Introduction
Colorectal cancer (CRC) is the third most common
malignant cancer and the second leading cause of mortality
worldwide (1). In China, the
incidence rate of CRC is increasing rapidly, particular in urban
areas, such as Shanghai, which was formerly considered a low-risk
area (2,3). Metastasis is the main cause of mortality
in CRC patients (4). The first sites
of metastatic CRC are the regional lymph nodes and the liver.
Investigations into the molecular mechanisms involved in CRC
metastasis are of major importance in order to develop novel
strategies for targeted therapies (5).
The mal, T-cell differentiation protein (MAL) gene
encodes the T-lymphocyte maturation-associated protein and
functions in T-cell differentiation (6). Recently, downregulation of MAL has been
associated with a variety of human epithelial malignancies. For
example, Mimori et al (7)
initially found that MAL was highly expressed in normal esophageal
epithelia, but silenced in esophageal tumors. Later, inactivation
of MAL was shown to be a common event in breast, head and neck, and
gastric cancer (8–11). However, the functional role and
mechanistic action of MAL in CRC remains largely unknown. The
present study aimed to determine the clinical significance of MAL
in colorectal cancer by measuring its expression level in CRC
tissues.
Materials and methods
Tissue samples and cell line
The present study enrolled a total of 30 CRC
patients who underwent surgery between 2010 and 2011 at Shanghai
First People's Hospital (Shanghai, China). The patients did not
receive any pre-operative cancer treatment prior to resection. Of
the 30 patients, 18 were male and 22 were female, and the age of
the patients ranged between 45 and 87 years (median, 69 years). The
clinicopathological data of the CRC patients are shown in Table I.
 | Table I.Clinical chracteristics of the
colorectal cancer patients. |
Table I.
Clinical chracteristics of the
colorectal cancer patients.
| Case | Gender | Age, years | Grade | Specimen site | TNM | Stage | Type |
|---|
| 1 | F | 67 | G1 | Right | T4aN1aM0 | IIIB | 2 |
| 2 | M | 66 | G1 | Left | T4aN0M0 | IIB | 1 |
| 3 | F | 73 | G3 | Left | T4aN1bM0 | IIIB | 1 |
| 4 | M | 72 | G2 | Left | T4aN1bM0 | IIIB | 1 |
| 5 | M | 77 | G2 | Right | T4aN0M0 | IIIB | 1 |
| 6 | M | 62 | G2 | Left | T4aN0M0 | IIB | 1 |
| 7 | M | 69 | G2 | Right | T4aN0M0 | IIB | 1 |
| 8 | M | 62 | G2 | Left | T4aN1aM0 | IIIB | 1 |
| 9 | F | 77 | G2 | Rectum | T4aN0M0 | IIB | 1 |
| 10 | M | 86 | G2 | Rectum | T2N0M0 | I | 1 |
| 11 | F | 64 | G2 | Rectum | T4aN1bM0 | IIIB | 1 |
| 12 | F | 77 | G2 | Left | T2N0M0 | I | 1 |
| 13 | F | 75 | G1 | Rectum | T1N0M0 | I | 1 |
| 14 | M | 69 | G2 | Right | T4aN0M0 | IIB | 1 |
| 15 | F | 73 | G3 | Right | T4aN2bM0 | IIIC | 1 |
| 16 | M | 69 | G1 | Rectum | T2N1aM0 | IIIA | 2 |
| 17 | M | 53 | G2 | Rectum | T4aN0M0 | IIB | 1 |
| 18 | M | 70 | G1 | Left | T4aN0M0 | IIB | 1 |
| 19 | F | 55 | G2 | Right | T4aN1bM0 | IIIB | 1 |
| 20 | M | 75 | G2 | Left | T2N0M0 | I | 1 |
| 21 | M | 45 | G2 | Rectum | T4aN0M0 | IIB | 2 |
| 22 | M | 87 | G2 | Right | T4aN0M0 | IIB | 1 |
| 23 | F | 78 | G2 | Right | T2N0M0 | I | 1 |
| 24 | F | 51 | G2 | Right | T4aN0M0 | IIB | 1 |
| 25 | M | 78 | G1 | Rectum | T4aN0M0 | IIB | 1 |
| 26 | F | 61 | G3 | Left | T4aN1bM0 | IIIB | 1 |
| 27 | M | 63 | G2 | Rectum | T2N2bM0 | IIIB | 1 |
| 28 | F | 59 | G2 | Right | T4aN1M0 | IIIB | 1 |
| 29 | M | 75 | G2 | Rectum | T4aN0M0 | IIB | 1 |
| 30 | M | 72 | G2 | Rectum | T4aN1M0 | IIIB | 1 |
Ethical approval
All clinical tissues were collected from the CRC
patients after obtaining informed consent according to an
established protocol approved by the Ethics Committee of Shanghai
First People's Hospital.
RNA isolation and semi-quantitative
polymerase chain reaction (PCR)
Total RNA was extracted from CRC patient samples
using the SQ DNA/RNA/Protein Tissue kit (#R8042-01; Sangon Biotech
Co., Ltd., Shanghai, China) and used for cDNA synthesis with the
Takara RNA PCR kit (AMV) Ver.3.0 (#RR019; Takara Biotechnology Co.,
Ltd., Dalian, China). MAL and β-actin were detected, and their
primers were as follows: MAL forward, 5′-TGGGTGATGTTCGTGTCTGTG-3′;
and reverse, 5′-TCAAGTTCTACTGCGGCTTTATG-3′; and β-actin forward,
5′-CTGGGACGACATGGAGAAAA-3′ and reverse,
5′-AAGGAAGGCTGGAAGAGTGC-3′.
Immunohistochemical staining
Immunohistochemical staining was performed to detect
the expression of MAL in the CRC and matched non-cancer tissues.
The primary antibody against MAL was obtained from Santa Cruz
Biotechnology Inc. (#sc-66978; Dallas, TX, USA). The intensity of
staining was scored as follows: 0, negative; 1, weak; 2, moderate;
or 3, strong. The extent of staining was based on the percentage of
positive tumor cells: 0, negative; 1, 1–25%; 2, 26–50%; 3, 51–75%;
and 4, 76–100%. The final score of each sample was assessed by the
sum of the results of the intensity and extent of staining.
Therefore, each case was considered negative if the final score was
0–1 (–) or 2–3 (±), and positive if the final score was 4–5 (+) or
6–7 (++), respectively. These scores were determined independently
by two senior pathologists.
Statistical analysis
Statistical analysis was performed using SPSS 16.0
(SPSS, Inc., Chicago, IL, USA) and values are expressed as the mean
± standard deviation. The χ2-test was used to evaluate
the differences in staining of MAL according to the patient and
tumor characteristics. The differences between groups were analyzed
by using Student's t-test. Spearman's rank test was used for
analysis of correlations between MAL RNA and protein levels.
P<0.05 was considered to indicate a statistically significant
difference.
Results
RNA expression level of MAL in CRC and
adjacent tissues
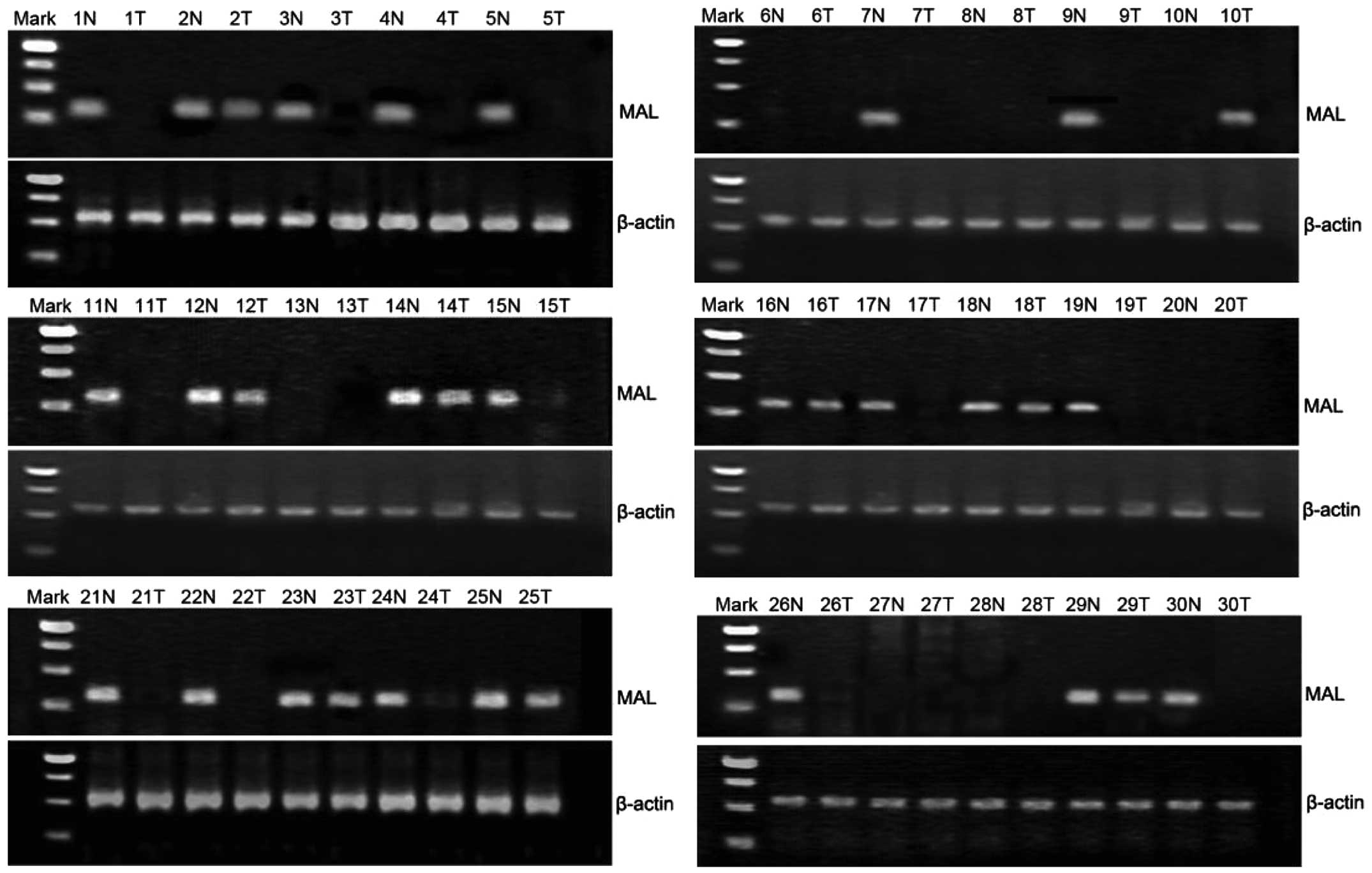
By performing a semi-quantitative PCR assay, MAL RNA
expression was detected in the 30 CRC tissues and the adjacent
non-cancer tissues. The expression of β-actin was considered as the
endogenous control. A total of 9 CRC specimens showed expression of
MAL, with a positive rate of 30% (9/30), and 23 adjacent tissues
showed expression of MAL, with a positive rate of 76.7% (23/30)
(Fig. 1). The RNA level of MAL was
significantly downregulated in the CRC tissues compared with the
adjacent tissues (χ2=13.125; P=0.001).
Protein expression level of MAL in CRC
and adjacent tissues
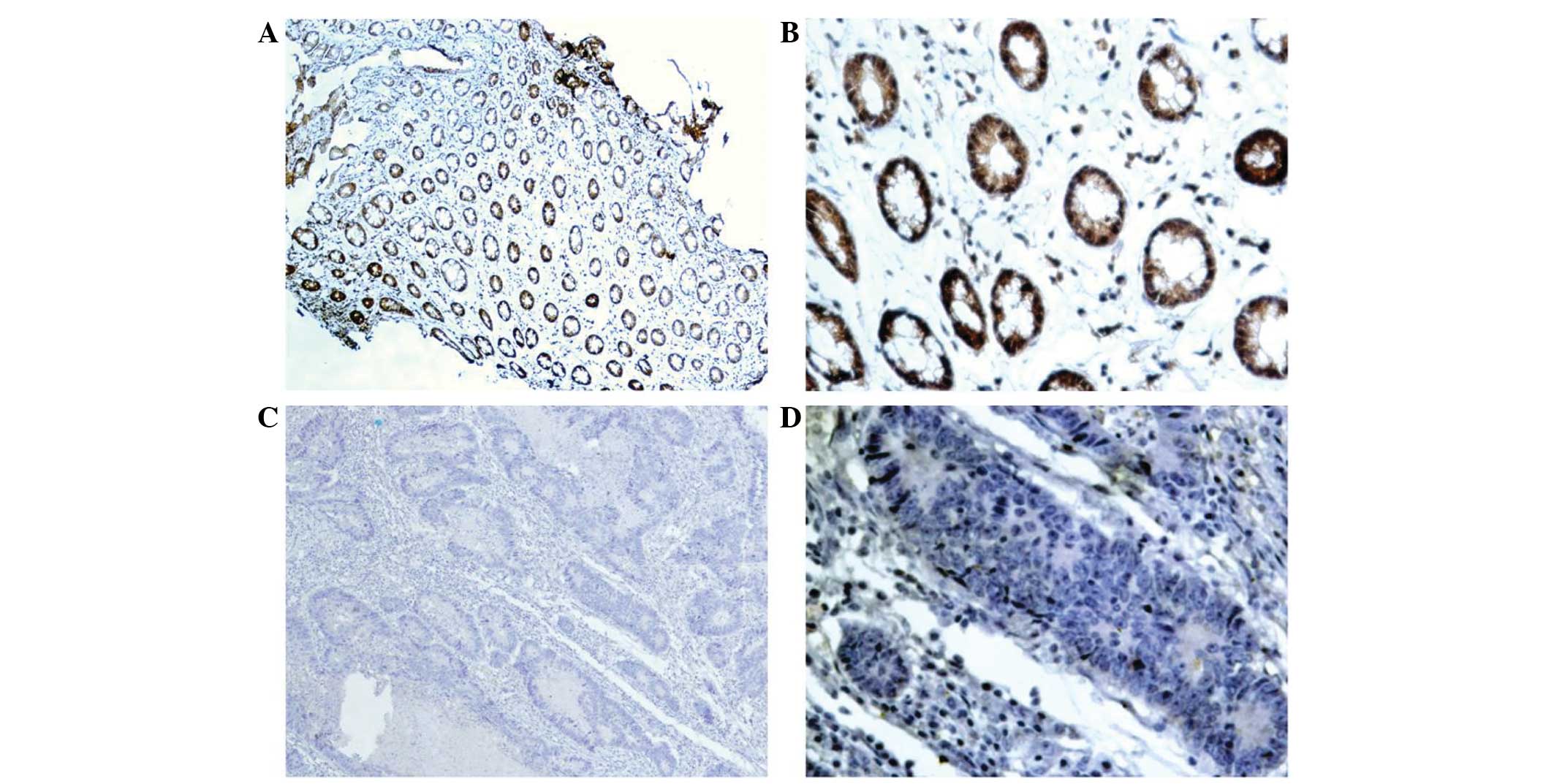
By performing immunohistochemical staining, MAL
protein was detected in the nucleus and cytoplasm of the CRC tumor
cells (Fig. 2). Immunohistochemical
staining results showed that 20% (6/30) of the CRC specimens
expressed MAL, while 66.7% (20/30) of the adjacent tissues
expressed MAL (Table II). The
protein level of MAL was significantly downregulated in the CRC
tissues compared with the adjacent tissues (χ2=13.303;
P=0.001). According to Spearman's rank correlation test, the RNA
level of MAL was significantly positively correlated with the
protein level of MAL (rs=0.818; P=0.0001).
 | Table II.Correlation between MAL protein level
and metastatic status in CRC patients. |
Table II.
Correlation between MAL protein level
and metastatic status in CRC patients.
|
|
| MAL expression |
|
|
|---|
|
|
|
|
|
|
|---|
| TNM stage | Cases, n | (+), n | (–), n | χ2 | P-value |
|---|
| CRC with
metastases | 12 | 0 | 12 | 5.000 | 0.031 |
| CRC without
metastases | 18 | 6 | 12 |
|
|
Correlation between MAL and
pathological staging, metastasis status and clinical
characteristics of the CRC patients
On the basis of the overall evaluation of the
immunohistochemical staining score, the present study investigated
whether MAL protein expression level was correlated with
pathological staging, metastasis status and the clinical
characteristics of the CRC patients. As shown in Table III, statistical analysis indicated
that the level of MAL expression was significantly lower in CRC
tumor-node-metastasis (TNM) stage III than in TNM stages I and II
(χ2=5.735; P=0.021). According to the analysis of lymph
node metastasis, the expression level of MAL was significantly
lower in the cases of CRCs with lymph node metastasis compared with
those without lymph node metastasis (χ2=5.000; P=0.031;
Table II). However, the expression
of MAL showed no significant correlation with the other
pathological factors of the CRC patients, including tumor location,
histological type and differentiation status (Table IV), and the clinical characteristics
of the CRC patients, including age and gender (Table V).
 | Table III.Correlation between MAL protein level
and different TNM stage in the colorectal cancer patients. |
Table III.
Correlation between MAL protein level
and different TNM stage in the colorectal cancer patients.
|
|
| MAL expression |
|
|
|---|
|
|
|
|
|
|
|---|
| TNM stage | Cases, n | (+), n | (–), n | χ2 | P-value |
|---|
| I,II | 17 | 6 | 11 | 5.735 | 0.021 |
| III | 13 | 0 | 13 |
|
|
 | Table IV.Correlation between MAL protein level
and pathological factors in the colorectal cancer patients. |
Table IV.
Correlation between MAL protein level
and pathological factors in the colorectal cancer patients.
|
|
| MAL expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Pathological
factors | Cases, n | (+), n | (–), n | χ2 | P-value |
|---|
| Tumor location | 30 | 6 | 24 | 1.818 | 0.403 |
| Right
colon | 10 | 2 | 8 |
|
|
| Left
colon | 9 | 3 | 6 |
|
|
|
Rectum | 11 | 1 | 10 |
|
|
| Histological
type |
|
|
| 0.833 | N/A |
|
Adenocarcinoma | 27 | 6 | 21 |
|
|
|
Mucinous adenocarcinoma | 3 | 0 | 3 |
|
|
| Differentiation
status |
|
|
| 1.429 | 0.490 |
|
Highly-differentiated | 6 | 2 | 4 |
|
|
|
Moderately-differentiated | 21 | 4 | 17 |
|
|
|
Poorly-differentiated | 3 | 0 | 3 |
|
|
 | Table V.Correlation between MAL protein level
and clinical characteristics in the colorectal cancer patients. |
Table V.
Correlation between MAL protein level
and clinical characteristics in the colorectal cancer patients.
|
|
| MAL expression |
|
|
|---|
|
|
|
|
|
|
|---|
| Clinical
characteristics | Cases, n | (+), n | (–), n | χ2 | P-value |
|---|
| Age, years | 30 | 6 | 24 | 1.500 | 0.553 |
|
<60 | 5 | 0 | 5 |
|
|
|
≥60 | 25 | 6 | 19 |
|
|
| Gender |
|
|
| 0.139 | N/A |
|
Male | 18 | 4 | 14 |
|
|
|
Female | 12 | 2 | 10 |
|
|
Discussion
The formation of polarity in epithelial cells
depends on the rigorous maintenance of the regulation of transport
and sorting process, which guarantees precise delivery of
biosynthetic cargo to varying areas of the plasma membrane
(13). The polarized transport of
lipids and proteins to the plasma membrane is vital for the
functions of the epithelial cells. MAL has been found to be a
pivotal component of the machinery for the direct transport route.
Loss of MAL leads to the loss of the polarized phenotype that
frequently accompanies the neoplastic transformation process
(14). Recent studies have suggested
that downregulation of MAL has been associated with a variety of
human epithelial malignancies, including esophageal (7), breast (8),
ovarian (9) and cervical (15) cancers. For example, Mimori et
al showed that overexpression of MAL in esophageal tumors
exhibited decreased cellular motility, a G1/S transition
block and increased levels of apoptosis via the Fas signaling
pathway (7). Overmeer et al
(9) further demonstrated the
repression of MAL tumor suppressor activity by promoter methylation
during cervical carcinogenesis, providing a predictive biomarker
for underlying high-grade lesions in cervical cancer. Beder et
al (10) further identified
potential genetic and epigenetic mechanisms associated with the
downregulation of MAL in head and neck squamous cell carcinomas,
including loss of heterozygosity, mutation and hypermethylation.
Buffart et al (11) indicated
that MAL promoter hypermethylation can be used as a novel
prognostic marker in gastric cancer.
For the first time, the present study evaluated the
expression of MAL in CRC. RNA and protein expression levels of MAL
were analyzed in 30 CRC patients and found to be significantly
downregulated in the CRC tissues compared with the adjacent tissue,
which was consistent with the previous studies in other epithelial
malignancies. To determine the clinical significance of MAL in CRC,
the correlation between its expression level and pathological
staging, metastasis status and the clinical characteristics of the
CRC patients was investigated. The results showed that the
expression levels of MAL were significantly associated with
different TNM stages and lymph node metastasis, but not with age,
gender, tumor site, differentiation status or pathological type.
These results indicate that MAL has a putative anti-metastasis
function in CRC, and that detection of MAL expression level may be
a potential biomarker for malignant CRC.
Acknowledgements
This study was supported by the Hunan Social
Development Support Program (grant number, 2012SK3190) and the
Hunan Graduate student research innovation project (grant number,
CX2012B090).
References
|
1
|
Jemal A, Siegel R, Ward E, Murray T, Xu J
and Thun MJ: Cancer statistics, 2007. CA Cancer J Clin. 57:43–66.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sung JJ, Lau JY, Goh KL and Leung WK: Asia
Pacific Working Group on Colorectal Cancer: Increasing incidence of
colorectal cancer in Asia: Implications for screening. Lancet
Oncol. 6:871–876. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ji BT, Devesa SS, Chow WH, Jin F and Gao
YT: Colorectal cancer incidence trends by subsite in urban
Shanghai, 1972–1994. Cancer Epidemiol Biomarkers Prev. 7:661–666.
1998.PubMed/NCBI
|
|
4
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou Y, Wan G, Spizzo R, et al: miR-203
induces oxaliplatin resistance in colorectal cancer cells by
negatively regulating ATM kinase. Mol Oncol. 8:83–92. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Alonso MA and Weissman SM: cDNA cloning
and sequence of MAL, a hydrophobic protein associated with human
T-cell differentiation. Proc Natl Acad Sci USA. 84:1997–2001. 1987.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Mimori K, Shiraishi T, Mashino K, et al:
MAL gene expression in esophageal cancer suppresses motility,
invasion and tumorigenicity and enhances apoptosis through the Fas
pathway. Oncogene. 22:3463–3471. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Horne HN, Lee PS, Murphy SK, Alonso MA,
Olson JA Jr and Marks JR: Inactivation of the MAL gene in breast
cancer is a common event that predicts benefit from adjuvant
chemotherapy. Mol Cancer Res. 7:199–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Overmeer RM, Henken FE, Bierkens M, et al:
Repression of MAL tumour suppressor activity by promoter
methylation during cervical carcinogenesis. J Pathol. 219:327–336.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Beder LB, Gunduz M, Hotomi M, et al:
T-lymphocyte maturation-associated protein gene as a candidate
metastasis suppressor for head and neck squamous cell carcinomas.
Cancer Sci. 100:873–880. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Buffart TE, Overmeer RM, Steenbergen RD,
et al: MAL promoter hypermethylation as a novel prognostic marker
in gastric cancer. Br J Cancer. 99:1802–1807. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edge SB and Compton CC: The American Joint
Committee on Cancer: The 7th edition of the AJCC cancer staging
manual and the future of TNM. Ann Surg Oncol. 17:1471–1474. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Schuck S and Simons K: Polarized sorting
in epithelial cells: raft clustering and the biogenesis of the
apical membrane. J Cell Sci. 117:5955–5964. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Marazuela M and Alonso MA: Expression of
MAL and MAL2, two elements of the protein machinery for
raft-mediated transport, in normal and neoplastic human tissue.
Histol Histopathol. 19:925–933. 2004.PubMed/NCBI
|
|
15
|
Lee PS, Teaberry VS, Bland AE, et al:
Elevated MAL expression is accompanied by promoter hypomethylation
and platinum resistance in epithelial ovarian cancer. Int J Cancer.
126:1378–1389. 2010.PubMed/NCBI
|