Introduction
Bladder cancer is one of the most frequently
diagnosed malignancies and is a common cause of cancer-related
mortality worldwide (1,2). In the United States in 2013, ~72,570 new
cases of bladder cancer were diagnosed and 15,210 mortalities were
ascribed to this malignancy (3). At
diagnosis, >70% of cases of bladder cancer are determined to be
superficial transitional cell carcinomas, although the majority of
these tumors relapse following transurethral resection.
Furthermore, ~15% will progress to muscle invasive disease
(4,5).
However, it is estimated that the morbidity rate of bladder cancer
will increase in the future due to changes in the exposure to
bladder cancer risk factors, as well as the aging global
population. In addition, the survival rate of affected patients is
decreased with tumor progression, despite the advancement of
surveillance and treatment strategies (6–10).
Conventional clinicopathological parameters are commonly used to
predict the course of bladder cancer, however, none are able to
accurately predict the prognosis of the majority of tumors. Thus,
the development of reliable prognostic biomarkers for bladder
cancer remains an important challenge (11,12).
It is known that bladder cancer, similar to other
types of human tumor, arises from accumulated genetic and
epigenetic changes that result in the inactivation of tumor
suppressor genes or the activation of proto-oncogenes (13). The epigenetic silencing of tumor
suppressor genes is of note in treatment and diagnosis, as
epigenetic changes may be reversed to restore gene function, as
well as be applied as useful biomarkers (14). For example, aberrant DNA methylation
is the most common epigenetic change in human malignancies, and may
be used as a diagnosis, surveillance and prognostic biomarker,
particularly when the methylation silences tumor suppressor genes
(15). In recent years, an
association between cadherin 11 (CDH11) expression and human tumors
has been proposed (16–19). CDH11 is a member of the cadherin
superfamily, a family of calcium-dependent intercellular adhesion
molecules that are crucial in cell adhesion, proliferation and
invasion. Recent studies have demonstrated that CDH11 functions as
a tumor suppressor gene, with the inactivation of CDH11 associated
with the malignant behavior of various types of human tumor
(16–19). The human CDH11 gene is located on
chromosome 16q22.1 and is frequently silenced by promoter
methylation in tumors. It is known that six classical cadherin
family members, CDH1, CDH3, CDH5, CDH8, CDH11 and CDH13, are
located at chromosome 16q22.1–16q24.3, as a feature termed a
six-cadherin cluster, and each member has similar functions
(18). In our previous studies, it
was demonstrated that aberrant methylation of CDH13 is a frequent
event in bladder cancer and may be applied as a useful biomarker
for patients with bladder cancer (20–22). In
consideration of the aforementioned findings, the present study
aimed to investigate the clinical significance of CDH11 methylation
in bladder cancer.
In the current study, the methylation status of
CDH11 was analyzed in the tumor tissues of patients with bladder
cancer by performing methylation-specific polymerase chain reaction
(PCR; MSP). Subsequently, this data was correlated with common
clinicopathological parameters and clinical outcomes to evaluate
its clinical significance.
Patients and methods
Patients and tissue samples
The present study was performed in accordance with
the Declaration of Helsinki and approved by the Ethics Committee of
the Third Hospital of Hebei Medical University (Shijiazhuang,
China). Written informed consent was obtained from each
participant. A total of 146 tumor samples were collected during
surgery at the Third Hospital of Hebei Medical University between
July 2003 and July 2007, including transurethral rescetion of
bladder tumor (n=95) and radical resection of bladder (n=51). The
criteria for the enrollment of patients with bladder cancer were as
follows: i) Histopathological diagnosis of bladder transitional
cell carcinoma for the first time; ii) no other malignant tumors;
iii) no anticancer therapy received prior to surgery; and iv)
availability of sufficient clinicopathological and follow-up data
(21). Tumor diagnosis, staging,
treatment and follow-up were performed according to the European
Association of Urology (EAU) Working Group on Oncological Urology:
Guidelines on bladder cancer (23–25). In
addition, healthy bladder epithelial tissues were obtained by
biopsy from 37 inpatients with bladder calculi and used as the
controls; these tissues were pathologically examined to exclude the
possibility of incidental tumors. Furthermore, all control patients
had no history of malignant tumors and had not previously received
anticancer therapy. All tissue samples were immediately frozen in
liquid nitrogen and stored at −80°C until use. The
clinicopathological and demographic characteristics of the patients
with bladder cancer are summarized in Table I.
 | Table I.Association between CDH11 methylation
and the clinicopathological features of bladder cancer (n=146). |
Table I.
Association between CDH11 methylation
and the clinicopathological features of bladder cancer (n=146).
| Variable | Patients, n | Unmethylated CDH11, n
(%) | Methylated CDH11, n
(%) | P-value |
|---|
| Age, years |
|
|
|
|
| ≤65 | 54 | 22 (40.7) | 32 (59.3) | 0.4716 |
|
>65 | 92 | 32 (34.8) | 60 (65.2) |
|
| Gender |
|
|
|
|
| Male | 102 | 37 (36.3) | 65 (63.7) | 0.7862 |
|
Female | 44 | 17 (38.6) | 27 (61.4) |
|
| Tumors |
|
|
|
|
|
Single | 52 | 25 (48.1) | 27 (51.9) | 0.0390 |
|
Multiple | 94 | 29 (30.9) | 65 (69.1) |
|
| Tumor diameter,
cm |
|
|
|
|
| ≤3 | 83 | 40 (48.2) | 43 (51.8) | 0.0013 |
|
>3 | 63 | 14 (22.2) | 49 (77.8) |
|
| Tumor shape |
|
|
|
|
|
Papillary | 97 | 37 (38.1) | 60 (61.9) | 0.6834 |
|
Non-papillary | 49 | 17 (34.7) | 32 (65.3) |
|
| Grade |
|
|
|
|
|
G1-G2 | 90 | 39 (43.3) | 51 (56.7) | 0.0440 |
|
G3 | 56 | 15 (26.8) | 41 (73.2) |
|
| Stage |
|
|
|
|
|
Ta-T1 | 95 | 41 (43.2) | 54 (56.8) | 0.0350 |
|
T2-T4 | 51 | 13 (25.5) | 38 (74.5) |
|
DNA extraction, bisulfite modification
and MSP
Genomic DNA was extracted from the preserved frozen
tissue samples using a DNeasy® Tissue kit (Qiagen, Inc., Valencia,
CA, USA), according to the manufacturer's instructions. The
extracted DNA was treated with bisulfite to convert unmethylated
cytosines to uracils prior to MSP using an EpiTect® Bisulfite kit
(Qiagen, Inc.), in accordance with the manufacturer's instructions
and our previous study (21). The
methylation status of the promoter region of CDH11 was examined by
performing MSP, using primers specific for methylated and
unmethylated CDH11 sequences. Each PCR reaction was carried out in
a total volume of 25 µl, including: 100 ng DNA template and 0.2 µl
(5 U/µl) Takara Taq DNA polymerase (Takara, Kyoto, Japan), 0.2 mM
of each primer, 2.5 mM magnesium chloride, 0.2 mM of each
deoxynucleotide triphosphate, 2.5 µl 10X PCR buffer (Takara). The
primers for the methylated reaction were as follows: Sense,
5′-TTATTTTTGTTATTAGCGCGTTC-3′ and antisense,
5′-CCATTCACAAATCAACGACG-3′, with a 123-bp amplification product.
The primers for the unmethylated reaction were as follows: Sense,
5′-TTTTTA TTTTTGTTATTAGTGTGTTT-3′ and antisense, 5′-TCCCAT
TCACAAATCAACAACA-3′, with a 128-bp amplification product (Shanghai
Sangon Biological Engineering Technology and Services Co., Ltd.,
Shanghai, China). PCR amplification of the modified DNA samples
consisted of one cycle at 94°C for 10 min, followed by 41 cycles at
94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec for the
methylated reaction, or 41 cycles at 94°C for 30 sec, 58°C for 30
sec and 72°C for 30 sec for the unmethylated reaction. A final
extension reaction was then performed at 72°C for 5 min. Normal
lymphocyte DNA, methylated in vitro with SssI methylase (New
England BioLabs, Inc., Beverly, MA, USA), was used as the
methylation-positive control and normal lymphocyte DNA was used as
the unmethylation-positive control (26). Water samples were included with each
assay as blank controls. PCR products were separated on 2% agarose
gel, stained with ethidium bromide and visualized under ultraviolet
illumination. Samples were scored as methylation-positive when
methylated alleles were present in the methylated DNA lane and as
methylation-negative when bands were present only in the
unmethylated DNA lane.
Quantitative PCR analysis
Total RNA was extracted from tissue samples using
TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
The mRNA expression level of CDH11 was determined by performing
quantitative PCR, as previously described (27). First-strand cDNA was synthesized from
1 ug of each purified RNA sample using ExScript RT-PCR kit
(Takara). Relative CDH11 mRNA expression was calculated using the
comparative cycle threshold method, with GAPDH as the internal
control. The primers for CDH11 were as follows: Sense,
5′-TCGCCTGCATCGTCATTC-3′; and antisense,
5′-GGCAATATCAAAGGCTTCTGTGTC-3′. The primers for GAPDH were as
follows: Sense, 5′-CGCTCT CTGCTCCTCCTGTTC-3′ and antisense,
5′-ATCCGTTGA CTCCGACCTTCAC-3′. The PCR conditions included a
denaturation step of 95°C for 2 min, followed by 40 cycles at 95°C
for 30 sec, 60°C for 30 sec and 72°C for 2 min, and a final
elongation step of 72°C for 10 min.
Statistical analysis
Fisher's exact test was used to assess the
difference in CDH11 methylation status between bladder cancer
samples and controls, and a χ2 test was performed to
determine the association between CDH11 methylation status and
clinicopathological features. Furthermore, the difference in CDH11
mRNA expression between the controls, patients with methylated
CDH11 and patients with unmethylated CDH11 was analyzed by one-way
analysis of variance. Kaplan-Meier survival analysis and log-rank
tests were performed to assess the difference in overall survival
between patients with methylated and unmethylated CDH11. In
addition, the multivariate Cox proportional hazard model analysis
was applied to determine the independent prognostic effect of CDH11
methylation. All statistical analyses were performed using SAS
software (version 8.0; SAS Institute, Cary, NC, USA) and two-sided
P<0.05 was considered to indicate a statistically significant
difference.
Results
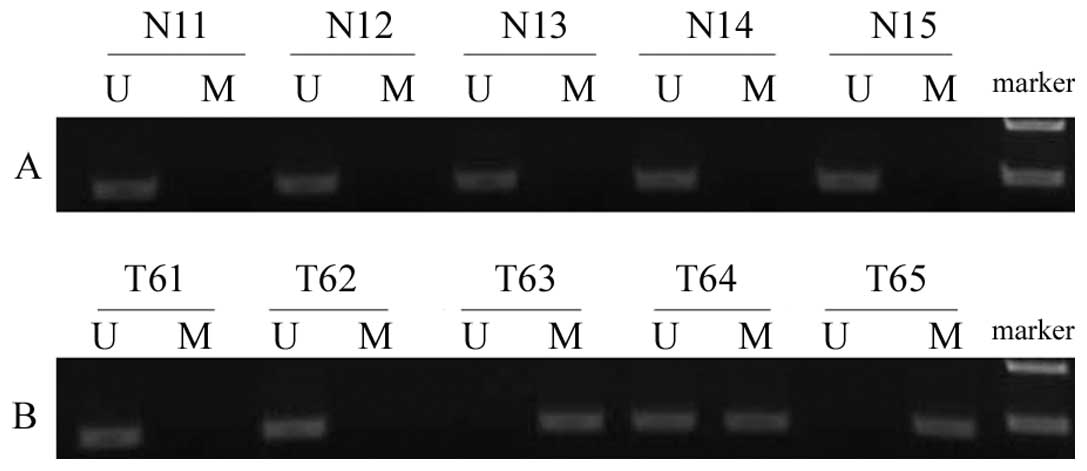
In the present study, the promoter hypermethylation
status of CDH11 was determined in bladder cancer tissues and normal
bladder epithelial tissues using MSP. It was identified that the
CDH11 promoter was hypermethylated in 63.0% (92/146) of bladder
cancer tissue samples, however, no methylation was detected in the
control samples. Thus, CDH11 promoter hypermethylation occurred
significantly more frequently in the bladder cancer tissues than in
the normal bladder epithelial tissues (P<0.05; Fig. 1).
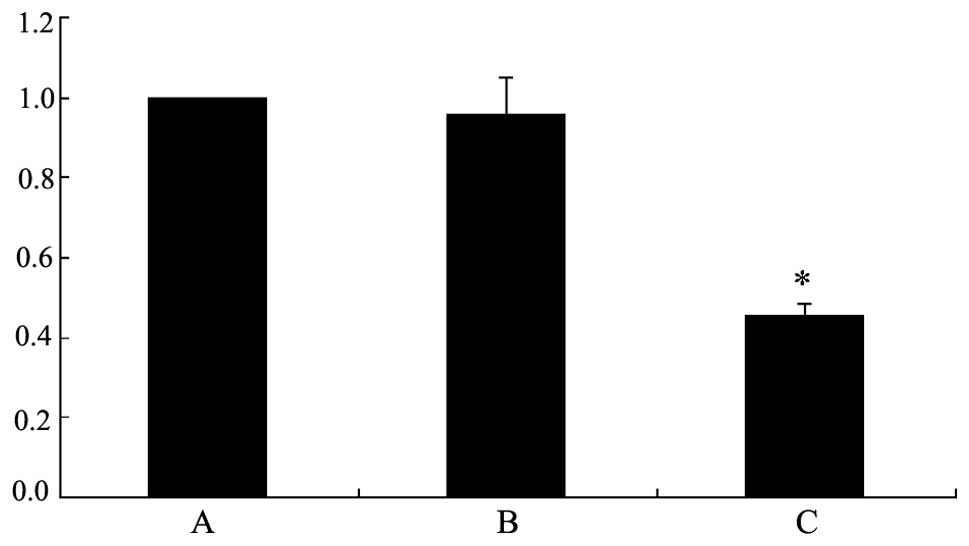
To clarify that hypermethylation of the CDH11
promoter region is correlated with the inactivation of its gene
expression, quantitative PCR was performed to detect the mRNA
expression levels of CDH11 in the bladder cancer and normal bladder
epithelial samples. The tumor samples were divided into two groups,
as samples with methylated or unmethylated CDH11. It was identified
that CDH11 mRNA expression was similar in the tumor samples with
unmethylated CDH11 and in the normal bladder epithelium. However,
CDH11 mRNA expression was significantly lower in the tumor samples
with methylated CDH11 compared with the normal bladder epithelium
and tumor samples with unmethylated CDH11 (P<0.05; Fig. 2). These results indicate that aberrant
CDH11 promoter hypermethylation is the major mechanism by which
CDH11 is inactivated in bladder cancer.
The aim of the present study was to evaluate the
clinical significance of CDH11 methylation in bladder cancer. In
addition, associations between the methylation status of CDH11 and
commonly used clinicopathological parameters in bladder cancer were
analyzed (Table II). It was
identified that aberrant hypermethylation of CDH11 in tumor tissues
was significantly associated with poor differentiation (P=0.0440),
an advanced disease stage (P=0.0350), a larger tumor size
(P=0.0013) and multiple tumors (P=0.0390). However, no association
was detected between CDH11 methylation and age, gender or tumor
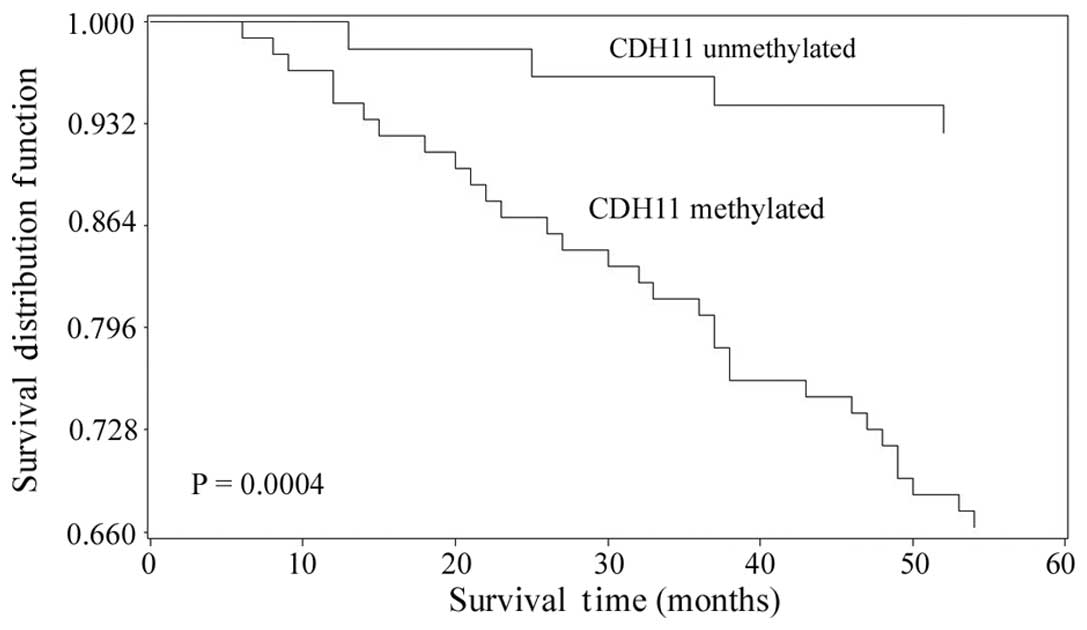
shape. The overall survival time of the patients with bladder
cancer was defined as the time from the date of diagnosis to the
date of mortality from any cause or the last contact if the patient
remained alive (21). Examination of
the overall survival of the patients with bladder cancer according
to the methylation status of CDH11 revealed that the patients with
methylated CDH11 had poorer outcomes than the patients with
unmethylated CDH11 (P=0.0004; Fig.
3). This finding indicated that CDH11 methylation in tumor
tissue samples may be associated with a poor prognosis of bladder
cancer. To further investigate the prognostic value of CDH11
methylation in bladder cancer, a multivariate Cox proportional
hazard model analysis was conducted. Notably, the results indicated
that CDH11 methylation may be independently associated with the
poor outcome of patients with bladder cancer, exhibiting a relative
risk of mortality of 6.852 (P=0.0082; 95% confidence interval,
3.461–16.177; Table II).
 | Table II.Multivariate Cox proportional hazard
analysis of potential prognostic factors for patients with bladder
cancer (n=146). |
Table II.
Multivariate Cox proportional hazard
analysis of potential prognostic factors for patients with bladder
cancer (n=146).
| Variable | HR (95% confidence
interval) | P-value |
|---|
| CDH11 methylation
(methylated vs. unmethylated) | 6.852
(3.461–16.177) | 0.0082 |
| Age (>65 vs. ≤65
years) | 1.312
(0.625–4.171) | 0.6429 |
| Gender (male vs.
female) | 1.125
(0.467–4.239) | 0.7063 |
| Number of tumors
(multiple vs. single) | 2.786
(0.879–9.418) | 0.4527 |
| Tumor diameter (>3
vs. ≤3 cm) | 3.054
(0.796–10.587) | 0.3786 |
| Shape (non-papillary
vs. papillary) | 3.613
(0.917–11.028) | 0.1891 |
| Grade (G3
vs. G1-G2) | 4.317
(0.936–15.068) | 0.0544 |
| Stage
(T2-T4 vs. Ta-T1) | 4.963
(2.567–14.622) | 0.0357 |
Discussion
The identification of prognostic and predictive
markers for bladder cancer is important, as it is a heterogeneous
disease with a clinical outcome that is difficult to predict
(5). Morphologically and
pathologically similar tumors may behave differently, therefore, it
is currently not possible to accurately predict the outcome of
bladder cancer following the receipt of initial adequate treatment.
Clinically, it is crucial to identify patients with a high risk of
mortality who require more aggressive treatment strategies and
patients with a low risk of mortality requiring less intensive
surveillance. Therefore, novel prognostic biomarkers must be
identified and applied in addition to common clinical and
pathological features. It is known that DNA methylation, one of the
most common epigenetic changes, frequently occurs in various types
of human cancer, including bladder cancer. Thus, the detection of
aberrant DNA methylation in primary bladder tumor specimens may
useful for predicting the outcome of patients with bladder cancer
(14).
In the current study, the methylation status of
CDH11 was investigated in bladder cancer tissues and normal bladder
epithelium. Aberrant CDH11 promoter hypermethylation occurred in
63.0% (92/146) of bladder tumor samples, however, no methylation
was detected in the normal bladder epithelium samples. These
results indicated that aberrant methylation of CDH11 is
tumor-specific, and that CDH11 may be used as potential biomarker
in bladder cancer. In addition, reduced CDH11 mRNA expression was
observed in the bladder tumors with aberrant promoter
hypermethylation. These findings indicate that epigenetic
inactivation of CDH11 by aberrant promoter hypermethylation may be
crucial in the formation of bladder cancer and thus, assessment of
the methylation status of CDH11 may be a useful biomarker in
determining the prognosis of patients with bladder cancer. To
verify this possibility, the methylation status of CDH11 was
subsequently correlated with specific clinicopathological
parameters of bladder cancer. Notably, aberrant methylation of
CDH11 occurred frequently in the tumors with an advanced stage,
high grade, larger tumor size and multiple tumors, factors that are
all risk factors of a poor prognosis in bladder cancer (28). This raises the possibility that CDH11
methylation status in tumors may be applied as a prognostic
biomarker that is as reliable or more reliable than currently used
clinicopathological factors. To investigate this hypothesis, the
overall survival of patients with bladder cancer was investigated
in terms of tumor tissue CDH11 methylation status. It was
identified that patients with methylated CDH11 had significantly
worse outcomes than patients with unmethylated CDH11. Furthermore,
multivariate Cox proportional hazard model analysis indicated that
aberrant CDH11 methylation was an independent prognostic factor for
the overall survival of patients with bladder cancer. Taken
together, the current findings suggested that aberrant methylation
of CDH11 in tumors indicates worse outcomes for patients with
bladder cancer. Therefore, it is recommended that patients with
bladder cancer and CDH11 methylation should receive aggressive
intervention following initial curative treatment to achieve more
favorable outcomes.
The findings of the present study are in accordance
with previous study about CDH11 in human cancers (16,18,19). DNA
methylation as a potential biomarker for human cancer is of
particular interest, since DNA may be collected conveniently from
tissues or body fluids. In addition, DNA methylation may be
reversed by demethylation agents and in the longer term may enable
more individualized therapies.
The present study identified a correlation between
CDH11 methylation status and the five-year overall survival time of
patients with bladder cancer. However, it was limited by the small
number of patients with bladder cancer that were analyzed and by
the use of a single center. It is proposed that future studies
continue to investigate the value of CDH11 methylation in
predicting the prognosis of bladder cancer using a greater number
of samples, to corroborate the findings of the present study.
In conclusion, aberrant methylation of CDH11
frequently occurs in bladder cancer and contributes to the
inactivation of its expression. In addition, CDH11 methylation
appears to be closely associated with malignant behavior in bladder
cancer and thus, may serve as an independent prognostic biomarker.
Considering the reversible nature of DNA methylation, CDH11
methylation may be a good therapeutic target for patients with
bladder cancer, however, future studies are required to verify this
hypothesis.
Acknowledgements
The present study was supported by the Xuzhou
Medical Talented Youth Project (grant no. 2014007).
References
|
1
|
Ploeg M, Aben KK and Kiemeney LA: The
present and future burden of urinary bladder cancer in the world.
World J Urol. 27:289–293. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Parkin DM: The global burden of urinary
bladder cancer. Scand J Urol Nephrol Suppl. 42:12–20. 2008.
View Article : Google Scholar
|
|
3
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim WJ, Kim EJ, Jeong P, et al: RUNX3
inactivation by point mutations and aberrant DNA methylation in
bladder tumors. Cancer Res. 65:9347–9354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kawamoto K, Enokida H, Gotanda T, et al:
p16INK4a and p14ARF methylation as a potential biomarker for human
bladder cancer. Biochem Biophys Res Commun. 339:790–796. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Montgomery JS, Miller DC and Weizer AZ:
Quality indicators in the management of bladder cancer. J Natl
Compr Canc Netw. 11:492–500. 2013.PubMed/NCBI
|
|
7
|
Shariat SF, Milowsky M and Droller MJ:
Bladder cancer in the elderly. Urol Oncol. 27:653–667. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mueller J, Schrader AJ, Schrader M,
Schnoeller T and Jentzmik F: Management of muscle-invasive bladder
cancer. Minerva Urol Nefrol. 65:235–248. 2013.PubMed/NCBI
|
|
9
|
Pal SK, Milowsky MI and Plimack ER:
Optimizing systemic therapy for bladder cancer. J Natl Compr Canc
Netw. 11:793–804. 2013.PubMed/NCBI
|
|
10
|
Kobeissi LH, Yassine IA, Jabbour ME,
Moussa MA and Dhaini HR: Urinary bladder cancer risk factors: A
Lebanese case-control study. Asian Pac J Cancer Prev. 14:3205–3211.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ghafouri-Fard S, Nekoohesh L and
Motevaseli E: Bladder cancer biomarkers: Review and update. Asian
Pac J Cancer Prev. 15:2395–2403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Ding MX, Wang HF, Wang JS, et al: ppGalNAc
T1 as a potential novel marker for human bladder cancer. Asian Pac
J Cancer Prev. 13:5653–5657. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yegin Z, Gunes S and Buyukalpelli R:
Hypermethylation of TWIST1 and NID2 in tumor tissues and voided
urine in urinary bladder cancer patients. DNA Cell Biol.
32:386–792. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim YK and Kim WJ: Epigenetic markers as
promising prognosticators for bladder cancer. Int J Urol. 16:17–22.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kandimalla R, van Tilborg AA and Zwarthoff
EC: DNA methylation-based biomarkers in bladder cancer. Nat Rev
Urol. 10:327–335. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Deng Z, Niu G, Cai L, Wei R and Zhao X:
The prognostic significance of CD44V6, CDH11 and β-catenin
expression in patients with osteosarcoma. BioMed Res Int.
2013:4961932013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Carmona FJ, Villanueva A, Vidal A, et al:
Epigenetic disruption of cadherin-11 in human cancer metastasis. J
Pathol. 228:230–240. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li L, Ying J, Li H, et al: The human
cadherin 11 is a pro-apoptotic tumor suppressor modulating cell
stemness through Wnt/β-catenin signaling and silenced in common
carcinomas. Oncogene. 31:3901–3912. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Nakajima G, Patino-Garcia A, Bruheim S, et
al: CDH11 expression is associated with survival in patients with
osteosarcoma. Cancer Genomics Proteomics. 5:37–42. 2008.PubMed/NCBI
|
|
20
|
Lin YL, He ZK, Li ZG and Guan TY:
Downregulation of CDH13 expression promotes invasiveness of bladder
transitional cell carcinoma. Urol Int. 90:225–232. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lin YL, Liu XQ, Li WP, Sun G and Zhang CT:
Promoter methylation of H-cadherin is a potential biomarker in
patients with bladder transitional cell carcinoma. Int Urol
Nephrol. 44:111–117. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lin YL, Sun G, Liu XQ, Li WP and Ma JG:
Clinical significance of CDH13 promoter methylation in serum
samples from patients with bladder transitional cell carcinoma. J
Int Med Res. 39:179–186. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Oosterlinck W, Lobel B, Jakse G, et al
European Association of Urology (EAU) Working Group on Oncological
Urology: Guidelines on bladder cancer. Eur Urol. 41:105–112. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Greene FL: The American Joint Committee on
Cancer: Updating the strategies in cancer staging. Bull Am Coll
Surg. 87:13–15. 2002.PubMed/NCBI
|
|
25
|
Babjuk M, Oosterlinck W, Sylvester R, et
al European Association of Urology (EAU): EAU guidelines on
non-muscle-invasive urothelial carcinoma of the bladder, the 2011
update. Eur Urol. 59:997–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Y, Zhang X, Lin L, et al: Aberrant
methylation of RASSF2A in tumors and plasma of patients with
epithelial ovarian cancer. Asian Pac J Cancer Prev. 15:1171–1176.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Nakajima G, Patino-Garcia A, Bruheim S, et
al: CDH11 expression is associated with survival in patients with
osteosarcoma. Cancer Genomics Proteomics. 5:37–42. 2008.PubMed/NCBI
|
|
28
|
Kaufman DS, Shipley WU and Feldman AS:
Bladder cancer. Lancet. 374:239–249. 2009. View Article : Google Scholar : PubMed/NCBI
|