Introduction
Hepatocellular carcinoma (HCC) is common malignant
form of cancer associated with high incidence (28.71 per 100,000 in
China) and mortality (26.04 per 100,000 in China) (1), which represents a critical threat to
public health. To date, there is no effective treatment for HCC,
and the lack of effective diagnostic techniques available, due to
the undefined etiology and pathogenesis and high-grade malignancy,
contributes to poor patient prognosis (2). Ascertaining the origin and development
of liver cancer and identification of novel targets for therapy
have been the focus of recent studies regarding HCC (3–7). MicroRNAs
(miRs) are single-stranded noncoding RNAs, containing 17–19
nucleotides, which are associated with the development and
progression of tumors (8–12) such as oropharyngeal squamous cell
carcinoma (8), non-small cell lung
cancer (9), thyroid cancer (10). miR-125a has previously been reported
to block the proliferation, invasion and migration of breast cancer
cells (13), and was also found to be
effective in preventing the invasion of ovarian cancer (14), glioma (15), lung cancer (16–18) and
gastric cancer (19). miR-125a has
been demonstrated to be implicated in hepatitis B virus duplication
and the progression of hepatitis B (20,21). Bi
et al (22) observed that
miR-125a inhibited the proliferation and migration of liver cancer
cells by targeting matrix metalloproteinase (MMP)11 and vascular
endothelial growth factor (VEGF). The phosphoinositide 3-kinase
(PI3K)/AKT pathway enhances not only cell proliferation, but also
cell invasion and migration (23). It
has been demonstrated that another miRNA, miR-21, is able to
control the proliferation of liver cancer cells by regulating the
PI3K/AKT pathway (24). To further
determine the effect of miR-125a in regulating the invasion of
liver cancer cells and elucidate the underlying mechanism, the
present study aimed to characterize the association between
miR-125a expression and the migration of human liver cancer cell
lines HepG2 and HCC-LM3 or non-malignant human epithelioid hepatic
cell line QZG and its role in modulating the PI3K/AKT pathway.
Materials and methods
Cell lines and antibodies
Human liver cancer cell lines (HepG2 and HCC-LM3)
and non-malignant human epithelioid hepatic cell line QZG were
purchased from the cell bank of the Chinese Academy of Sciences
(Beijing, China) and kept in liquid nitrogen in the central
laboratory of the 187th Hospital of Chinese PLA (Haikou, China).
Cells were grown in modified RPMI-1640 supplemented with 10% FBS
(Hyclone; GE Healthcare Life Sciences, Logan, UT, USA). Rat
anti-human PI3K, AKT and mammalian target of rapamycin (mTOR)
polyclonal antibodies were obtained from Santa Cruz Biotechnology,
Inc. (Dallas, TX, USA).
miR-125a gene transfection
Using the methods described by Jiang et al
(16), 2′-O-methyl (2′-O-Me)
oligonucleotides containing miR-125a were synthesized by Shanghai
Gene Pharma Co., Ltd (Shanghai, China) as follows: 2′-O-Me-3p
sense, 5′-ACAGGUGAGGUUCUUGGGAGCC-3′ and 2′-O-Me-3p antisense,
5′-GGCUCCCAAGAACCUCACC UGU-3′; 2′-O-Me-scramble-3p, 5′-GGUCGGUGCUCG
AUGCAGGUAA-3′; 2′-O-Me-5p sense, 5′-UCCCUGAGACCC UUUAACCUGUGA-3′
and 2′-O-Me-5p antisense, 5′-UCA CAGGUUAAAGGGUCUCAGGGA-3′;
2′-O-Me-scramble-5p, 5′-GGACGGCGAUCAGAUAAGAGUUCU-3′. In addition,
the fluorochrome FAM (Apeptide Co., Ltd., Shanghai, China) was used
as a fluorescent tracer for the oligonucleotides. HepG2, HCC-LM3
and QZG cells (at a concentration of 1×105/ml) were
transfected with the aforementioned sequences using Lipofectamine®
2000 transfection reagent (Invitrogen Life Technologies, Carlsbad,
CA, USA). The protocol used was as follows: On the day prior to
transfection, 1–3×104 cells were seeded on a 24-well
plate containing 500 µl modified RPMI-1640 medium supplemented with
10% FBS and maintained at 37°C in 5% CO2 until 70–90%
confluence was reached. Oligonucleotides (100 pmol) were added to
50 µl of Opti-MEM serum-free medium (Hyclone) and mixed gently.
Subsequently, 1 µl Lipofectamine 2000 was diluted in thoroughly
mixed Opti-MEM serum-free medium (50 µl), mixed gently and
maintained at room temperature for 5 min. The diluted
oligonucleotides and 1 µl Lipofectamine 2000 were combined and this
mixture was added to each well containing cells and medium and
mixed gently by rocking the plate back and forth. The cells were
then incubated at 37°C in a CO2 incubator for 24 h prior
to observation under a fluorescence microscope (CX41-32RFL, Olympus
Corporation, Tokyo, Japan). The medium was replaced with 500 µl
fresh modified RPMI-1640 medium following 5 h of incubation. In
addition, a blank control group transfected with isometric
phosphate-buffered saline (PBS; Beyotime Institute of
Biotechnology, Jiangsu, China) and Lipofectamine 2000 using an
identical protocol was set up. In order to block the PI3K/AKT/mTOR
signaling pathway, PI3K inhibitor LY294002 (Sigma-Aldrich, St.
Louis, MO, USA) was added into the culture medium at a
concentration of 10 nmol/l.
Quantitative polymerase chain reaction
(qPCR) analysis
Total RNA was extracted with TRIzol (Life
Technologies, Grand Island, NY, USA). First-strand complementary
(c)DNA was generated using RevertAid First Strand cDNA Synthesis
kit (Invitrogen Life Technologies). Specifically, 2 µl RNA was
mixed with 1 µl oligo (dT) and 10 µl RNase-free deionized water,
incubated in the PCR machine at 70°C for 5 min and chilled
immediately on ice. cDNA synthesis was induced by adding 4 µl 5X
buffer, 2 µl 10 mM deoxyribose nucleotide triphosphates, 1 µl RNA
inhibitor and 1 µl reverse transcriptase, prior to incubation in a
PCR machine at 42°C for 1 h. The reaction was terminated by
incubation at 70°C for 5 min. Quantitative measurements were
performed using the THUNDERBIRD SYBR® qPCR Mix kit (Toyobo Co.,
Ltd, Tokyo, Japan). For PCR, 12.5 µl 2X qPCR Mix, 2.0 µl each
primer (2.5 µM), 2.0 µl cDNA and 8.5 µl double distilled
H2O were added to a 0.2 ml PCR tube. Amplification
conditions were comprised of 40 cycles at 95°C for 15 min, 95°C for
15 sec, and 55°C for 30 sec, followed by 72°C for 25 sec. The qPCR
primers are listed in Table I.
 | Table I.Quantitative polymerase chain reaction
primers. |
Table I.
Quantitative polymerase chain reaction
primers.
| Gene | Primer, 5′→3′ |
|---|
| miR-125a | Sense:
CTATGTTTGAATGAGGCTTCAG |
|
| Antisense:
CGCGTCGCCGCGTGTTTAAACG |
| PI3K | Sense:
GCCCAGGCTTACTACAGAG |
|
| Antisense:
AAGTAGGGAGGCATCTCG |
| AKT | Sense:
CTCATTCCAGACCCACGAC |
|
| Antisense:
ACAGCCCCGAAGTCCGTTA |
| mTOR | Sense:
ATGACGAGACCCAGGCTAA |
|
| Antisense:
GCCAGTCCTCTACAATACGC |
| β-actin | Sense:
ATCATGTTTGAGACCTTCAACA |
|
| Antisense:
CATCTCTTGGTCGAAGTCCA |
Western blot analysis
Cells were collected by centrifugation at 4,000 × g
at 37°C for 5 min and 1×106 cells were lysed in 250 µl
radioimmunoprecipitation assay buffer (Beyotime Institute of
Biotechnology). Subsequently, 50 µg cellular protein was separated
by 10% SDS-PAGE (Sigma-Aldrich, St. Louis, MO, USA) and transferred
to a 0.45 µm polyvinylidene difluoride membrane (Sigma-Aldrich),
which was incubated overnight at 4°C with rat anti-human monoclonal
antibodies against PI3K, AKT and mTOR (1:3,000 dilution). Following
three washes with PBS, horseradish peroxidase-conjugated goat
anti-rat secondary antibody (Boster Bio, Pleasanton, CA, USA) at
1:3,000 dilution was added for 30 min at room temperature, and any
non-conjugated antibodies were washed away. Proteins were
visualized with enhanced chemiluminescence detection reagents
(Pierce Biotechnology, Co., Thermo Fisher Scientific, Rockford, IL,
USA) and were exposed to X-ray film (Beyotime Institute of
Biotechnology). Developed films were processed with BandScan
software version 5.0 (Glyko, Novato, CA, USA) to determine optical
densities.
Colony formation assay
Exponentially growing cells were diluted to
1×103 cells/ml. Soft agar (5%; Beyotime Institute of
Biotechnology) was mixed thoroughly with the medium at a ratio of
1:9, added to the plates, and set aside at room temperature to
allow agar to solidify prior to seeding a mixture of cell
suspension (1.5 ml) with an equal volume of 0.5% soft agar. The
plates were placed in an incubator at 37°C under 5% CO2
and following two weeks of incubation, colonies (defined as
containing >50 cells) were counted using the following formula:
Colony formation rate (%)=(number of colonies/number of cells
incubated) x100%.
In vitro invasion assay
Cell invasion was measured using a Transwell chamber
model (Chemicon International, Temecula, CA, USA). Cells were
suspended at a concentration of 1×105 cells/ml and 50 µl
of the suspension was seeded into the upper chamber. The lower
chamber contained RPMI-1640 medium with 10% FBS. Following
incubation for 24 h, cells attached to the lower chamber were fixed
with 10% formalin (Nanjing Chemical Technology Co., Ltd., Nanjing,
China) and stained with Giemsa (Solarbio Science and Technology
Co., Ltd., Beijing, China) to quantify cell migration.
Statistical analysis
Data are expressed as the mean ± standard deviation
and were analyzed with paired Student's t-test using the SPSS 16.0
software (SPSS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
miR-125a expression is downregulated
in HCC cells
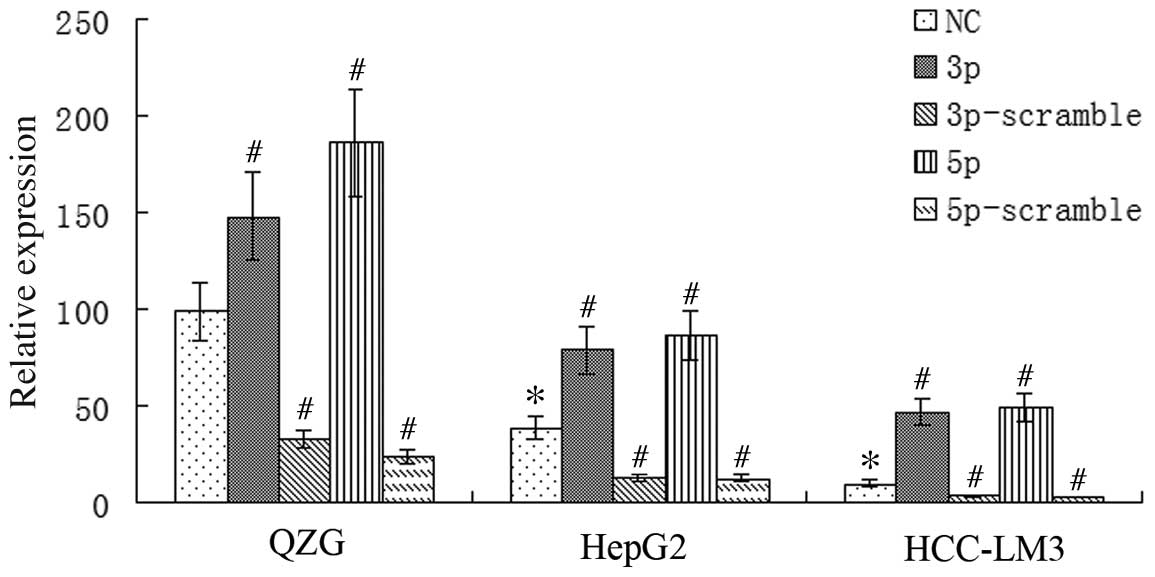
qPCR analysis was used to measure miR-125a mRNA
expression in HepG2, HCC-LM3 and QZG cells. miR-125a expression was
found to be downregulated in HepG2 and HCC-LM3 cells when compared
with that of the immortal cell line QZG (P<0.01). Furthermore,
the lowest expression was observed in HCC-LM3 cells, which are
associated with a high metastatic ability (25). miR-125a mRNA expression was
significantly upregulated in HepG2, HCC-LM3 and QZG cells
transfected with miR-125a-3p or -5p when compared with that of the
negative control group (P<0.01), whereas transfection with
miR-125a-3p-scramble (3p-s) or 5p-s resulted in significantly lower
expression of miR-125a mRNA (P<0.01). Given that the
non-malignant QZG cells exhibited the highest miR-125a expression,
whilst the malignant cell line HCC-LM3 exhibited the lowest
expression, the results indicated that miR-125a may be associated
with the migration of HCC cells and that the expression of miR-125a
may be regulated in liver cancer cells via transfection with miR
mimics/scrambled sequences (Fig.
1).
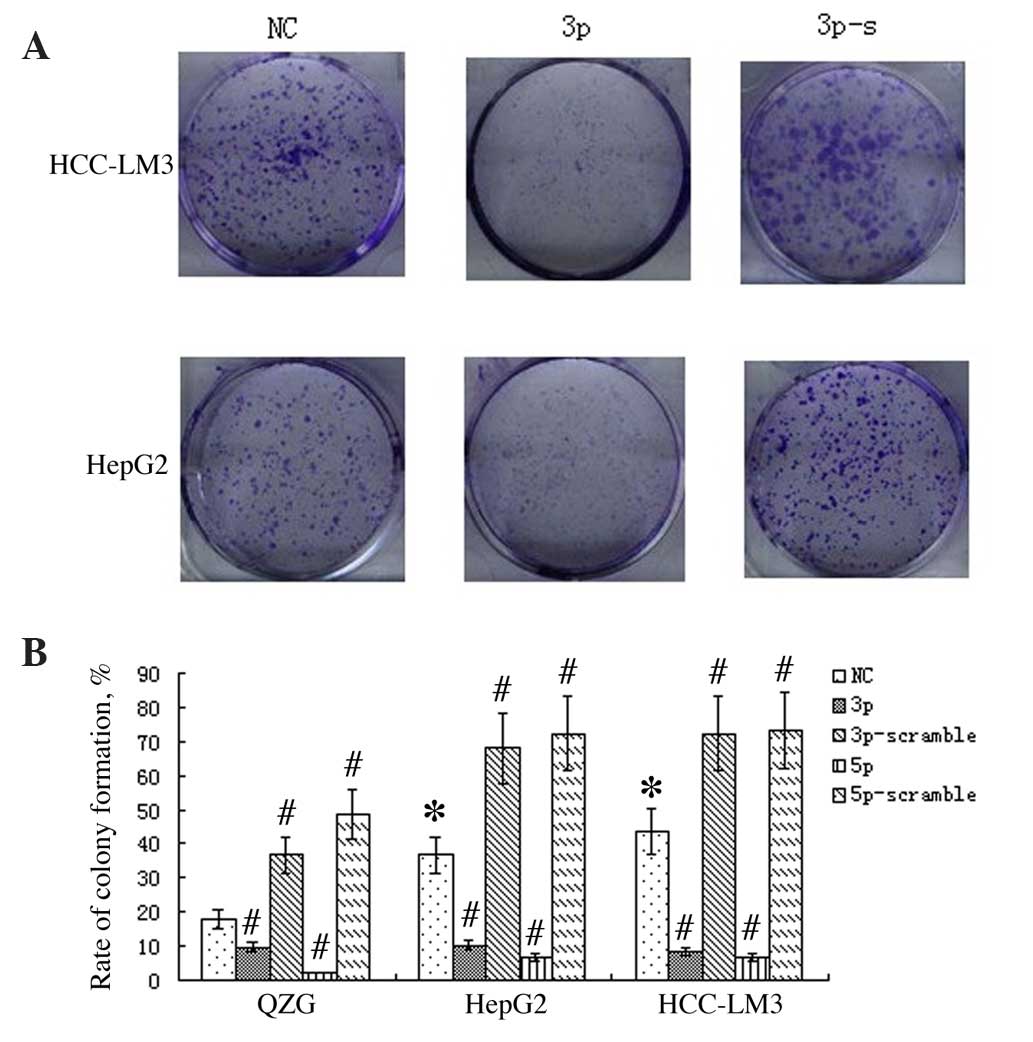
miR-125a suppresses liver cancer cell
proliferation
Following transfection of HepG2, HCC-LM3 and QZG
cells with miR-125a-3p, -5p, -3p-s and -5p-s, respectively, a
colony formation assay was performed on exponentially growing cells
to evaluate cell proliferation ability. The results indicated that
the colony formation rate was reduced following transfection with
miR-125a-3p or -5p (P<0.01), but increased following
transfection of miR-125a-3p-s or -5p-s (P<0.01; Fig. 2).
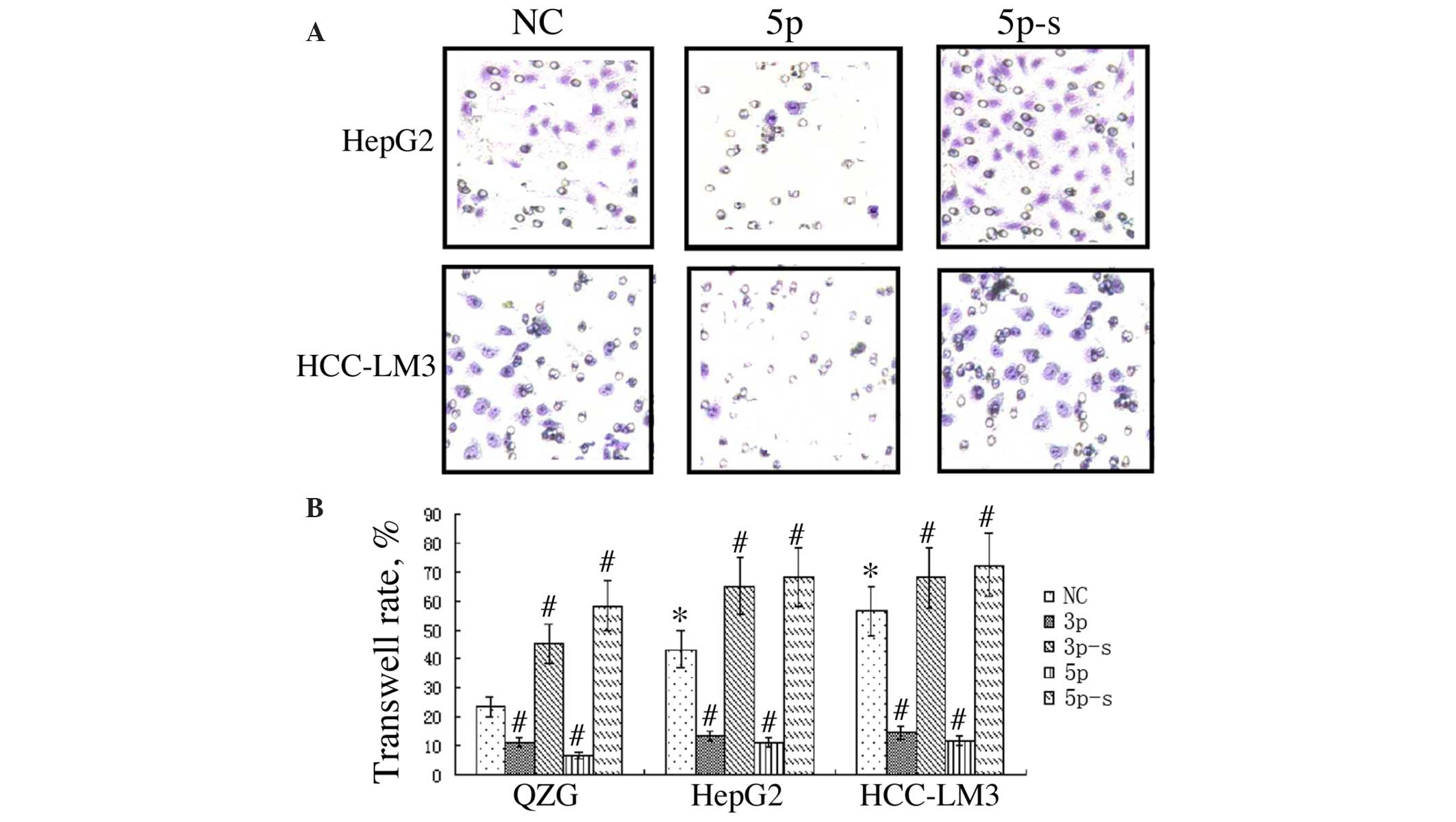
miR-125a inhibits liver cancer cell
migration
QZG, HepG2 and HCC-LM3 cells were transfected with
the various miR-125a sequences and a Transwell assay was performed
at 48 h post transfection to study cell migration ability. The
results indicated that HepG2 and HCC-LM3 cells exhibited a
significantly greater rate of migration than that of QZG cells
(P<0.05). Transfection with miR-125a-3p or -5p significantly
inhibited migration in comparison with that of the negative
controls, while miR-125a-3p-s or -5p-s exerted the opposite effect
(P<0.01; Fig. 3).
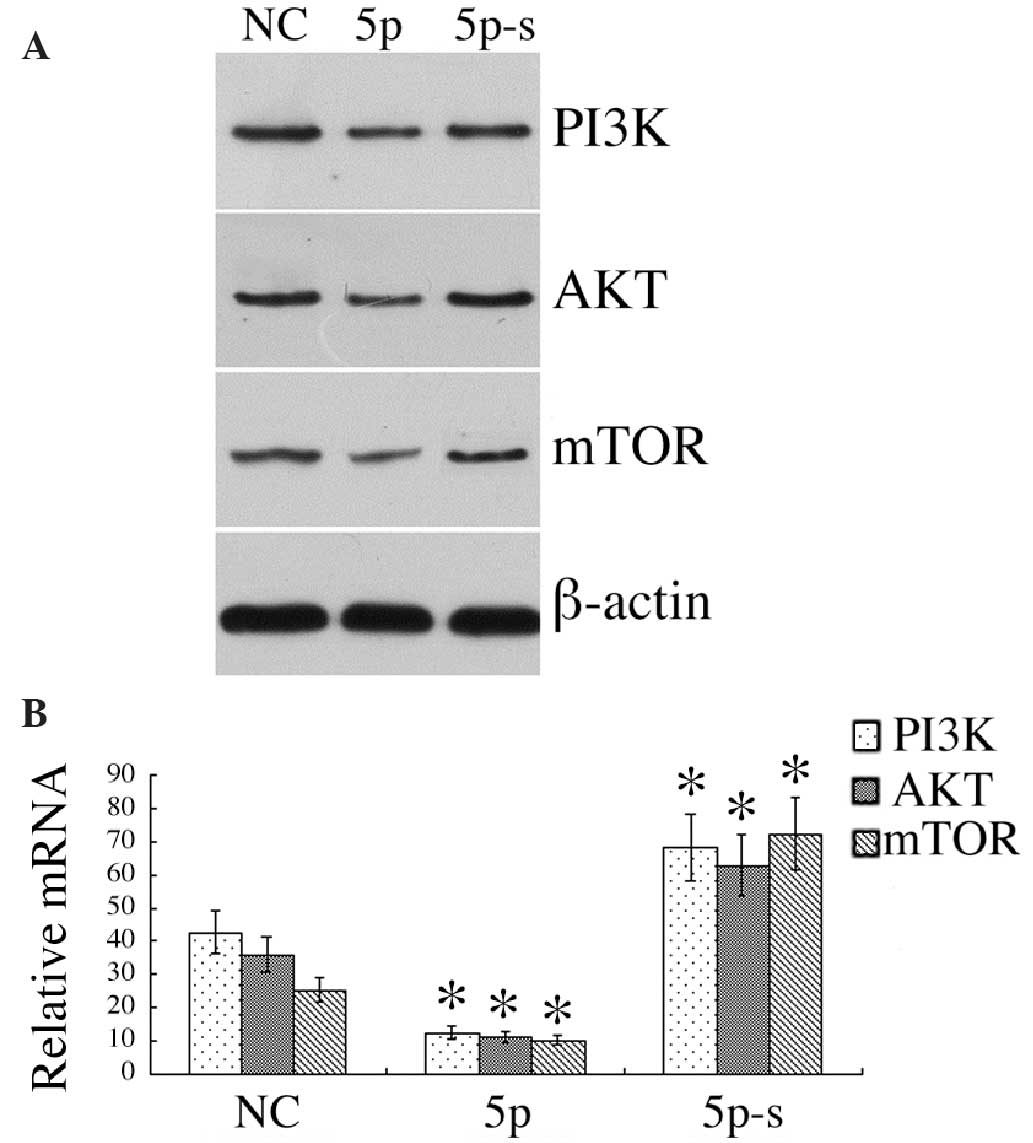
miR-125a modulates the expression of
PI3K/AKT/mTOR
Western blot analysis identified the levels of
PI3K/AKT/mTOR in HCC-LM3 cells to be 0.12±0.25/0.11±0.23/0.10±0.24,
respectively, following transfection of miR-125a-5p, significantly
lower than the 0.43±8.68/0.36±0.72/0.26±0.55 in the negative
control group (P<0.01). However, expression levels were
significantly increased following transfection of miR-125a-5p-s
(0.68±0.13/0.63±0.12/0.72±0.14), significantly higher than those of
the negative control group (P<0.01). qPCR analysis identified
downregulated PI3K/AKT/mTOR mRNA expression in HCC-LM3 cells
following transfection of miR-125a-5p, while the opposite effect
was observed following the transfection of miR-125a-5p-s
(P<0.01; Fig. 4).
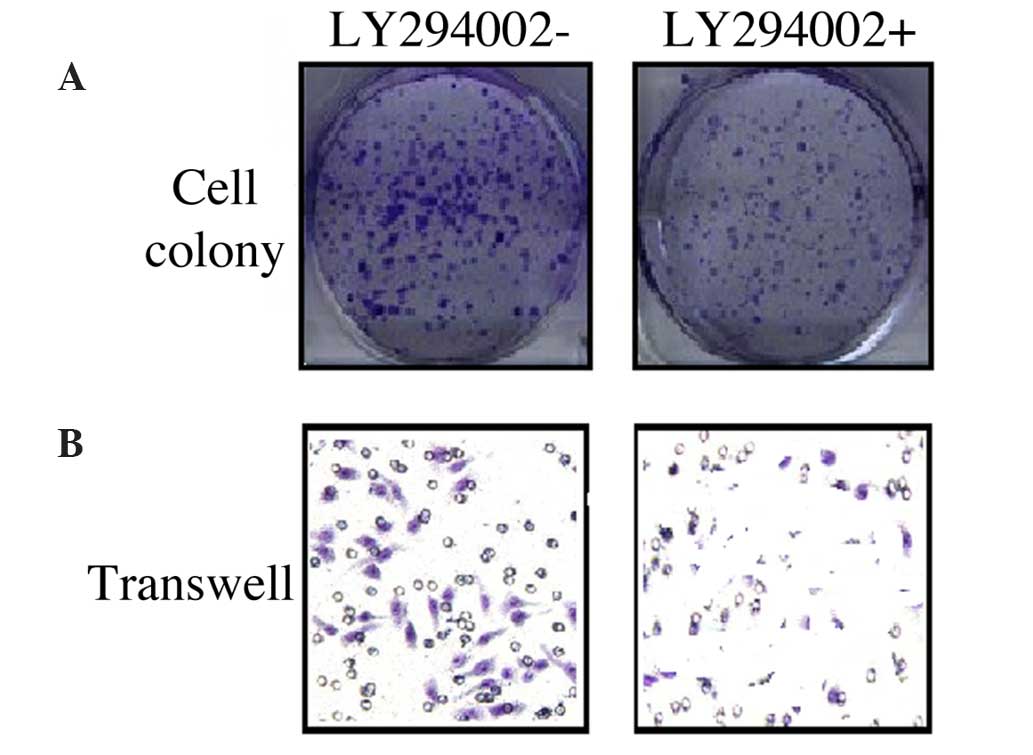
PI3K/AKT knockdown suppresses the
proliferation and migration of liver cancer cells
In order to elucidate the correlation between the
PI3K/AKT pathway and the proliferation and migration of liver
cancer cells, LY294002 was used to block the PI3K/AKT pathway. The
results revealed that LY294002 inhibited HCC-LM3 cell colony
formation (Fig. 5A), which further
attenuated cell migration ability (Fig.
5B).
Discussion
HepG2 and HCC-LM3 are two liver cancer cell lines
with differing metastatic potentials. HCC-LM3 cells have a
relatively greater migration and invasion ability. The present
study identified that the expression of miR-125a was significantly
reduced in HCC-LM3 cells, when compared with that in HepG2 cells.
QZG are hepatocyte-specific cells, which are not malignant.
miR-125a was expressed at significantly higher levels in QZG cells
than those of the other two hepatic cell lines evaluated. These
findings suggested that miR-125a may be associated with conferring
the invasion and migration ability of liver cancer cells. Bi et
al (22) reported expression of
miR-125a in liver cancer cell lines with varying invasive ability
via immnunohistochemical, western blot and qPCR analyses. miR-125a
was demonstrated to be significantly downregulated in liver cancer
tissues, particularly in cells with high invasive ability, which
indicated that the expression of miR-125a was associated with the
invasion and migration of liver cancer cells, and that miR-125a may
be used as a marker for predicting the prognosis of liver cancer
patients. To further validate the function of miR-125a in
modulating the invasion and migration of liver cancer cells,
miR-125a was altered by transfection of miR-125a-3p/5p and
miR-125a-3p-s/5p-s. Transfection with miR-125a-3p/5p upregulated
miR-125a expression, while miR-125a-3p-s/5p-s inhibited its
expression in liver cancer cells.
In the present study, soft agar and Transwell assays
were used to validate the role of miR-125a in regulating the
proliferation and migration of liver cancer cells. A soft agar
colony formation assay may be used to monitor tumor
anchorage-independence growth and tumor malignancy, where a
stronger invasion ability of tumor cells is associated with a
greater number of cell colonies (24,26,27). Tumor
migration and invasion ability is dependent on the microenvironment
for growth and the extracellular matrix (ECM), therefore a
Transwell chamber model that imitates the ECM represents a reliable
method for assaying cell invasion ability (28). A marked decrease in liver cancer cell
colony formation was detected following miR-125a overexpression in
the present study, while a significant increase was observed
following miR-125a silencing. Furthermore, overexpression of
miR-125a resulted in reduced migration of liver cancer cells, while
the opposite effect was observed following miR-125a silencing.
These findings suggested that overexpression of miR-125a may
suppress the proliferation and invasion of liver cancer cells.
miR-125a was demonstrated to regulate the invasion
and migration of liver cancer cells; however, the underlying
mechanism remained to be elucidated. Bi et al (22) reported that miR-125a mediated the
expression of MMP11 and VEGF in liver cancer cells. The
PI3K/AKT/mTOR pathway is involved in the occurrence of liver cancer
and the subsequent invasion and migration, and has therefore been a
major therapeutic target in the treatment of liver cancer (28–30). The
present study demonstrated reduced levels of PI3K/AKT/mTOR mRNA and
protein following miR-125a overexpression, but upregulated levels
following miR-125a knockdown. Therefore, miR-125a may suppress the
proliferation and migration of liver cancer cells through
inhibition of PI3K/AKT/mTOR pathway. In order to verify the
regulatory role of the PI3K/AKT/mTOR pathway in the proliferation
and migration of liver cancer cells, cells were treated with
inhibitor LY294002. The results confirmed that the proliferation
and migration of hepatic cancer cells were reduced following
LY294002-mediated inhibition of the PI3K/AKT/mTOR pathway.
In conclusion, miR-125a is involved in the
proliferation and migration of liver cancer cells, and the
underlying mechanism is associated with interference with the
PI3K/AKT/mTOR pathway. miR-125a may therefore represent a novel
therapeutic target for the treatment of hepatic cancer.
References
|
1
|
Hao J and Chen WQ: The 2012 Chinese cancer
registry annual report. Military Medical Science Press; Beijing,
China: pp. 27–60. 2012, (In Chinese).
|
|
2
|
Giannini EG, Farinati F, Ciccarese F, et
al Italian Liver Cancer (ITA.LI.CA.) group: Prognosis of untreated
hepatocellular carcinoma. Hepatology. 61:184–190. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Deng GL, Zeng S and Shen H: Chemotherapy
and target therapy for hepatocellular carcinoma: New advances and
challenges. World J Hepatol. 7:787–798. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu X, Zhou Y, Liu X, et al: MPHOSPH1: A
potential therapeutic target for hepatocellular carcinoma. Cancer
Res. 74:6623–6634. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou D, Huang C, Kong L and Li J: Novel
therapeutic target of hepatocellular carcinoma by manipulation of
macrophage colony-stimulating factor/tumor-associated macrophages
axis in tumor microenvironment. Hepatol Res. 44:E318–E319. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Galuppo R, Ramaiah D, Ponte OM and Gedaly
R: Molecular therapies in hepatocellular carcinoma: What can we
target? Dig Dis Sci. 59:1688–1697. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lee TK, Cheung VC, Lu P, et al: Blockade
of CD47-mediated cathepsin S/protease-activated receptor 2
signaling provides a therapeutic target for hepatocellular
carcinoma. Hepatology. 60:179–191. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao G, Gay HA, Chernock RD, et al: A
microRNA expression signature for the prognosis of oropharyngeal
squamous cell carcinoma. Cancer. 119:72–80. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen S, Xue Y, Wu X, et al: Global
microRNA depletion suppresses tumor angiogenesis. Genes Dev.
28:1054–1067. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vriens MR, Weng J, Suh I, et al: MicroRNA
expression profiling is a potential diagnostic tool for thyroid
cancer. Cancer. 118:3426–3432. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
van Kouwenhove M, Kedde M and Agami R:
MicroRNA regulation by RNA-binding proteins and its implications
for cancer. Nat Rev Cancer. 11:644–656. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Scott GK, Goga A, Bhaumik D, et al:
Coordinate suppression of ERBB2 and ERBB3 by enforced expression of
micro-RNA miR-125a or miR-125b. J Biol Chem. 282:1479–1486. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cowden DK, Dahl R, Kruichak JN and Hudson
LG: The epidermal growth factor receptor responsive miR-125a
represses mesenchymal morphology in ovarian cancer cells.
Neoplasia. 11:1208–1215. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cortez MA, Nicoloso MS, Shimizu M, et al:
miR-29b and miR-125a regulate podoplanin and suppress invasion in
glioblastoma. Genes Chromosomes Cancer. 49:981–990. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jiang L, Huang Q, Zhang S, et al:
Hsa-miR-125a-3p and hsa-miR-125a-5p are downregulated in non-small
cell lung cancer and have inverse effects on invasion and migration
of lung cancer cells. BMC Cancer. 10:3182010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang G, Mao W, Zheng S and Ye J: Epidermal
growth factor receptor-regulated miR-125a-5p - a metastatic
inhibitor of lung cancer. FEBS J. 276:5571–5578. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang L, Zhang Q, Chang H, et al:
hsa-miR-125a-5p enhances invasion in non-small cell lung carcinoma
cell lines by upregulating rock-1. Zhongguo Fei Ai Za Zhi.
12:1069–1073. 2009.(In Chinese). PubMed/NCBI
|
|
19
|
Hashiguchi Y, Nishida N, Mimori K, et al:
Down-regulation of miR-125a-3p in human gastric cancer and its
clinicopathological significance. Int J Oncol. 40:1477–1482.
2012.PubMed/NCBI
|
|
20
|
Coppola N, Potenza N, Pisaturo M, et al:
Liver microRNA hsa-miR-125a-5p in HBV chronic infection:
Correlation with HBV replication and disease progression. PLoS One.
8:e653362013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Potenza N, Papa U, Mosca N, et al: Human
microRNA hsa-miR-125a-5p interferes with expression of hepatitis B
virus surface antigen. Nucleic Acids Res. 39:5157–5163. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bi Q, Tang S, Xia L, et al: Ectopic
expression of MiR-125a inhibits the proliferation and metastasis of
hepatocellular carcinoma by targeting MMP11 and VEGF. PLoS One.
7:e401692012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Martini M, De Santis MC, Braccini L, et
al: PI3K/AKT signaling pathway and cancer: An updated review. Ann
Med. 46:372–383. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yan-nan B, Zhao-yan Y, Li-xi L, et al:
MicroRNA-21 accelerates hepatocyte proliferation in vitro via
PI3K/AKT signaling by targeting PTEN. Biochem Biophys Res Commun.
443:802–807. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang RY, Chen L, Chen HY, et al: MUC15
inhibits dimerization of EGFR and PI3K-AKT signaling and is
associated with aggressive hepatocellular carcinomas in patients.
Gastroenterology. 145:1436–1448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Abbud-Antaki RA, Marhefka JN, DeLuca AL
and Zuromskis MP: The cancer biochip system: A functional genomic
assay for anchorage-independent three-dimensional breast cancer
cell growth. Horm Cancer. 3:261–270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guadamillas MC, Cerezo A and Del PM:
Overcoming anoikis-pathways to anchorage-independent growth in
cancer. J Cell Sci. 124:3189–3197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Okabe H, Ishimoto T, Mima K, et al: CD44s
signals the acquisition of the mesenchymal phenotype required for
anchorage-independent cell survival in hepatocellular carcinoma. Br
J Cancer. 110:958–966. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marshall J: Transwell invasion assays.
Methods Mol Biol. 769:97–110. 2011.PubMed/NCBI
|
|
30
|
Janku F, Kaseb AO, Tsimberidou AM, et al:
Identification of novel therapeutic targets in the PI3K/AKT/mTOR
pathway in hepatocellular carcinoma using targeted next generation
sequencing. Oncotarget. 5:3012–3022. 2014.PubMed/NCBI
|