
|
47
|
Zeng XL, Zhang SY, Zheng JF, Yuan H and
Wang Y: Altered miR-143 and miR-150 expressions in peripheral blood
mononuclear cells for diagnosis of non-small cell lung cancer. Chin
Med J (Engl). 126:4510–4516. 2013.PubMed/NCBI
|
|
48
|
Bi N, Cao J, Song Y, et al: A microRNA
signature predicts survival in early stage small-cell lung cancer
treated with surgery and adjuvant chemotherapy. PLoS One.
9:P12014.
|
|
49
|
Di Masi A, Viganotti M, Antoccia A, et al:
Characterization of HuH6, Hep3B, HepG2 and HLE liver cancer cell
lines by WNT/β-catenin pathway, microRNA expression and protein
expression profile. Cell Mol Biol (Noisy-le-grand). 56(Suppl):
OL1299–OL1317. 2010.PubMed/NCBI
|
|
50
|
Zhang J, Luo N, Luo Y, Peng Z, Zhang T and
Li S: MicroRNA-150 inhibits human CD133-positive liver cancer stem
cells through negative regulation of the transcription factor
c-Myb. Int J Oncol. 40:747–756. 2012.PubMed/NCBI
|
|
51
|
Weiland M, Gao XH, Zhou L and Mi QS: Small
RNAs have a large impact: circulating microRNAs as biomarkers for
human diseases. RNA Biol. 9:850–859. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lulla RR, Costa FF, Bischof JM, et al:
Identification of differentially expressed microRNAs in
osteosarcoma. Sarcoma. 2011:7326902011.PubMed/NCBI
|
|
53
|
Namløs HM, Meza-Zepeda LA, Barøy T, et al:
Modulation of the osteosarcoma expression phenotype by microRNAs.
PLoS One. 7:e480862012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Mosakhani N, Guled M, Leen G, et al: An
integrated analysis of miRNA and gene copy numbers in xenografts of
ewing's sarcoma. J Exp Clin Cancer Res. 31:242012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Magrelli A, Azzalin G, Salvatore M, et al:
Altered microRNA expression patterns in hepatoblastoma patients.
Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Amaral FC, Torres N, Saggioro F, et al:
MicroRNAs differentially expressed in ACTH-secreting pituitary
tumors. J Clin Endocrinol Metab. 94:320–323. 2009. View Article : Google Scholar : PubMed/NCBI
|















