Introduction
It is well-known that lung cancer is the leading
cause of cancer-associated mortality worldwide. In 2011, ~221,000
novel cases of lung and bronchial cancer were diagnosed, and
~156,900 mortalities occurred due to lung cancer in the USA. The
five-year survival rate of all lung cancer patients subsequent to
diagnosis is only ~15.6%. Delayed diagnosis is a fundamental
obstacle in improving lung cancer outcomes (1). A number of studies have focused on the
molecular and cellular changes that occur during the initiation and
progression of lung cancer. However, the gene polymorphisms
involved, which are closely associated with the tumorigenesis and
invasion of lung cancer, remain unclear (2–4).
Therefore, it is essential to investigate effective lung cancer
biomarkers at the early stages of disease and identify novel
targets for the treatment of the disease.
The receptor for advanced glycation end products
(RAGE) is a multi-ligand transmembrane receptor that belongs to the
immunoglobulin superfamily. The extracellular domain of RAGE is
termed soluble RAGE (sRAGE) and acts as an endogenous inhibitor of
RAGE by binding circulating ligands and inhibiting RAGE-induced
cellular signaling, tissue damage and dysfunction (5). RAGE is a pattern-recognition receptor
that binds a variety of ligands, including the S100/calgranulin
family, high mobility group box-1 (HMGB-1), cluster of
differentiation (CD)11b, amyloid-α peptide and β-sheet fibrils.
RAGE is involved in several pathological processes, including those
in diabetes, Alzheimer's disease and systemic amyloidosis (5).
In contrast to healthy adult tissues in the body, in
which RAGE and sRAGE are expressed at a low level, the expression
levels of endogenous RAGE and sRAGE have been reported to be high
in normal adult lung tissue (6).
However, the expression of RAGE and sRAGE has been identified as
downregulated in non-small cell lung carcinoma (NSCLC) (7) and idiopathic pulmonary fibrosis
(8). This reveals that RAGE is
involved in lung pathogenesis.
Previous studies have indicated that RAGE expression
is also closely associated with the invasive and metastatic
activity of cancer, including gastric (9) and colon cancer (10). Upregulation of RAGE has been
identified in breast (11), colon and
pancreatic cancers (12), but
downregulation of the expression of RAGE and sRAGE has been
reported in lung and esophageal cancer (12,13).
Studies investigating RAGE polymorphism have
demonstrated that the genetic background of RAGE is associated with
NSCLC (14,15). Schenk et al identified six
novel sequence variants of RAGE in primary NSCLC lesions compared
with the corresponding normal tissues, but no somatic
tumor-associated mutations were identified in the sequence analysis
(14). Therefore, additional
investigation of the expression of RAGE and polymorphism of the
RAGE gene in various stages of lung cancer is required.
In the present study, the expression levels of
sRAGE, RAGE and the HMGB1 and S100 RAGE ligands were measured in
serum and tissue obtained from patients with small cell lung cancer
(SCLC) and NSCLC. The presence of several types of RAGE
polymorphism was assessed in these lung cancer tissue samples, and
the current study aimed to identify the association between various
types of RAGE gene polymorphism and lung cancer. Furthermore, a
comparison was made between RAGE expression and the presence of
polymorphisms and variables comprising gender variance and various
histological subtypes and stages of lung cancer. The aim of the
present study was to identify aspects of the biological role of the
RAGE gene and to determine a biomarker for the diagnosis of lung
cancer and evaluation of the prognosis of a patient.
Patients and methods
Patients and samples
Lung cancer patients and healthy volunteers were
recruited from Qingdao Municipal Hospital and The Affiliated
Hospital of Qingdao University Medical School (Qingdao, Shandong,
China), between January 2010 and December 2012. All subjects were
aged between 30 and 75 years old and did not possess a current
diagnosis of high blood pressure, diabetes mellitus, liver or
kidney dysfunction, or Alzheimer's disease (AD). Venous blood
samples were collected from patients by venipuncture subsequent to
a 12-h fasting period, and the samples were drawn into sterile
vacutainers (Hunan Liuyang Medical Instrument Factory, Liuyang,
China) that either contained 1% heparin as an anticoagulant for
blood cell isolation or lacked anticoagulant for serum isolation.
The lung tissue specimens used for immunohistochemical (IHC)
staining were collected from tumor and paired non-tumor tissues
obtained from a target number of ≥20 patients diagnosed with each
type of lung cancer at various stages. The tissues were excised
during pulmonary resection. In total, 5 ml of venous blood was also
collected from each patient during a lung biopsy procedure for the
measurement of sRAGE expression.
For the polymorphism study, 275 lung cancer patients
and 126 healthy volunteers were recruited in total. The lung cancer
patients comprised 113 patients with adenocarcinoma, 99 patients
with squamous cell carcinoma and 63 patients with small cell
carcinoma (Table I). The patients
diagnosed with adenocarcinoma and squamous cell carcinoma were
grouped according to the guidelines for tumor-node-metastasis (TNM)
stages I–IV, based on the size of the primary tumor, lymphatic
nodal involvement and distance between the metastasis and primary
tumor (16). Patients possessing
tumors classified as stage I, II or IIIA were classified as
early/middle stage, while patients possessing tumors classified as
stages IIIB and IV were classified as late stage lesions.
 | Table I.Clinical characteristics of lung
cancer patients compared with healthy control individuals. |
Table I.
Clinical characteristics of lung
cancer patients compared with healthy control individuals.
|
| Group, n (%) |
|---|
|
|---|
| Patient
characteristics | Lung cancer | Healthy control |
|---|
| Total, n (%) | 275 (100.00) | 126 (100.00) |
| Male | 170 (61.82) | 84 (66.67) |
|
Female | 105 (38.18) | 42 (33.33) |
| Age, years ± SD | 59.8±10.4 | 57.1±11.2 |
| Adenocarcinoma, n
(%) |
|
|
|
Total | 113 (100.00) |
|
|
Stage |
|
|
|
Early/middle | 47 (41.59) |
|
|
Late | 66 (58.41) |
|
|
Gender |
|
|
|
Male | 59 (52.21) |
|
|
Female | 54 (47.79) |
|
| Squamous cell
carcinoma, n (%) |
|
|
|
Total | 99 (100.00) |
|
|
Stage |
|
|
|
Early/middle | 48 (48.48) |
|
|
Late | 51 (51.52) |
|
|
Gender |
|
|
|
Male | 70 (70.71) |
|
|
Female | 29 (29.29) |
|
| Small cell carcinoma,
n (%) |
|
|
|
Total | 63 (100.00) |
|
| Male | 41 (65.08) |
|
|
Female | 22 (34.92) |
|
The present study was conducted in accordance with
the Declaration of Helsinki (17),
and was performed with approval from the Ethics Committee of the
Affiliated Hospital of Qingdao University. Written informed consent
was obtained from all participants.
Cytokine measurement
To obtain serum samples, the blood samples were left
undisturbed to clot at room temperature for 15–30 min. The clot was
removed by centrifugation at 1,000–2,000 × g for 10 min, at 4°C.
The serum was separated and stored at −80°C for subsequent testing
of cytokine production. sRAGE production was assayed using an ELISA
kit (R&D Systems, Minneapolis, MN, USA) according to the
manufacturer's instructions.
IHC analysis
Lung tissue specimens were obtained from resected
tumor tissues and adjacent non-tumor tissues excised during the
pulmonary resection procedure. The lung tissues were fixed in 4%
formalin, embedded in paraffin, sliced and placed on slides prior
to staining. The slides were deparaffinized with xylene and graded
ethanol solutions. All slides were quenched in hydrogen peroxide,
blocked with 10% normal rabbit serum and digested with proteinase
K. The slides were stained with polyclonal rabbit anti-human RAGE
(cat. no. ab65965), monoclonal mouse anti-pig HMGB1 (cat. no.
ab11354) or monoclonal mouse anti-human S100 A (cat. no. ab52272)
antibodies, which were purchased from Abcam (Cambridge, MA, USA)
and then detected using the Universal Vectastain Elite ABC kit,
anti-mouse IgG/rabbit IgG kit (cat. no. PK-6200; Vector
Laboratories, Burlingame, CA, USA). Five images were captured from
each slide using an Olympus IX50 inverted microscope (original
magnification, x100; Olympus, Tokyo, Japan). The slides were
quantified by histogram analysis using Adobe Photoshop CS (Adobe
Systems, Mountain View, CA, USA), as previously described (18,19). The
data were expressed as the percentage of positive cells per
high-power field (HP) in 10 images and were expressed as the mean ±
standard error of the mean (SEM).
RAGE polymorphism analysis
Blood was collected from donors and DNA was isolated
using a Genomic DNA Isolation kit (Jiamei Biotech Co., Ltd.) for
whole blood according to the manufacturer's instructions. Four
polymorphisms of the RAGE gene, comprising −429 T/C, −374 T/A,
Gly82Ser and 2184A/G, were tested using polymerase chain reaction
(PCR) and the restriction digestion method (20,21).
Enzyme digestion products were separated on 3% agarose gels. The
PCR primer sequences, annealing temperatures, size of PCR
fragments, restriction enzymes and expected size of the fragments
from various genotypes of RAGE polymorphism are listed in Table II.
 | Table II.Polymerase chain reaction conditions
and expected products for various polymorphisms of the RAGE
gene. |
Table II.
Polymerase chain reaction conditions
and expected products for various polymorphisms of the RAGE
gene.
| Polymorphism | Primer squence | Tm, °C | Product length,
bp | Enzyme | Expected band size,
bp |
|---|
| Gly82Ser | F,
5′-GTAAGCGGGGCTCCTGTTGCA-3′ | 63.0 | 397 | AluI | GGa: 248, 149 |
|
| R,
5′-GGCCAAGGCTGGGGTTGAAGG-3′ |
|
|
| SSb: 181, 67, 149 |
|
|
|
|
|
| GSb: 248, 149, 181, 67 |
| −374T/A | F,
5′-GGGGCAGTTCTCTCCTCACT-3′ | 59.5 | 250 | MfeI
(MunI) | TTa: 215, 35 |
|
| R,
5′-GGTTCAGGCCAGACTGTTGT-3′ |
|
|
| AAb: 250 |
|
|
|
|
|
| TAb: 250, 215, 35 |
| −429T/C | F,
5′-GGGGCAGTTCTCTCCTCACT-3′ | 59.5 | 250 | AluI | TTa: 250 |
|
| R,
5′-GGTTCAGGCCAGACTGTTGT-3′ |
|
|
| CCb: 162, 88 |
|
|
|
|
|
| TCb: 250, 162, 188 |
| 2184A/G | F,
5′-GGGGCAGTTCTCTCCTCACT-3′ | 59.5 | 402 | BfaI | AAa: 266, 136 |
|
| R,
5′-GGTTCAGGCCAGACTGTTGT-3′ |
|
|
| GGb: 174, 92, 136 |
|
|
|
|
|
| AGb: 266, 136, 174, 92 |
Statistical analysis
The results are expressed as the mean ± SEM. The
number of independent donors in each experiment is indicated in the
figure legend. Statistical analysis of IHC staining and cytokine
analysis was performed by independent sample t-tests, which
were conducted using SPSS software, version 17 (SPSS, Chicago, IL,
USA). For the analysis of the polymorphisms of RAGE, the incidence
of the wild-type, minor homozygous and heterozygous alleles were
expressed as a percentage of the total number of patient samples
analyzed. Differences between the allelic and genotypic frequencies
observed in the control group and those identified in lung cancer
patients were assessed for significance using Fisher's exact test
or χ2 test. P≤0.05 was considered to indicate a
statistically significant difference. Statistical analysis was
performed using the StatView software (Apple, Inc., Cupertino, CA,
USA).
Results
Clinical characteristics of the
patients with lung cancer
The clinical characteristics of the lung cancer
patients enrolled for the assessment of polymorphisms, including
the age, gender and smoking history, were obtained from medical
records and are presented in Table I.
For the assessment of polymorphisms, age-matched lung cancer
patients (59.8±10.4 years old) and control donors (57.1±11.2 years
old) were recruited. The percentage of males and females was also
maintained between the lung cancer and control groups. The lung
cancer patients consisted of 170 male and 105 female patients.
Overall, 51.2% of male patients with lung cancer were cigarette
smokers and 28.6% of female patients were cigarette smokers. By
contrast, in the healthy control group, the percentage of smokers
was decreased compared with the lung cancer group, as 27.4% of
males were smokers and 16.7% of females were smokers. The present
study defined smokers as active smokers and patients that had
completely discontinued smoking for <15 years. The incidence of
passive smoking was not considered.
Lung cancer patients were classified based on the
clinical pathological diagnosis and allocated to the
adenocarcinoma, squamous cell carcinoma, including adenosquamous
cell carcinoma, and small cell carcinoma groups. Patients with
adenocarcinoma and squamous cell carcinoma were sub-classified into
the early/middle and late stages of disease, as aforementioned. Out
of the 113 adenocarcinoma patients, 39.8% of patients demonstrated
early/middle-stage and 60.2% demonstrated late-stage disease. Out
of the 99 patients with squamous cell carcinoma, 48.3% of patients
demonstrated early/middle-stage and 51.7% demonstrated late-stage
disease.
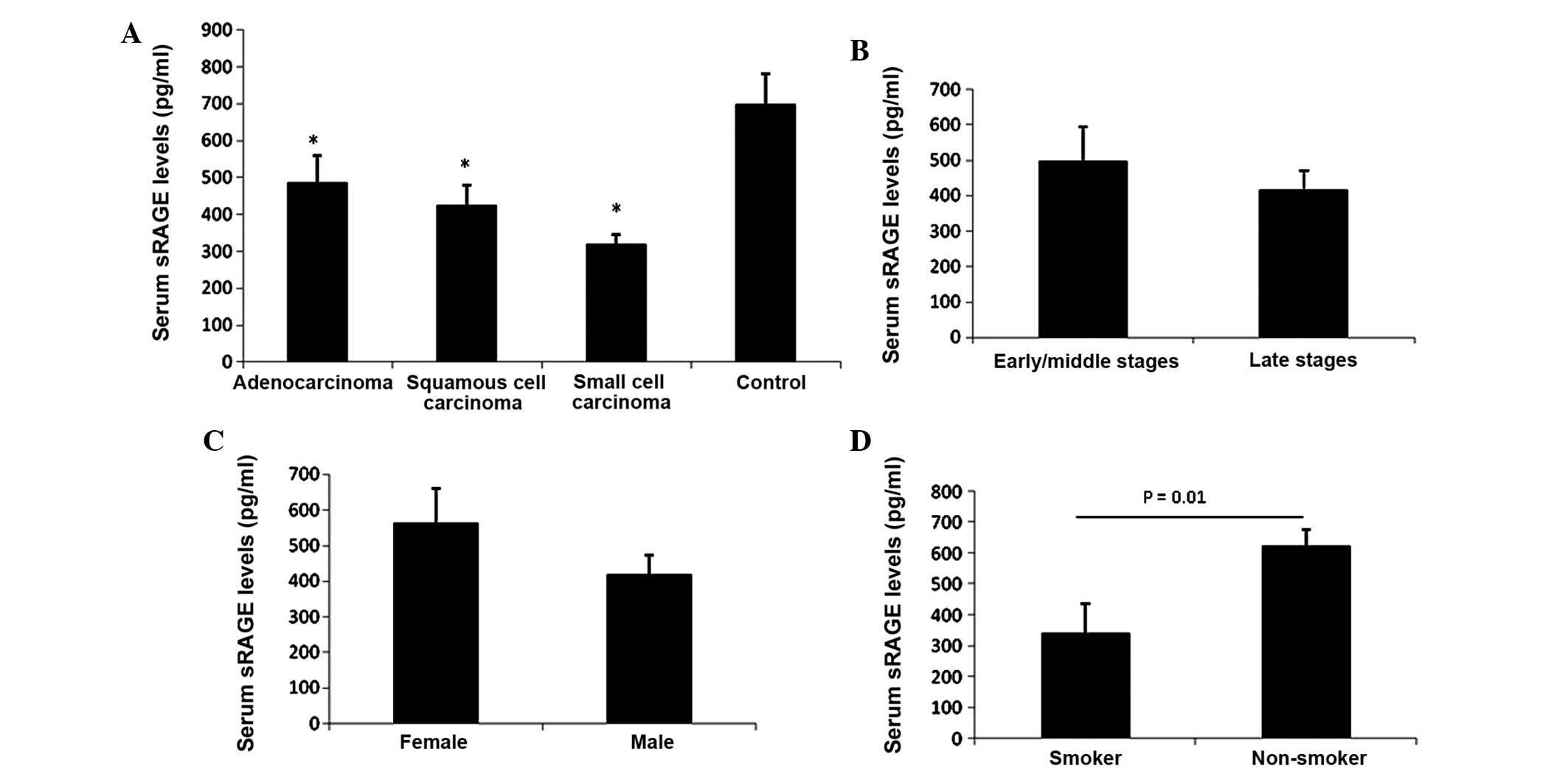
Serum sRAGE level decreased in all
tested lung cancer patients
It has been reported that the serum sRAGE level can
be used as a predictive biomarker for diabetes (22), sepsis (23) and acute lung injury (24). Therefore, the sRAGE level in the serum
of lung cancer patients was examined using ELISA. The data revealed
that the serum sRAGE level was significantly decreased in
adenocarcinoma (485.6±73.1 pg/ml), squamous cell carcinoma
(424.5±55.2 pg/ml) and small cell carcinoma patients (318.9±25.9
pg/ml) compared with the healthy control group (697.6±83.2 pg/ml)
(Fig. 1A). However, detailed analysis
did not reveal a stage-dependent change in serum sRAGE level, as no
significant change between late stage cancer and early/middle stage
cancer patients was identified (Fig.
1B). Notably, a significant decrease in the sRAGE level was
identified in the serum of smokers (338.9±22.9 pg/ml) compared with
non-smokers (623.4±92 pg/ml) in the lung cancer group (Fig. 1C). Furthermore, a comparison was made
between the expression of sRAGE in male (339.4±36.3 pg/ml) and
female patients (564.6±89.1 pg/ml), and it was revealed that male
patients possessed a decreased sRAGE level compared with female
patients (Fig. 1D). This may possibly
be due to 51.2% of male patients being smokers, while only 28.6% of
female patients were smokers.
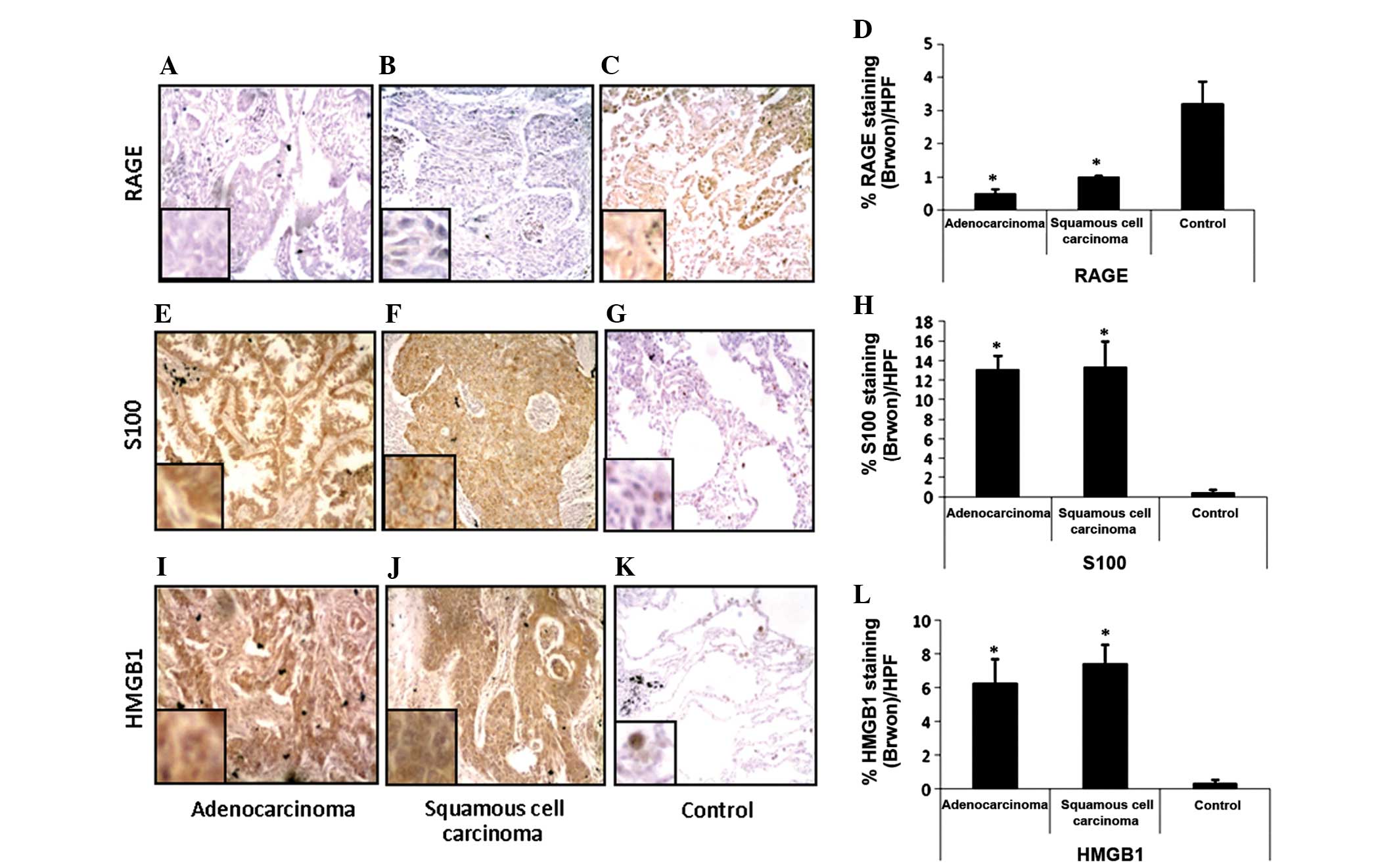
Expression of RAGE decreased in lung
cancer tissues, while the expression of the RAGE ligands
increased
Since the serum level of sRAGE was reduced
significantly in all lung cancer patients, RAGE expression was then
examined in the lung tissue samples using an IHC method. As
expected, RAGE expression was markedly decreased in adenocarcinoma
(P<0.05) and squamous cell carcinoma tissue samples (P<0.05)
compared with the non-cancerous tissue obtained from the same
patient (Fig. 2A–D). However, there
was no significant difference in RAGE expression between
adenocarcinoma and squamous cell carcinoma tissues (Fig. 2A, B and D), which indicates that the
downregulation of RAGE expression is not dependent on lung cancer
types. This finding was consistent with Bartling's finding that
RAGE expression was reduced in lung cancer patients at the mRNA and
protein level (7).
Subsequently, the expression of the RAGE ligands
HMGB-1 and S100 was examined in lung cancer tissues and the
presence of a correlation between the expression of RAGE and the
ligands was assessed. As expected, the expression of S100 (Fig. 2G) and HMGB-1 (Fig. 2G) in normal lung tissue was revealed
to be extremely low and almost undetectable. However, in
adenocarcinoma (Fig. 2E and I) and
squamous cell carcinoma (Fig. 2F and
G) tissues, the expression of S100 and HMGB-1 was markedly
upregulated and there was a significant difference between the
expression of the two RAGE ligands in cancerous and normal lung
tissue samples (Fig. 2H and L).
Gly82Ser and −374T/A RAGE
polymorphisms were not associated with lung cancer, but the −429T/C
and 2184A/G RAGE polymorphisms were associated with lung
cancer
In the present study, the occurrence of four
different types of RAGE gene polymorphisms was assessed in lung
cancer patients and healthy control individuals, with the
investigated polymorphisms consisting of Gly82Ser, −374T/A, −429T/C
and 2184A/G. For each polymorphism, the percentage frequency of the
wild-type and minor alleles, homozygous type, and minor and
heterozygous alleles of the polymorphisms were calculated for the
lung cancer and control groups. The frequency of these alleles in
samples obtained from various stages of adenocarcinoma and squamous
cell carcinoma were also calculated. Statistical analysis of the
differences between the stages of small cell carcinoma was not
performed due to the low number of patient samples.
Statistical analysis of the differences between the
allelic and genotype frequencies of the Gly82Ser (Table III) and −374T/A (Table IV) polymorphisms of RAGE did not
reveal any variation in distribution between the patients with lung
cancer and healthy control individuals. Detailed analysis did not
reveal any statistical difference in the occurrence of these
polymorphisms between the early/middle stage and late stage cancer
patients, indicating that the Gly82Ser and −374T/A polymorphisms of
the RAGE gene were not associated with lung cancers in the present
study. Additionally, the Gly82Ser and −374T/A polymorphisms were
not associated with the stage of lung cancer.
 | Table III.Frequency of the alleles of the
Gly82Ser polymorphism of the RAGE gene in lung cancer patients and
healthy controls. |
Table III.
Frequency of the alleles of the
Gly82Ser polymorphism of the RAGE gene in lung cancer patients and
healthy controls.
| Patient
characteristics | GG, % | GS, % | SS, % | G, % | S, % |
|---|
| Adenocarcinoma | 64.83 | 30.77 | 4.40 | 80.22 | 19.78 |
|
Early/middle stage | 52.72 | 41.67 | 5.56 | 73.61 | 26.39 |
| Late
stage | 72.72 | 23.64 | 3.64 | 84.55 | 15.45 |
| Squamous cell
carcinoma | 78.18 | 20.00 | 1.82 | 88.18 | 11.82 |
|
Early/middle stage | 81.48 | 18.52 | 0.00 | 90.74 |
9.26 |
| Late
stage | 75.00 | 21.43 | 3.57 | 85.71 | 14.29 |
| Small cell
carcinoma | 76.92 | 23.08 | 0.00 | 88.46 | 11.54 |
| Healthy
controls | 67.53 | 28.57 | 3.90 | 81.82 | 18.18 |
 | Table IV.Frequency of the alleles of the
−374T/A polymorphisms of the RAGE gene in lung cancer patients and
healthy controls. |
Table IV.
Frequency of the alleles of the
−374T/A polymorphisms of the RAGE gene in lung cancer patients and
healthy controls.
| Patient
characteristics | TT, % | TA, % | AA, % | T, % | A, % |
|---|
| Adenocarcinoma | 71.59 | 23.86 | 4.55 | 83.52 | 16.48 |
|
Early/middle stage | 77.14 | 17.14 | 5.71 | 85.71 | 14.29 |
| Late
stage | 67.92 | 28.30 | 3.77 | 82.08 | 17.92 |
| Squamous cell
carcinoma | 63.79 | 31.03 | 5.17 | 79.31 | 20.69 |
|
Early/middle stage | 60.71 | 35.71 | 3.57 | 78.57 | 21.43 |
| Late
stage | 65.52 | 27.59 | 6.90 | 79.31 | 20.69 |
| Small cell
carcinoma | 53.85 | 38.46 | 7.69 | 73.08 | 26.92 |
| Healthy
controls | 72.73 | 24.68 | 2.60 | 85.06 | 14.94 |
In the case of the −429T/C polymorphism (Table V), the present results revealed that
out of the patients with squamous cell carcinoma, 79.66% possessed
the T-allele, 20.34% possessed the C-allele, 62.71% possessed the
TT allele and 3.39% possessed the CC allele. In the control group,
89.61% possessed the T-allele, 15.05% possessed the C-allele,
79.22% possessed the TT allele and 0% possessed the CC allele. The
frequency of the TT genotype and T allele was significant lower in
patients with squamous cell carcinoma compared with the healthy
control group (P=0.047). A detailed analysis of patients with
squamous cell carcinoma at various stages revealed that there was
no significant difference between the occurrence of polymorphism in
the early/middle stage and that in the healthy controls. However,
there was a notable increase in the incidence of polymorphism in
the late stage of squamous cell carcinoma, where the T- and
C-alleles were present in 70.97 and 29.03% of patients,
respectively, and the TT, TC and CC variants were present in 48.39,
45.16 and 6.45% of patients, respectively, which was a significant
increase compared with the healthy controls (P=0.05). In the
patients with adenocarcinoma, no significant difference in the
occurrence of the −479T/C polymorphism was observed between the
lung cancer patients and healthy controls, but the patients with
late-stage adenocarcinoma demonstrated a significantly decreased
genotypic frequency of the wild-type TT allele (57.14%) and
increased genotypic frequency of the minor type CC allele (41.07%)
compared with patients in the early/middle stage of disease, who
demonstrated an incidence of 89.47 and 10.53% for the TT and CC
alleles, respectively (P=0.001).
 | Table V.Frequency of the alleles of the
−429T/C polymorphisms of the RAGE gene in lung cancer patients and
healthy controls. |
Table V.
Frequency of the alleles of the
−429T/C polymorphisms of the RAGE gene in lung cancer patients and
healthy controls.
| Patient
characteristics | TT, % | TC, % | CC, % | T, % | C, % |
|---|
| Adenocarcinoma | 70.97 | 27.96 | 1.08 | 84.95 | 15.05 |
|
Early/middle
stagea | 89.47 | 10.53 | 0.00 | 94.74 |
5.26 |
| Late
stage | 57.14 | 41.07 | 1.79 | 77.68 | 22.32 |
| Squamous cell
carcinomab | 62.71 | 33.90 | 3.39 | 79.66 | 20.34 |
|
Early/middle
stagec | 78.57 | 21.43 | 0.00 | 89.29 | 10.71 |
| Late
stage | 48.39 | 45.16 | 6.45 | 70.97 | 29.03 |
| Small cell
carcinoma | 84.62 | 15.38 | 0.00 | 92.31 |
7.69 |
| Healthy
controls | 79.22 | 20.78 | 0.00 | 89.61 | 10.39 |
Notably, the most significant association between
the RAGE polymorphism and lung cancer was in the occurrence of the
2184A/G polymorphism (Table VI). In
the healthy control population, only the wild-type A allele was
identified, but analysis of all three types of lung cancer revealed
the appearance of the minor G-allele. The G-allele was present in
6.98% of adenocarcinoma patients, 6.12% of squamous cell carcinoma
patients and 7.69% of small cell carcinoma patients. The present
data revealed a statistical difference between the occurrence of
this allele in all cancer groups and the healthy control group
(P<0.05). Additionally, no patients were identified as
homozygous for the minor GG allele in the present study. In the
comparisons of genotypic frequencies between various stages of lung
cancer, it was found that the AG variant was present in 13.95% of
adenocarcinoma patients, while the AG variant was present in 3.23%
of early/middle stage patients and 20.37% in late stage patients,
demonstrating a significant increase between the two groups
(P=0.048). The AG variant was present in 12.24% of squamous cell
carcinoma patients, and the genotypic frequency of AG increased
between the early/middle stage incidence of 8% and the late stage
incidence of 16.67% (P=0.05). In small cell carcinoma patients, the
AG variant was determined to be present in 15.38% of patients.
Overall, the 2184A/G polymorphism was the only RAGE polymorphism in
the present study that demonstrated the ability to distinguish
between the small cell carcinoma patients and the healthy control
individuals.
 | Table VI.Frequency of the alleles of the
2184A/G polymorphism of the RAGE gene in lung cancer patients and
healthy controls. |
Table VI.
Frequency of the alleles of the
2184A/G polymorphism of the RAGE gene in lung cancer patients and
healthy controls.
| Patient
characteristics | AA, % | AG, % | GG, % | A, % | G, % |
|---|
|
Adenocarcinomaa | 86.05 | 13.95 | 0.00 | 93.02 | 6.98 |
|
Early/middle
stageb | 96.77 | 3.23 | 0.00 | 98.39 | 1.61 |
| Late
stage | 79.63 | 20.37 | 0.00 | 89.81 | 10.19 |
| Squamous cell
carcinomac | 87.76 | 12.24 | 0.00 | 93.88 | 6.12 |
|
Early/middle stage | 92.00 | 8.00 | 0.00 | 96.00 | 4.00 |
| Late
stage | 83.33 | 16.67 | 0.00 | 91.67 | 8.33 |
| Small cell
carcinomad | 84.62 | 15.38 | 0.00 | 92.31 | 7.69 |
| Healthy
controls | 100.00 | 0.00 | 0.00 | 100.00 | 0.00 |
Incidence of the −429T/C and 2184A/G
polymorphisms of RAGE was significantly different between male and
female lung cancer patients
Subsequent to the allelic frequency of the four
types of polymorphism of the RAGE gene being elucidated in patients
with adenocarcinoma, squamous cell carcinoma and small cell
carcinoma, a clarification of whether gender played a role in RAGE
gene polymorphism in various cancers was attempted. Table VII reports the allelic frequency of
the Gly82Ser polymorphism of the RAGE gene in male and female lung
cancer patients, which did not demonstrate any statistically
significant difference between the genders in all three types of
cancer. Similarly, no difference was observed in the incidence of
the −374T/A polymorphism between males and females with
adenocarcinoma or squamous cell carcinoma (Table VIII). However, a significant change
was observed in the incidence of the −374T/A polymorphism between
males and females with small cell carcinoma (P<0.05), as 100% of
female patients expressed the heterozygous TA allele, while in male
patients, 50% expressed the TT allele, 30% expressed the TA allele
and 20% expressed the AA allele. In female patients, 50% were
identified as possessing the T allele and 50% possessed the A
allele, while in male patients, 65% possessed the T allele and 35%
possessed the A allele.
 | Table VII.Frequency of the alleles of the
Gly82Ser polymorphism of the RAGE gene in male and female lung
cancer patients. |
Table VII.
Frequency of the alleles of the
Gly82Ser polymorphism of the RAGE gene in male and female lung
cancer patients.
| Gender | GG, % | GS, % | SS, % | G, % | S, % |
|---|
| Adenocarcinoma | 66.32 | 29.47 | 4.21 | 81.05 | 18.95 |
|
Male | 63.83 | 34.04 | 2.13 | 80.85 | 19.15 |
|
Female | 66.67 | 27.08 | 6.25 | 80.21 | 19.79 |
| Squamous cell
carcinoma | 78.57 | 19.64 | 1.79 | 88.39 | 11.61 |
|
Male | 78.85 | 19.23 | 1.92 | 88.46 | 11.54 |
|
Female | 75.00 | 25.00 | 0.00 | 87.50 | 12.50 |
| Small cell
carcinoma | 76.92 | 23.08 | 0.00 | 88.46 | 11.54 |
|
Male | 77.78 | 22.22 | 0.00 | 88.89 | 11.11 |
|
Female | 75.00 | 25.00 | 0.00 | 87.50 | 12.50 |
 | Table VIII.Frequency of the alleles of the
−374T/A polymorphism of the RAGE gene in male and female lung
cancer patients. |
Table VIII.
Frequency of the alleles of the
−374T/A polymorphism of the RAGE gene in male and female lung
cancer patients.
| Gender | TT, % | TA, % | AA, % | T, % | A, % |
|---|
| Adenocarcinoma | 23.86 | 71.59 | 4.55 | 59.66 | 40.34 |
|
Male | 22.73 | 70.45 | 6.82 | 57.95 | 42.05 |
|
Female | 25.00 | 72.73 | 2.27 | 61.36 | 38.64 |
| Squamous cell
carcinoma | 31.03 | 63.79 | 5.17 | 62.93 | 37.07 |
|
Male | 32.08 | 64.15 | 3.77 | 64.15 | 35.85 |
|
Female | 25.00 | 75.00 | 0.00 | 62.50 | 37.50 |
| Small cell
carcinoma | 38.46 | 53.85 | 7.69 | 65.38 | 34.62 |
|
Malea | 50.00 | 30.00 | 20.00 | 65.00 | 35.00 |
|
Female | 0.00 | 100.00 | 0.00 | 50.00 | 50.00 |
Notably, the 429T/C (Table IX) and 2184A/G (Table X) polymorphisms exhibited a
significant difference between the distribution of the
polymorphisms in male and female patients in all three types of
lung cancer (P<0.05). It was observed that in adenocarcinoma,
the male patients demonstrated a decreased frequency of the
wild-type TT allele of the −429T/C polymorphism (61.70%) and a
decreased incidence of the wild-type AA allele of the 2184A/G
polymorphism (81.40%) compared with the female patients (80.43 and
90.70%, respectively). In small cell carcinoma, male patients
possessed an increased frequency of the wild-type TT allele of the
−429T/C polymorphism (100.00%) and an increased frequency of the
wild-type AA allele of the 2184A/G polymorphism (100.00%) compared
with the female patients (50.00 and 50.00%, respectively). By
contrast, in squamous cell carcinoma, the male patients
demonstrated an increased frequency of the wild-type TT allele of
the −429T/C polymorphism (63.16%) and a decreased frequency of the
wild-type AA allele of the 2184A/G polymorphism (81.25%) compared
with the female patients (50.00 and 100.00%, respectively).
 | Table IX.Frequency of the alleles of the
−429T/C polymorphism of the RAGE gene in male and female lung
cancer patients. |
Table IX.
Frequency of the alleles of the
−429T/C polymorphism of the RAGE gene in male and female lung
cancer patients.
| Gender | TT, % | TC, % | CC, % | T, % | C, % |
|---|
| Adenocarcinoma | 70.97 | 27.96 | 1.08 | 86.00 | 14.00 |
|
Malea | 61.70 | 36.17 | 2.13 | 90.50 | 9.50 |
|
Female | 80.43 | 19.57 | 0.00 | 95.50 | 4.50 |
| Squamous cell
carcinoma | 62.71 | 33.90 | 3.39 | 88.00 | 12.00 |
|
Malea | 63.16 | 33.33 | 3.51 | 88.50 | 11.50 |
|
Female | 50.00 | 50.00 | 0.00 | 99.50 | 0.50 |
| Small cell
carcinoma | 84.62 | 15.38 | 0.00 | 99.00 | 1.00 |
|
Malea | 100.00 | 0.00 | 0.00 | 100.00 | 0.00 |
|
Female | 50.00 | 50.00 | 0.00 | 99.00 | 1.00 |
 | Table X.Frequency of the alleles of the
2184A/G polymorphism of the RAGE gene among male and female lung
cancer patients. |
Table X.
Frequency of the alleles of the
2184A/G polymorphism of the RAGE gene among male and female lung
cancer patients.
| Gender | AA, % | AG, % | GG, % | A, % | G, % |
|---|
| Adenocarcinoma | 86.05 | 13.95 | 0.00 | 93.02 | 6.98 |
|
Malea | 81.40 | 18.60 | 0.00 | 90.70 | 9.30 |
|
Female | 90.70 | 9.30 | 0.00 | 95.35 | 4.65 |
| Squamous cell
carcinoma | 87.76 | 12.24 | 0.00 | 93.88 | 6.12 |
|
Malea | 81.25 | 18.75 | 0.00 | 90.63 | 9.38 |
|
Female | 100.00 | 0.00 | 0.00 | 100.00 | 0.00 |
| Small cell
carcinoma | 84.62 | 15.38 | 0.00 | 92.31 | 7.69 |
|
Malea | 100.00 | 0.00 | 0.00 | 100.00 | 0.00 |
|
Female | 50.00 | 50.00 | 0.00 | 75.00 | 25.00 |
Discussion
Gene profiling in tissues derived from lung
carcinomas has previously revealed large numbers of differentially
expressed genes in these tissues compared with the histologically
normal tissues obtained from the same individual or tissue obtained
from healthy individuals (25). In
the present study, it was demonstrated that the multi-ligand
receptor RAGE was one of the genes that is differentially expressed
in lung cancer patients. Initially, the serum sRAGE level was found
to be decreased in NSCLC and SCLC patients. Subsequently, it was
demonstrated that the expression of RAGE was decreased in patients
with lung cancer while the expression of the RAGE ligands HMGB1 and
S100 were increased in lung cancer tissue compared with
non-cancerous tissue excised from NSCLC patients. In addition, the
occurrence of four types of polymorphism of the RAGE gene,
consisting of Gly82Ser, −374T/A, −429T/C and 2184A/G, was measured
in the patients with lung cancer and the healthy control
individuals. It was demonstrated that the Gly82Ser and −374T/A RAGE
polymorphisms were not associated with lung cancer, while the
−429T/C and 2184A/G polymorphisms were associated with lung
cancer.
The mechanism behind the association between sRAGE
and disease remains controversial. One study has reported that
sRAGE was elevated in patients with sepsis and was associated with
patient outcome (23). Another study
demonstrated that the sRAGE level was elevated during acute lung
injury or acute respiratory distress syndrome, regardless of the
presence or absence of severe sepsis (26). It has also been revealed that the
level of serum sRAGE was elevated in breast cancer patients
(27), and the elevated serum sRAGE
levels were reported to be associated with an increased risk of
cardiovascular disease in Japanese patients with type 2 diabetes
(28). In the present study, it was
revealed that the serum sRAGE level was significantly decreased in
NSCLC and SCLC patients compared with healthy controls. In
addition, a notable decrease in the level of serum sRAGE was
identified in smokers compared with non-smokers. It is well-known
that cigarette smoking is a major cause of lung cancer, and the
decrease in the expression of sRAGE in lung cancer patients and
smokers provides a simple supportive tool for predicting the
development of lung cancer. Therefore, the serum sRAGE level may be
used as potential biomarker for the occurrence of lung cancer.
The biological function of RAGE and its ligands in
cancer also remain unclear. Increased RAGE activation has been
observed in a variety of pathological disorders. In certain
cancers, the expression of RAGE and its ligands is highly
upregulated and in others the expression is downregulated. For
example, in pancreatic and prostate cancer, enhanced expression of
RAGE and HMGB1 were found to be associated with metastases
(12). In colon cancer, joint
expression of RAGE and HMGB1 was found to lead to enhanced
migration in the cancer cell line, and the level of expression of
RAGE and HMGB1 increased with the stage of cancer (12). By contrast, in lung cancer, a higher
tumor stage was reported to be characterized by a downregulation in
RAGE expression at the mRNA and protein levels (7). The present study confirmed that the
expression of RAGE was downregulated in lung cancer tissue compared
with noncancerous tissue from the same patient. The downregulation
of sRAGE and RAGE was found to not be dependent on the type or
stage of lung cancer. Investigation of the RAGE binding ligands
revealed the upregulation of the RAGE ligands S100 and HMGB1 in
cancer tissues. It is possible that in normal tissues, RAGE binds
and leads to the degradation of the ligands, but in lung cancer
tissue the downregulation of RAGE results in more ligands being
present in the tissue.
It has been revealed that S100 calcium binding
protein P (S100P) was expressed at greater levels in colon cancer
compared with matched normal tissue, and S100P has also been
demonstrated to stimulate colon cancer cell growth and migration,
extracellular signal-regulated kinase phosphorylation, and nuclear
factor-κB activation in vitro. However, in contrast to the
expression in lung cancers, RAGE was also upregulated in colon
cancer tissues (10). As a result, it
was challenging to apply the simple ligand/receptor binding and
degradation theory as an explanation for the phenomena of RAGE
expression. Notably, RAGE is a receptor that is capable of binding
multiple ligands in all cell types, and thus RAGE and the RAGE
ligands HMGB1 and S100 are able to affect each other. These ligands
also bind other receptors in addition to RAGE. Thus, RAGE and the
HMGB1 and S100 ligands are able to function independently of each
other.
In the present study, the incidence of four types of
RAGE gene polymorphism was assessed. It was revealed that the
Gly82Ser and −374T/A RAGE polymorphisms were not associated with
lung cancer, while the −429T/C polymorphism was associated with
squamous cell carcinoma and the 2184A/G RAGE polymorphism was
associated with all three types of lung cancer. Notably, the
healthy control individuals demonstrated an increased incidence of
the wild-type TT allele of the −429T/C polymorphism and all
individuals possessed the wild-type AA allele of the 2184A/G
polymorphism. Patients in a late stage of disease demonstrated a
reduced frequency of the wild-type allele and an increased
frequency of the minor allele. Consistent with the present results,
a study investigating RAGE polymorphism and breast cancer performed
by Tesarová et al found no difference in the incidence of
the −374T/A polymorphism between breast cancer patients and healthy
controls (13). It was reported that
the −374T/A polymorphism demonstrated no effect on the sRAGE level.
However, it was also observed that the serum sRAGE level was higher
in patients that possessed the wild-type TT variant instead of the
−429 T/C polymorphism, wild-type GG variant instead of the
Gly82/Ser polymorphism, and wild-type AA variant instead of the
2184A/G polymorphism compared with patients that possessed the
minor allele (13). However, a study
by Wang et al revealed that the presence of the homozygous
minor-type SS variant at the location of the Gly82Ser polymorphism
resulted in a lower response to chemotherapy and a poorer clinical
outcome in the advanced stages of NSCLC (15).
Notably, the −429T/C polymorphism of RAGE was
associated with squamous cell carcinoma and the 2184A/G
polymorphism was associated with the development of NSCLC and SCLC.
These variations in genotype cannot be used to explain the common
transcriptional downregulation of RAGE in lung cancer patients, as
not all lung cancer patients possessed the minor allele of the RAGE
gene.
In summary, the present findings reveal that the
expression of RAGE was reduced in tissues from human lung cancer
patients, and that the polymorphisms of RAGE, in particular the
−429T/C and 2184A/G polymorphisms, were associated with the genesis
and progression of lung cancer. The levels of serum sRAGE and
tissue RAGE may serve as an effective and convenient diagnostic
biomarker for lung cancer, and the presence of RAGE polymorphism
may aid the diagnosis of lung cancer and the clinical assessment of
prognosis.
Acknowledgements
The authors would like to thank Haiyan Wang, the
technician of the Asthma Laboratory (Qingdao Key Laboratory of
Common Disease, Qingdao Municipal Hospital) for her skillful
technical expertise.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pulford DJ, Falls JG, Killian JK and
Jirtle RL: Polymorphisms, genomic imprinting and cancer
susceptibility. Mutat Res. 436:59–67. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Galvani E, Peters GJ and Giovannetti E:
EGF receptor-targeted therapy in non-small-cell lung cancer: role
of germline polymorphisms in outcome and toxicity. Future Oncol.
8:1015–1029. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Habbous S, Pang V, Eng L, et al: p53
Arg72Pro polymorphism, HPV status and initiation, progression and
development of cervical cancer: a systematic review and
meta-analysis. Clin Cancer Res. 18:6407–6415. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herold K, Moser B, Chen Y, Zeng S, Yan SF,
Ramasamy R, Emond J, Clynes R and Schmidt AM: Receptor for advanced
glycation end products (RAGE) in a dash to the rescue: Inflammatory
signals gone awry in the primal response to stress. J Leukoc Biol.
82:204–212. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Brett J, Schmidt AM, Yan SD, et al: Survey
of the distribution of a newly characterized receptor for advanced
glycation end products in tissues. Am J Pathol. 143:1699–1712.
1993.PubMed/NCBI
|
|
7
|
Bartling B, Hofmann HS, Weigle B, Silber
RE and Simm A: Down-regulation of the receptor for advanced
glycation end-products (RAGE) supports non-small cell lung
carcinoma. Carcinogenesis. 26:293–301. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Englert JM, Hanford LE, Kaminski N, et al:
A role for the receptor for advanced glycation end products in
idiopathic pulmonary fibrosis. Am J Pathol. 172:583–591. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kuniyasu H, Oue N, Wakikawa A, Shigeishi
H, Matsutani N, Kuraoka K, Ito R, Yokozaki H and Yasui W:
Expression of receptors for advanced glycation end-products (RAGE)
is closely associated with the invasive and metastatic activity of
gastric cancer. J Pathol. 196:163–170. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fuentes MK, Nigavekar SS, Arumugam T,
Logsdon CD, Schmidt AM, Park JC and Huang EH: RAGE activation by
S100P in colon cancer stimulates growth, migration, and cell
signaling pathways. Dis Colon Rectum. 50:1230–1240. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsieh HL, Schäfer BW, Sasaki N and
Heizmann CW: Expression analysis of S100 proteins and RAGE in human
tumors using tissue microarrays. Biochem Biophys Res Commun.
307:375–381. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sparvero LJ, Asafu-Adjei D, Kang R, et al:
RAGE (Receptor for Advanced Glycation Endproducts), RAGE ligands,
and their role in cancer and inflammation. J Transl Med. 7:172009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tesarová P, Kalousová M, Jáchymová M,
Mestek O, Petruzelka L and Zima T: Receptor for advanced glycation
end products (RAGE) - soluble form (sRAGE) and gene polymorphisms
in patients with breast cancer. Cancer Invest. 25:720–725. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Schenk S, Schraml P, Bendik I and Ludwig
CU: A novel polymorphism in the promoter of the RAGE gene is
associated with non-small cell lung cancer. Lung Cancer. 32:7–12.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang X, Cui E, Zeng H, Hua F, Wang B, Mao
W and Feng X: RAGE genetic polymorphisms are associated with risk,
chemotherapy response and prognosis in patients with advanced
NSCLC. PLoS ONE. 7:e437342012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mountain CF: A new international staging
system for lung cancer. 1986. Chest. 136:(Suppl).
e252009.PubMed/NCBI
|
|
17
|
World Medical Association, . World Medical
Association Declaration of Helsinki-Ethical Principles for Medical
Research Involving Human Subjects. http://www.wma.net/en/30publications/10policies/b3/index.htmlAccessed.
March 11–2015
|
|
18
|
Lehr HA, Mankoff DA, Corwin D, et al:
Anpplication of photoshop-based image analysis to quantification of
hormone receptor expression in breast cancer. J Histochem Cytochem.
45:1559–1565. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang Y, Wang H, Piper MG, et al: sRAGE
induces human monocyte survival and differentiation. J Immunol.
185:1822–1835. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beránek M, Kanková K, Kolár P and Znojil
V: Polymorphisms in the von Willebrand factor gene are not
associated with proliferative retinopathy in non-insulin-dependent
diabetes mellitus. Ophthalmic Res. 34:327–330. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanková K, Márová I, Záhejský J, Muzík J,
Stejskalová A, Znojil V and Vácha J: Polymorphisms 1704G/T and
2184A/G in the RAGE gene are associated with antioxidant status.
Metabolism. 50:1152–1160. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tam XH, Shiu SW, Leng L, Bucala R,
Betteridge DJ and Tan KC: Enhanced expression of receptor for
advanced glycation end-products is associated with low circulating
soluble isoforms of the receptor in Type 2 diabetes. Clin Sci
(Lond). 120:81–89. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bopp C, Hofer S, Weitz J, Bierhaus A,
Nawroth PP, Martin E, Büchler MW and Weigand MA: sRAGE is elevated
in septic patients and associated with patients outcome. J Surg
Res. 147:79–83. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu X, Chen Q, Shi S, Shi Z, Lin R, Tan L,
Yu J, Shu Q and Fang X: Plasma sRAGE enables prediction of acute
lung injury after cardiac surgery in children. Crit Care.
16:R912012. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Garber ME, Troyanskaya OG, Schluens K, et
al: Diversity of gene expression in adenocarcinoma of the lung.
Proc Natl Acad Sci USA. 98:13784–13789. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jabaudon M, Futier E, Roszyk L, et al:
Soluble form of the receptor for advanced glycation end products is
a marker of acute lung injury but not of severe sepsis in
critically ill patients. Crit Care Med. 39:480–488. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Piperis M, Provatopoulou X, Sagkriotis A,
Kalogera E, Ampatzoglou E, Zografos GC, Athanasiou E and Gounaris
A: Effect of breast cancer adjuvant therapies on potential
biomarkers of pulmonary inflammation. Anticancer Res. 32:4993–5002.
2012.PubMed/NCBI
|
|
28
|
Fujisawa K, Katakami N, Kaneto H, et al:
Circulating soluble RAGE as a predictive biomarker of
cardiovascular event risk in patients with type 2 diabetes.
Atherosclerosis. 227:425–428. 2013. View Article : Google Scholar : PubMed/NCBI
|