Introduction
Hepatocellular carcinoma is a common malignant tumor
with poor prognosis. RNA interference (RNAi) can reduce oncogene
expression and alter the biology of tumor cells. Therefore,
application of RNAi to enhance the chemosensitivity of
hepatocellular carcinoma to cisplatin, a commonly used anticancer
drug that inhibits cancer cell DNA replication, is a potentially
important therapeutic strategy (1).
Livin is a member of the inhibitor of the apoptosis
(IAP) family and the baculoviral IAP repeat-containing 7
(BIRC7) gene that encodes Livin is located on human
chromosome 20q13.3, is 46 kb in length and comprises 7 exons and 6
introns (2). Overexpression of Livin
has been observed in certain types of cancer tissues and cell lines
(3). This may inhibit the cellular
apoptosis induced by several anticancer agents. It is also
associated with drug resistance in cancer cells (2,4,5). RNA interference (RNAi) is the phenomenon
by which double-stranded RNA molecules homologous to a target gene
are processed by the enzyme Dicer to produce active small
interfering RNA (siRNA) molecules. siRNAs form a complex called the
RNA-induced silencing complex (RISC) that includes endonucleases
termed argonaut proteins. The RISC, guided by the siRNA, binds to
and specifically cleaves mRNA from the target gene, leading to its
degradation. This is one form of post-translational gene silencing
(2).
In the present study, a eukaryotic expression vector
for Livin was constructed. The use of short hairpin (sh)RNA vectors
is a popular approach for delivering siRNA. Using the RNAi
approach, Livin expression in the HepG2 hepatocellular carcinoma
cells was inhibited and the effects of this on the chemosensitivity
of the cells were investigated.
Materials and methods
Primary reagents
Fetal bovine serum (FBS), Dulbecco's modified Eagle's
medium (DMEM), Opti-MEM, and a Lipofectamine 2000 transfection kit
were purchased from Invitrogen Life Technologies (Carlsbad, CA,
USA). RNAiso Plus, a PrimeScript™ Reverse Transcription (RT)
Reagent Kit with gDNA Eraser, and an LA PCR Amplification Kit
Version 2.1 were purchased from Takara Biotechnology Co., Ltd.
(Dalian, China). Radioimmunoprecipitation assay (RIPA) buffer,
phenylmethylsulfonyl fluoride (PMSF) protease inhibitor and a
BCA-100 protein assay kit were purchased from Genechem Co., Ltd.
(Shanghai, China). Monoclonal mouse anti-human antibodies against
Livin (catalog no. ab118026) and GAPDH (catalog no. ab8245) were
purchased from Abcam (Shanghai, China). Horseradish peroxidase
(HRP)-labeled goat anti-mouse IgG antibody (catalog no. IH-0041)
was purchased from Beijing Dingguo Changsheng Co., Ltd. (Beijing,
China). MTT was purchased from Sigma-Aldrich (St. Louis, MO,
USA).
Cell culture
The human hepatocellular carcinoma cell line HepG2
was obtained from the central laboratory of Provincial Hospital
Affiliated to Shandong University (Shandong, China). HepG2 cells
were seeded at a density of 4×105 cells/well in 6-well
plates with DMEM containing 10% FBS in an incubator at 37°C and 5%
CO2. These cells became adherent and 0.25% Trysin-EDTA
(Gibco Life Technologies, Carlsbad, CA, USA) was used for
passage.
Construction of the Livin shRNA
vector
The full-length mRNA sequence of the Livin gene
(BIRC7; NCBI gene ID, 79444) was identified in the NCBI gene
database. Based on shRNA design principles and relevant literature
(6), two 19-nucleotide (nt) sequences
were selected for use as target sequences for the siRNA vector
construction as follows: Livin 1, 5′-GTG GTT CCC CAG CTG TCAG-3′
(611–629) and Livin 2, 5-GGA AGA GAC TTT GTC CACA-3′ (648–666).
BLAST analysis confirmed that the selected sequences were not
homologous to other human gene sequences. The sequences were
flanked by BamHI and HindIII restriction sites at
each end. Sense and antisense strands were connected by a 9-nt loop
sequence (TTC AAG AGA). Clone construction involved direct
annealing ligation; the shRNA coding fragments flanked by
restriction sites were annealed and then directly ligated (T4 DNA
Ligase; Invitrogen Life Technologies) with the vector prepared by
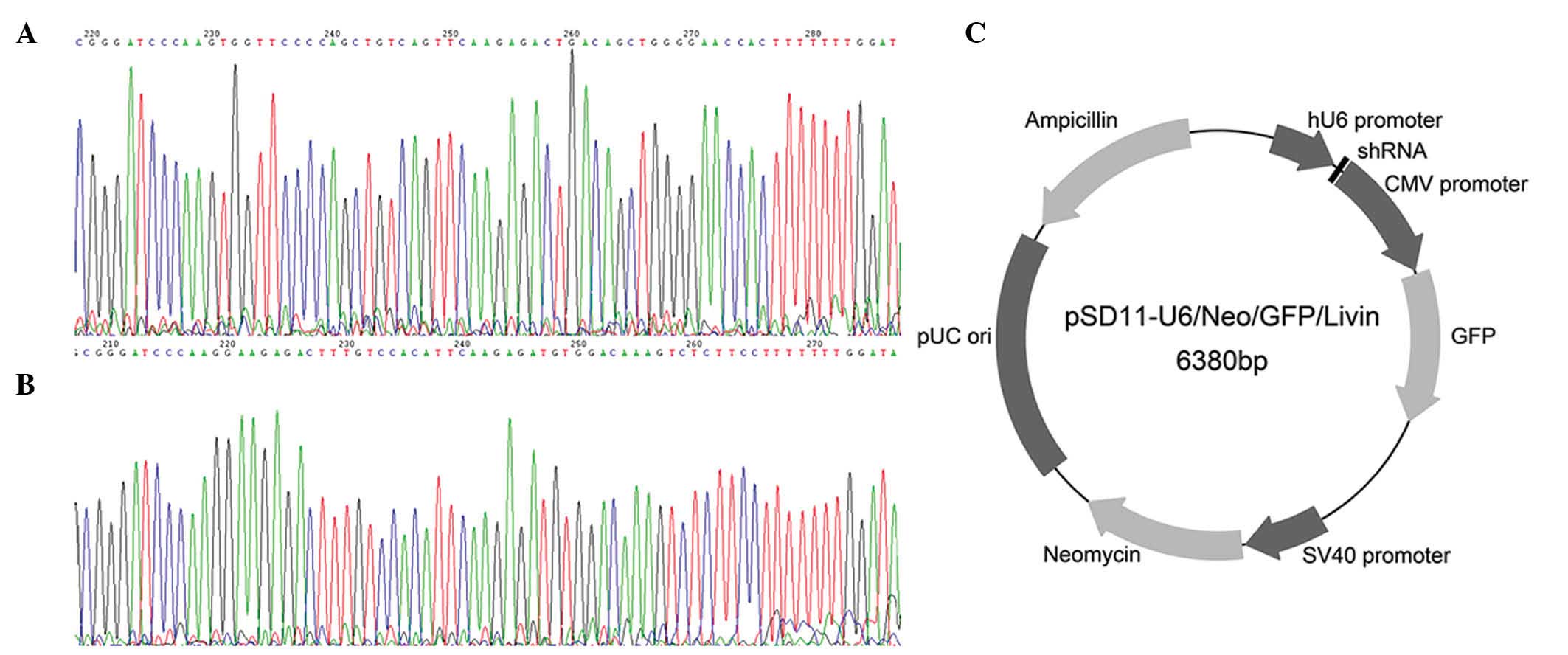
restriction digestion. The structure of the recombinant plasmid
pSD11-U6/Neo/GFP/Livin is presented in Fig. 1. The 2 expression vectors produced in
the present study are designated pSD11-Livin1 and pSD11-Livin2. A
negative control vector, termed pSD11-NC, was also constructed. It
was constructed by re-ligation following the restriction
endonuclease digestion. The construction of the Livin shRNA vector
was performed at Shanghai Jikai Biotechnology Co., Ltd. (Shanghai,
China) and sequencing was performed at Sangon Biotech Co., Ltd.
(Shanghai, China).
Cell transfection
At 24 h prior to transfection, the HepG2 cells were
trypsinized (0.25% trypsin in DMEM) and seeded in 6-well plates at
a density of 2×105 cells/well. The cells were cultured
in DMEM for 24 h, reaching 80% confluence, which was confirmed by
inverted microscopy (CKX31 Inverted Microscope, Olympus
Corporation, Tokyo, Japan). Cell transfection was performed in
accordance with the manufacturer's instructions using the
Lipfectamine 2000 kit. The experiments were performed in 4 groups
as follows: The Livin 1 group (cells transfected with
pSD11-Livin1); the Livin 2 group (cells transfected with
pSD11-Livin2); the NC group (cells transfected with pSD11-NC); and
the control group (untransfected cells).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
At 48 h following transfection, the total RNA was
extracted from cells in each group. RT was performed using the
RNAiso Plus kit according to the manufacturer's instructions. The
reaction conditions were as follows: 37°C for 15 min and 85°C for 5
sec and then the mix was stored at 4°C. Primer Premier software
(version 5.0; http://www.premierbiosoft.com/primerdesign/index.html)
was used to design primers for the fluorescent qPCR. The sequences
of the primers used were as follows: Livin, F
5′-CCATCAGCCCCCATTTCT-3′ and R 5′-CCATCAGCCCCCATTTCT-3′ (the
amplicon was 79 bp in length); GAPDH, F 5′-TGCACCACCAACTGCTTAGCA-3′
and R 5′-TGCACCACCAACTGCTTAGCA-3′ (the amplicon was 87 bp in
length). The primers were synthesized by Shanghai Sangon Biotech
Co., Ltd. The qPCR was performed on a LightCycler 480 Instrument II
using SYBR Green I (Roche Diagnostics, Shanghai, China). The
thermal cycling conditions were as follows: Pre-denaturation at
95°C for 30 sec followed by 40 cycles of amplification at 95°C for
5 sec and 60°C for 20 sec. A melting curve analysis was performed
following PCR. To exclude false positive results, PCR for the Livin
gene and GAPDH internal reference gene were conducted using blank
controls to which no cDNA had been added.
Western blot analysis
At 72 h following transfection, cells were collected
from each group. RIPA lysis buffer and PMSF (at a ratio of 100:1)
were added to the cell pellets and incubated in an ice bath for 30
min. The samples were centrifuged at 2,000 × g for 30 min at 4°C.
The supernatant was collected and mixed with protein loading buffer
(Shenergy Biocolor Bioscience & Technology Company, Shanghai,
China) and incubated at 100°C for 10 min. Protein samples were
resolved by 10% polyacrylamide gel electrophoresis and then
transferred to a polyvinylidene fluoride membrane (Merck Millipore,
Darmstadt, Germany). The membranes were then blocked with
Tris-buffered saline and 0.05% Tween-20 (TBS-T) containing 5%
skimmed milk powder at 37°C for 60 min. The mouse anti-human Livin
(dilution, 1:1,000) and GAPDH (dilution, 1:3,000) antibodies were
added and incubated at 4°C overnight. The membranes were then
washed with TBS-T and incubated with the HRP-labeled goat
anti-mouse IgG antibody (dilution, 1:3,000) with shaking at room
temperature for 1 h. Following thorough washing with TBS-T, the
membranes were immersed in a solution of enhanced chemiluminescence
liquid (Immobilon Western Chemiluminescent HRP Substrate, Merck
Millipore) and incubated for 5 min at room temperature. The images
were visualized using gel imaging equipment; bands from the
electrophoresis were analyzed with an AlphaImager® 2200 image
analyzer (ProteinSimple Bioscience & Technology Co., Ltd.,
Shanghai, China).
MTT assay
One day prior to transfection, HepG2 cells were
seeded in 96-well plates at a density of 5×105
cells/well. Grouping was as described above and each group
consisted of 5 replicates. In addition, a blank group was set up
containing HepG2 cells only and no added reagents. The transfection
procedure was performed using the Lipofectamine 2000 kit, according
to the manufacturer's instructions. At 48 h following transfection,
DMEM containing 10% FBS and 2.0 mg/l cisplatin (central laboratory
of Provincial Hospital Affiliated to Shandong University) was added
to the cells. The cells were cultured for an additional 24, 36 or
48 h. The culture medium was removed and replaced with 20 µl MTT (5
mg/ml) solution at the time points indicated. The cells were
incubated at 37°C with 5% CO2 for 4 h and then 150 µl
dimethyl sulfoxide was added to each well and oscillated for 10
min. The optical density (OD) was measured using a Microplate
Reader (DNM-9602; Nanjing Perlove Eadial-Video Equipment Co., Ltd.,
Nanjing, China) at 490 nm. The rate of cell growth inhibition (IR)
was calculated using the following formula: IR = (1 -
ODExperimental/ODBlank) × 100.
Statistical analysis
SPSS statistical software, version 15.0 (SPSS, Inc.,
Chicago, IL, USA) was used for analysis. Data are expressed as the
mean ± standard error. The statistical significance was evaluated
by a one-way analysis of variance with Tukey's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Verification of construction of
recombinant plasmid
DNA sequencing confirmed that the shRNA coding
sequences were correctly inserted into the plasmid vector
pSD11-U6/Neo/GFP and that recombinant plasmids pSD-11-Livin1 and
pSD11-Livin2 were as designed. Thus, the recombinant plasmids were
successfully prepared.
HepG2 cellular mRNA expression changes
following transfection
RT-qPCR was performed to quantify the changes in
Livin mRNA expression levels. The melting curve for the Livin PCR
product peaked at 89.9°C, and the melting curve for the GAPDH PCR
product peaked at 88.1°C. The PCR products produced a single peak,
excluding the possibility of non-specific amplification. The
negative control group without cDNA did not produce a melting
curve, excluding the possibility of false positive results. GAPDH
served as an internal reference gene and relative quantification
(Livak method) (7) was used to
calculate the relative expression level, presented as ΔΔCt. The
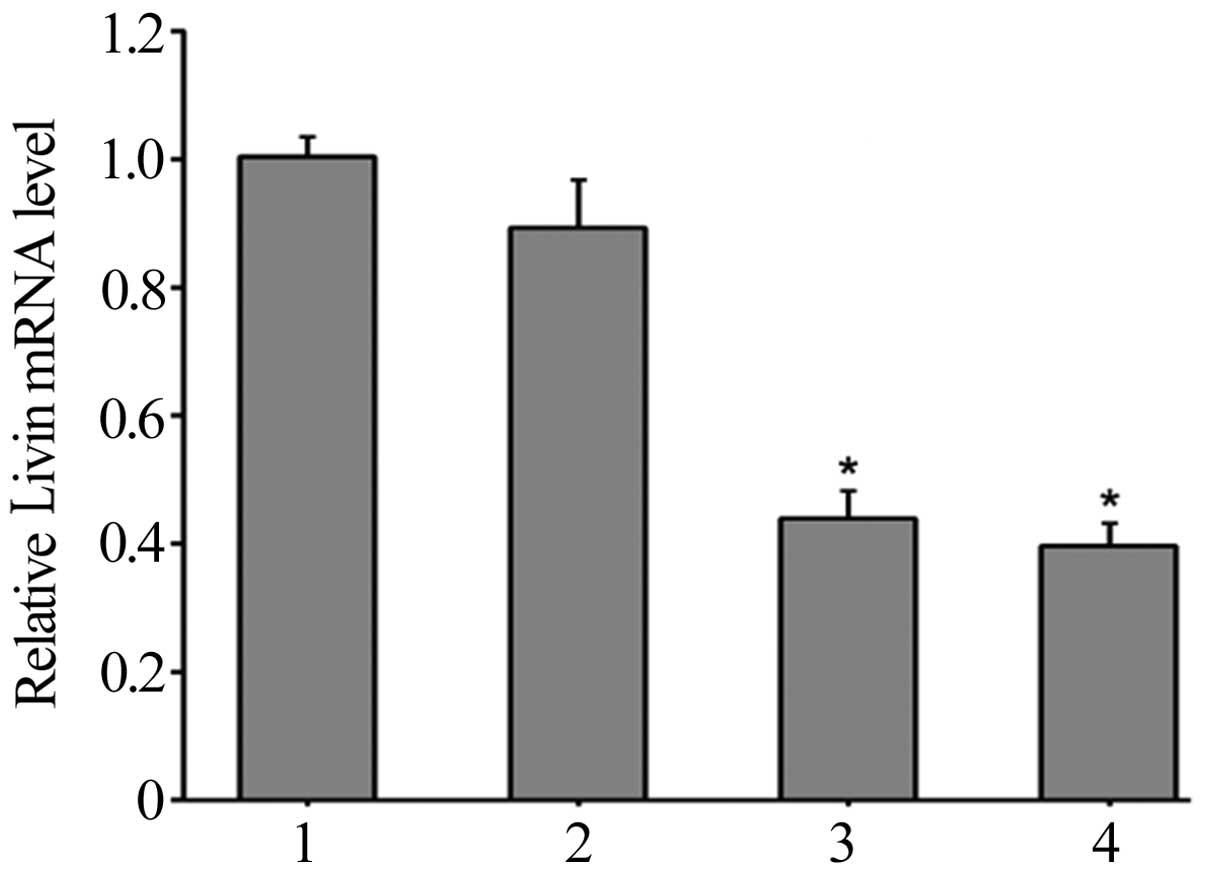
results demonstrated that 48 h following transfection, the relative
levels of Livin mRNA expression in HepG2 cells in the Livin 1,
Livin 2, NC and control groups were 0.44±0.04, 0.40±0.03, 0.90±0.07
and 1.00±0.03, respectively (Fig. 2).
The expression levels in the Livin 1 and Livin 2 groups were
significantly lower than those of the NC and control groups
(P<0.05). These results indicated that pSD11-Livin1 and
pSD-11-Livin2 transfection in HepG2 cells specifically reduced the
expression levels of Livin mRNA (Fig.
2).
Changes in Livin protein expression in
HepG2 cells following transfection
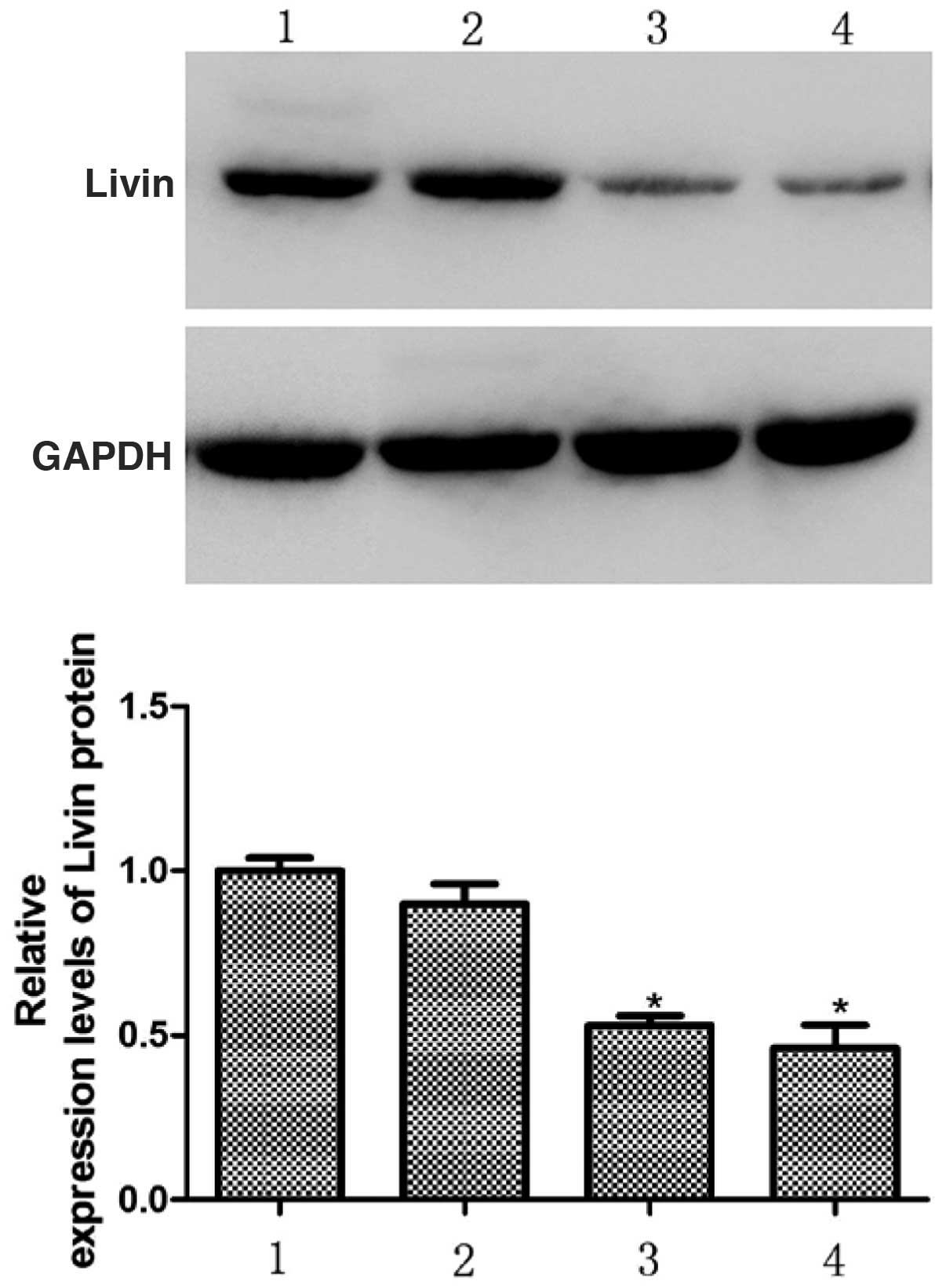
Bands from the western blots were analyzed using an
Alpha Imager 2200. The results of this analysis demonstrated that
72 h following transfection, the relative expression levels of
Livin protein in the HepG2 cells in the Livin 1, Livin 2 and NC
groups were 0.53±0.03, 0.46±0.07, and 0.90±0.06, respectively
relative to the control group, which was set to 1. There were
significantly lower protein levels in the Livin 1 and Livin 2
groups compared with the NC and control groups (P<0.05). These
results indicate that pSD-11-Livin1 and pSD11-Livin2 transfection
in HepG2 cells specifically downregulates the expression of Livin
protein (Fig. 3).
Inhibition of HepG2 cell growth by
cisplatin
The results of the MTT assay demonstrated that at a
cisplatin concentration of 2 mg/l, each group exhibited various
degrees of growth inhibition. This was recorded at 24, 36 and 48 h,
and the rate of inhibition gradually increased over time. The rates
of cell growth inhibition in the Livin 1 and Livin 2 groups were
significantly higher than in the NC and control groups (P<0.05;
Table I). No statistically
significant differences were identified between the Livin 1 and
Livin 2 groups, or between the NC and control groups (Table I).
 | Table I.Rate of cell growth inhibition (%) by
cisplatin in HepG2 cells following transfection with Livin shRNA
expression vectors at different time points. |
Table I.
Rate of cell growth inhibition (%) by
cisplatin in HepG2 cells following transfection with Livin shRNA
expression vectors at different time points.
|
| Percentage cell
growth inhibition at different time points |
|---|
|
|
|
|---|
| Group | 24 h | 36 h | 48 h |
|---|
| Control | 6.12±0.88 | 14.75±1.10 | 23.03±1.07 |
| NC | 6.51±1.03 | 15.34±1.11 | 23.59±1.32 |
| Livin 1 |
15.87±0.84a |
28.00±0.85a |
36.76±1.20a |
| Livin 2 |
16.57±1.03a |
28.10±0.96a |
37.10±1.48a |
Discussion
Hepatocellular carcinoma is a common type of
malignant tumor in gastroenterological clinical practice. The
efficacy of traditional therapeutics for hepatocellular carcinoma,
including surgical resection and chemotherapy, is often limited and
the disease is difficult to cure; this may be related to the
presence of chronic liver conditions, such as hepatitis and
cirrhosis. The relationship between changes in gene expression in
cancer cells and their chemosensitivity has become an important
part of anticancer research (8,9). RNAi
provides considerable potential in this line of research. Ashhab
et al (10) described 6
approaches for introducing siRNAs into mammalian cells. Of these
approaches, the design and introduction of shRNA expression vectors
into mammalian cells that produce siRNA against a target gene
allows relatively stable and specific RNAi without any of the
effects of directly introducing siRNA into cells. This strategy
also permits inhibition of multiple variants of a single gene or a
sequence shared among multiple genes (11).
The post-transcriptional processing of the Livin
gene forms 2 types of mature Livin mRNA, Livin α and Livin β, which
contain 1,351 and 1,297 nts, respectively. The difference between
these 2 types of mRNA is a fragment of 54 nts; the rest of the
sequence is the same. Livin α and β encode a protein with the
required structure for anti-apoptotic function, a baculovirus IAP
repeat (BIR) domain. Although the 2 proteins differ in 18 amino
acid residues in this domain, their functions are essentially
similar; to bind caspase via the BIR domain, inhibiting its
activity and thus inhibiting cancer cell mitochondrial apoptosis
(12,13). A number of studies have demonstrated
that the overexpression of Livin in certain types of cancer tissues
and cell lines can inhibit the apoptosis induced by multiple
anticancer agents (14,15). Other previous studies have
demonstrated Livin overexpression to be associated with drug
resistance in cancer cells (16–18).
Previous studies have also produced shRNA expression vectors and
used them to stably silence the Livin gene in human bladder cancer
T24 cells (19), HeLa cervical cancer
(20) and Lovo colon cancer (3) cell lines. The impact of shRNA expression
vectors on cancer cell growth and apoptosis has also been studied.
Liang et al (21) demonstrated
that Livin expression in HepG2 cells increased significantly
following the addition of a chemotherapeutic agent, contributing to
resistance to chemotherapy. To the best of our knowledge, there
have not yet been any studies on the use of RNAi to reduce Livin
expression in HepG2 hepatocellular carcinoma cells, or the effects
on chemosensitivity.
In the present study, 2 target sequences were
selected from the sequence shared by Livin α and β mRNAs, and were
used to construct shRNA eukaryotic vectors. Following transfection
of the hepatocellular carcinoma cell line HepG2 with these vectors,
the expression levels of Livin mRNA and protein were evaluated with
RT-qPCR and western blot analysis. The results demonstrated that
Livin expression levels were downregulated in HepG2 cells
transfected with pSD11-Livin1 and pSD-11-Livin2. The differences
from cells transfected with pSD11-NC and control cells were
statistically significant, indicating the effectiveness of the
selected target sequences. Further experiments were performed on
HepG2 cells 48 h following Livin transfection. These cells were
treated with 2.0 mg/l cisplatin for various lengths of time and
then an MTT assay was performed to determine the absorbance (OD
values) and to calculate the rates of cell growth inhibition. The
results demonstrated that the cell growth inhibition rate
attributable to cisplatin in HepG2 cells transfected with
pSD11-Livin1 and pSD11-Livin2 were significantly higher than those
in cells transfected with pSD11-NC and untransfected control cells.
The present study also indicated that transfection with an shRNA
eukaryotic expression vector against Livin mRNA may inhibit Livin
gene expression and effectively increase the chemosensitivity of
hepatocellular carcinoma cells. The present study provides novel
experimental evidence that manipulating the expression of the Livin
gene in hepatocellular carcinoma cells can affect their
chemosensitivity.
It has been previously demonstrated that
chemotherapeutic agents affect the changes made by Livin gene
silencing on apoptotic pathways in cancer cells (21,22).
Crnkovic-Mertens et al (7)
demonstrated that in HeLa cells treated with Livin shRNA and a
reduced dose of chemotherapeutic agent, Livin gene expression
reduced and caspase-3 expression increased significantly. Quintieri
et al (23) stated that the
activation of caspases is key to the cancer cell apoptosis that is
induced by chemotherapeutic agents, and that the reduction in
caspase activity caused by the factors in cancer cells is a notable
cause of the reduction in their chemosensitivity. In the present
study, Livin gene expression was increased in hepatocellular
carcinoma cells, which may alter the apoptotic pathways, leading to
the inhibition of apoptosis induced by chemotherapeutic agents. The
combined use of chemotherapeutic agents and RNAi technology to
investigate the changes in apoptotic pathways in cancer cells
warrants further study. The potential results may aid in the
improvement of canonical chemotherapeutic agents and the
development of novel adjuvant anticancer drugs.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Shandong province (grant no. Y2008C110,
awarded to Professor Hong Chang).
References
|
1
|
Sibley CR, Seow Y and Wood MJ: Novel
RNA-based strategies for therapeutic gene silencing. Mol Ther.
18:466–476. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen X, Wang T, Yang D, et al: Expression
of thr IAP protein family acts cooperatively to predict prognosis
in human bladder cancer patients. Oncol Lett. 5:1278–1284.
2013.PubMed/NCBI
|
|
3
|
Liu X, Wang A, Gao H, et al: Expression
and role of the inhibitor of apoptosis protein livin in
chemotherapy sensitivity of ovarian carcinoma. Int J Oncol.
41:1021–1028. 2012.PubMed/NCBI
|
|
4
|
Wang X, Xu J, Ju S, et al: Livin gene
plays a role in drug resistance of colon cancer cells. Clin
Biochem. 43:655–660. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dubrez-Daloz L, Dupoux A and Cartier J:
IAPs: more than just inhibitors of apoptosis proteins. Cell Cycle.
7:1036–1046. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Crnkovic-Mertens I, Hoppe-Seyler F and
Butz K: Induction of apoptosis in tumor cells by siRNA-mediated
silencing of the livin/ML-IAP/KIAP gene. Oncogene. 22:8330–8336.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kempkensteffen C, Hinz S, Krause H, et al:
Expression of splicing variants of the inhibitor of apoptosis livin
in testicular germ cell tumors. Tumour Biol. 29:76–82. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Augello C, Caruso L, Maggioni M, et al:
Inhibitors of apoptosis proteins (IAPs) expression and their
prognostic significance in hepatocellular carcinoma. BMC Cancer.
9:1252009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashhab Y, Alian A, Polliack A, et al: Two
splicing variants of a new inhibitor of apoptosis gene with
different biological properties and tissue distribution pattern.
FEBS Lett. 495:56–60. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brummelkamp TR, Bernards R and Agami R: A
system for stable expression of short interfering RNAs in mammalian
cells. Science. 296:550–553. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Takeuchi H, Kim J, Fujimoto A, et al:
X-Linked inhibitor of apoptosis protein expression level in
colorectal cancer is regulated by hepatocyte growth factor/C-met
pathway via Akt signaling. Clin Cancer Res. 11:7621–7628. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuan D, Liu L, Xu H and Gu D: The effects
on cell growth and chemosensitivity by livin RNAi in non-small cell
lung cancer. Mol Cell Biochem. 320:133–140. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang R, Lin F, Wang X, et al: Silencing
Livin gene expression to inhibit proliferation and enhance
chemosensitivity in tumor cells. Cancer Gene Ther. 15:402–412.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xu M, Xia LP, Fan LJ, et al: Livin and
caspase-3 expression are negatively correlated in cervical squamous
cell cancer. Eur J Gynaecol Oncol. 34:152–155. 2013.PubMed/NCBI
|
|
16
|
Myung DS, Park YL, Chung CY, et al:
Expression of Livin in colorectal cancer and its relationship to
tumor cell behavior and prognosis. PLoS One. 8:e732622013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li F, Yin X, Luo X, et al: Livin promotes
progression of breast cancer through induction of
epithelial-mesenchymal transition and activation of AKT signaling.
Cell Signal. 25:1413–1422. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li X, Fan S, Li L, et al: RNA
interference-mediated knockdown of Livin suppresses cell
proliferation and invasion and enhances the chemosensitivity to
cisplatin in human osteosarcoma cells. Int J Oncol. 43:159–168.
2013.PubMed/NCBI
|
|
19
|
Yang D, Song X, Zhang J, et al:
Suppression of livin gene expression by siRNA leads to growth
inhibition and apoptosis induction in human bladder cancer T24
cells. Biosci Biotechnol Biochem. 74:1039–1044. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yu L and Wang Z: Effects of Livin gene RNA
interference on apoptosis of cervical cancer HeLa cells and
enhanced sensitivity to cisplatin. J Huazhong Univ Sci Technolog
Med Sci. 29:625–630. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liang SR, Hu GR, Fang LJ, et al: CpG
oligodeoxynucleotides enhance chemosensitivity of 5-fluorouracil in
HepG2 human hepatoma cells via downregulation of the antiapoptotic
factors survivin and livin. Cancer Cell Int. 13:1062013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Choi J, Hwang YK, Sung KW, et al:
Expression of Livin, an antiapoptotic protein, is an independent
favorable prognostic factor in childhood acute lymphoblastic
leukemia. Blood. 109:471–477. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Quintieri L, Fantin M and Vizler C:
Identification of molecular determinants of tumor sensitivity and
resistance to anticancer drugs. Adv Exp Med Biol. 593:95–104. 2007.
View Article : Google Scholar : PubMed/NCBI
|