Introduction
Breast cancer is the most prominent health problem
for women worldwide, affecting one in eight women (1–3). Over the
past three decades, there have been numerous advances for breast
cancer patients, including earlier detection, increased treatment
options and more prevention strategies. However, breast cancer
remains to be the second highest cause of cancer-associated
mortality in women (1,2). Breast cancer progression, such as
metastasis to distant organs, is significantly associated with
cancer-associated mortality (1,3). Thus, it
is crucial to identify biomarkers to predict prognosis and
treatment response, and elucidate novel therapeutic targets of
breast cancer.
Proteolytic activity has been reported to be
significantly associated with cancer metastasis and altered
proteolytic activities was also found to cause a wide range of
diseases, including cancer (4). To
date, ~500–600 proteases have been identified in the human genome
(5). Among them, up to 60 are
lysosomal proteases and cathepsins; cathepsins are a group of
lysosomal or cysteine proteases, which include 16 members in humans
(6–9).
Expression of cathepsins has been shown to be upregulated in
various types of human cancers (10–12); in
addition, altered expression of cathepsins has been associated with
tumor angiogenesis, proliferation and invasion (13,14). For
example, during cancer progression, translocated or secreted
cathepsins were reported to promote tumor cell invasion and
metastasis (14).
However, different cathepsins may have different
roles in human cancers. Cathepsins B and D were often found to be
overexpressed and associated with invasive and metastatic
phenotypes of various types of human cancers (15–18).
However, the role of each member of the cathepsin family in breast
cancer remains to be fully elucidated and to date there have been a
limited number of studies on the function of different members of
the cathepsin family in breast cancer. A small number of previous
studies have reported the altered expression of cathepsins D, B and
E in breast carcinogenesis (19–23). In
addition, previous studies have reported that cathepsin D was
overexpressed in invasive ductal carcinoma compared with its
expression in lobular carcinoma of the mammary glands (21,24,25).
Furthermore, overexpression of cathepsin D was associated with
lymph node metastasis of breast invasive ductal carcinoma (21).
Notably, the design of small molecular inhibitors
targeting cathepsins is a strategy that has been used previously
against human cancer, among other diseases (26). Each cathepsin has a different cleavage
bond-specificity for substrate proteins, thus allowing for the
development of cathepsin-specific inhibitors for targeting
different family members (26,27). Such
inhibitors include E-64, leupeptin and antipain (26,28,29). These
inhibitors are able to selectively suppress cathepsin activity and
inhibit tumor growth. For example, anti-cathepsin B and D have been
demonstrated to have efficacy in cancer therapy (16,27,30).
Detailed comparison studies of the expression of
different members of the cathepsin family in breast cancer are
limited. Therefore, the aim of the present study was to analyze the
expression of six important members of cathepsin family, including
cathepsins B, D, G, K, L and V, in breast cancer tissue specimens.
In addition, the expression of these cathepsins was compared with
that of reversion-inducing cysteine-rich protein with Kazal motifs
(RECK) and vascular endothelial growth factor (VEGF). The
expression of these proteins was then analyzed for associations
with the clinicopathological data of patients and breast cancer
patient survival rates. These data may allow for the development of
novel therapies for the treatment of breast cancer patients. In
addition, the results of the present study may elucidate cathepsins
that have a more dominant role in breast cancer compared with other
family members, thus providing evidence for their use in predicting
the prognosis of breast cancer patients as well as the development
of novel cathepsin-specific treatment options.
Materials and methods
Patient specimens
In the present study, tissue specimens were
retrospectively collected from 188 breast cancer patients who
underwent treatment at the Liaoning Cancer Hospital and Institute
(Liaoning, China). All patients were pathologically diagnosed with
breast invasive ductal carcinoma. The present study was approved by
the hospital review board of the Liaoning Cancer Hospital and
Institute and each patient provided written informed consent for
their participation in the study. In addition, follow-up data
regarding survival and adverse effects were also collected for the
present study at follow-up clinic visits or via phone interview.
The detailed clinicopathological data, including age, gender,
pathological type, clinical symptoms and stage, lymph node and
distant metastasis, expression of estrogen receptor (ER),
progesterone receptor (PR) and erythroblastic leukemia viral
oncogene homolog 2 (erbB2), menstrual status and laboratory tests,
were collected from the patients' medical records.
Immunohistochemistry
All tissue specimens were fixed in 4%
paraformaldehyde (Sigma-Aldrich, St. Louis, MO, USA) and then
dehydrated and embedded in paraffin (Sigma-Aldrich). For
immunohistochemistry, the tissue blocks were cut into 4-cm-thick
sections and placed in vertical plastic boxes until further use.
The sections were dewaxed in analytical reagent (AR) grade xylene
AR and rehydrated through graded alcohol series (AR grade; 100, 95,
90, 80 and 70%). For antigen retrieval, the sections were heated in
a citrate buffer (0.01 M; pH 6.0) using a microwave oven (800 W;
Haier, Qingdao, China) for 30 min at 93°C. Following washing three
times in phosphate-buffered saline (PBS; Abcam, Cambridge, MA, USA)
for 10 min each, the sections were incubated with 3% bovine serum
albumin (Abcam) at 37°C for 30 min. Tissues were then incubated
with primary antibodies, including antibodies against cathepsin B
(rabbit anti-human polyclonal; 1:50 dilution; cat no. 12216-1-AP),
cathepsin K (rabbit anti-human polyclonal; 1:100 dilution; cat no.
11239-1-AP), cathepsin D (mouse anti-human monoclonal; 1:200
dilution; cat no. 2926S), cathepsin G (rabbit anti-human
polyclonal; 1:600 dilution; cat no. ab64891), cathepsin L (mouse
anti-human monoclonal; 1:100 dilution; cat no. ab6314), cathepsin V
(mouse anti-human monoclonal; 1:200 dilution; cat no. ab24508),
VEGF (mouse anti-human monoclonal; 1:200 dilution; cat no.
ab464154) and RECK (mouse anti-human monoclonal; 1:40 dilution; cat
no. 611512) overnight at 4°C. Cathepsin B and K were obtained from
Proteintech (Chicago, IL, USA); cathepsin D, G, L and V, and VEGF
were obtained from Abcam; and RECK was obtained from Becton
Dickinson (Franklin lakes, New Jersey, USA). The following day, the
sections were washed three times with PBS and then further
incubated with the corresponding secondary antibodies (Abcam) at
37°C for 30 min. For color reactions, the sections were washed
again with PBS for three times of 5 min each and then stained with
a diaminobenzidine solution (Abcam) for up to 5 min. Protein
expression in tumor cells was assessed for proportion and intensity
score independently by two experienced pathologists. Specifically,
the intensity score was obtained by the average intensity of
positive cells, which was scored as follows: 0, none; 1, weak; 2,
intermediate; and 3, strong. The proportion score was determined
according to the proportion of positive cells as follows: 0, none;
1, ≤10%; 2, 11–25%; 3, 26–50%; and 4, >50%. The final score for
each protein was calculated by adding the scores for proportion and
intensity, then statistically analyzed as low (total score, 0–2)
vs. high (total score, 3–8) expression.
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 statistical software package (SPSS Inc., Chicago, IL,
USA). The Fisher exact test or χ2 test was used to
analyze the difference between two groups, while the hazard ratios
and 95% confidence intervals were used to calculate the difference
between low and high expression groups. Cumulative survival curves
were generated using the Kaplan-Meier method and analyzed using a
Cox regression test. Univariate and multivariate analyses were
performed to analyze the association between clinicopathological
parameters and patient outcomes. P<0.05 was considered to
indicate a statistically significant difference between values.
Results
Expression of cathepsins B, D, G, K, L
and V as well as RECK and VEGF in breast cancer tissue
specimens
In the present study, the expression of cathepsins
B, D, G, K, L and V was assessed using immunohistochemistry. In
addition, cathepsin expression levels were compared with those of
RECK, a metastasis suppressor with protease inhibitor-like domains,
and VEGF, which is involved in the proliferation and metastasis of
cancer cells. As shown in Fig. 1, all
members of these cathepsin families as well as RECK and VEGF
demonstrated positive cytoplasmic staining in these breast cancer
tissues. Of note, cathepsin B was expressed in 109/142 (76.76%),
cathepsin D in 91/155 (58.71%), cathepsin G in 99/171 (57.89%),
cathepsin K in 97/143 (67.83%), cathepsin L in 95/166 (57.23%),
cathepsin V in 45/164 (27.44%), RECK in 67/175 (38.29%) and VEGF in
100/164 (60.98%) samples of breast cancer tissues. Due to
unsuccessful staining in certain samples, staining information
could not be obtained from the total 188 samples.
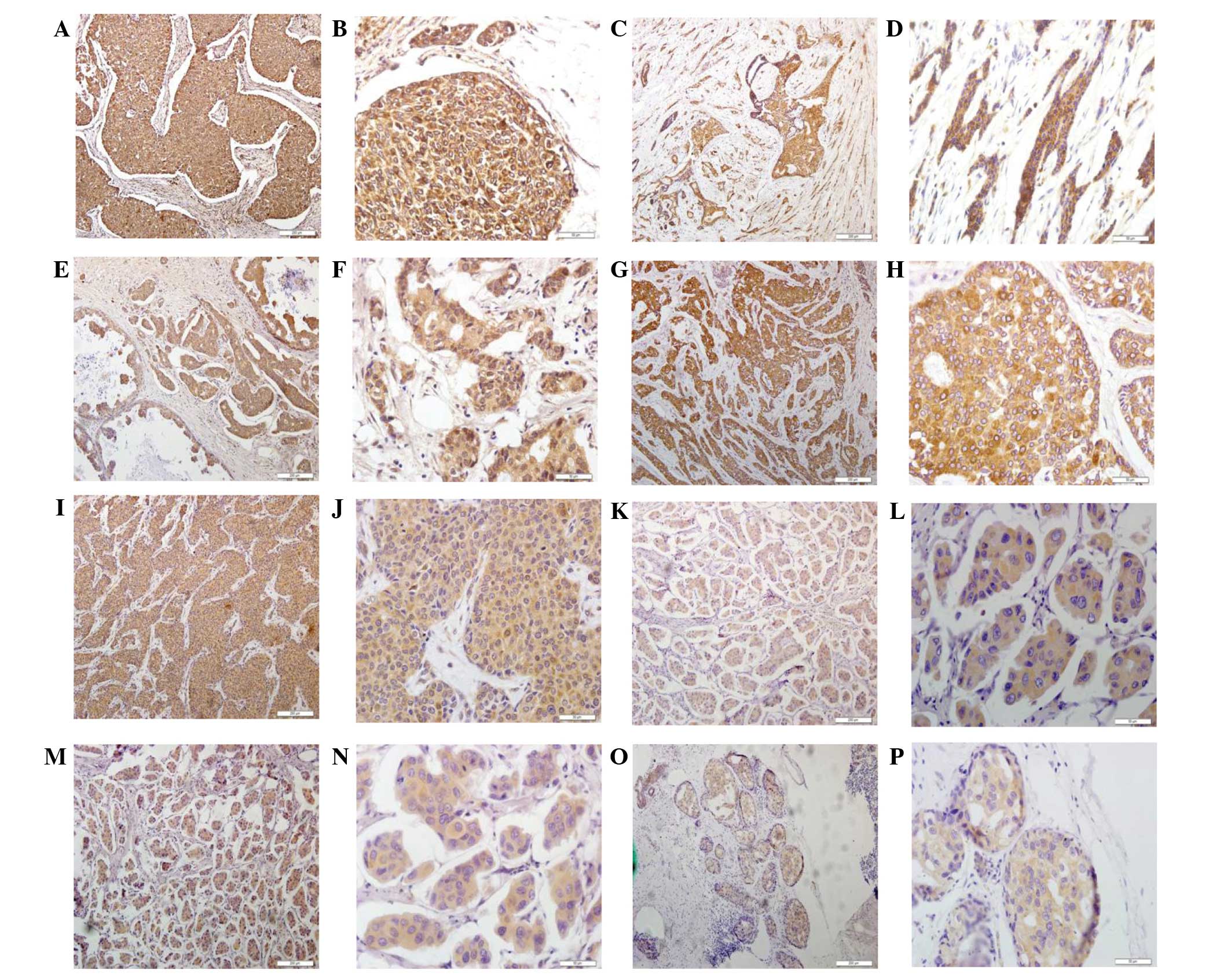 | Figure 1.Detection of cathepsins B, D, G, K, L
and V as well as RECK and VEGF proteins in breast cancer tissue
specimens using immunohistochemistry. Representative images were
selected to reflect the expression of (A and B) cathepsin B, (C and
D) cathepsin C, (E and F) cathepsin D, (G and H) cathepsin K, (I
and J) cathepsin L, (K and L) RECK, (M and N) cathepsin V
expression and (O and P) VEGF in breast cancer tissues.
Magnification, ×200. RECK, reversion-inducing cysteine-rich protein
with Kazal motifs; VEGF, vascular endothelial growth factor. |
Associations between cathepsins B, D,
G, K, L and V as well as RECK and VEGF expression in breast cancer
tissue specimens
The expression of certain proteins may have an
effect on the expression of other proteins in the same family. In
order to determine whether cathepsins interact to effect each
others expression, the dependence in expression of different
members of the cathepsin family, as well as RECK and VEGF, was
analyzed. The results revealed that the expression of cathepsin B
was associated with the expression of cathepsins D (P=0.036) and L
(P=0.002), while cathepsin D expression was associated with the
expression of cathepsins G (P=0.016), L (P=0.003) and V (P=0.006)
(Table I). In addition, cathepsin G
expression was associated with the expression of cathepsins K
(P=0.023), L (P=0.035) and V (P=0.025) (Table I). Furthermore, cathepsin L expression
was associated with that of VEGF (P=0.032), while VEGF expression
was associated with RECK expression (P=0.001) (Table I).
 | Table I.Association of these protein
expressions in breast cancer tissues. |
Table I.
Association of these protein
expressions in breast cancer tissues.
|
| Cathepsin D | Cathepsin G | Cathepsin K | Cathepsin L | Cathepsin V | VEGF | RECK |
|---|
|
|
|
|
|
|
|
|
|
|---|
| Variable | L | H | P | L | H | P | L | H | P | L | H | P | L | H | P | L | H | P | L | H | P |
|---|
| Cathepsin B |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 55 | 8 | 0.036 | 49 | 17 | 0.105 | 36 | 16 | 0.830 | 52 | 14 | 0.002 | 42 | 18 | 0.439 | 30 | 38 | 0.100 | 43 | 29 | 0.625 |
| + | 41 | 16 |
| 39 | 25 |
| 37 | 15 |
| 33 | 29 |
| 48 | 15 |
| 17 | 40 |
| 35 | 28 |
|
| Cathepsin D |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | 89 | 28 | 0.016 | 67 | 26 | 0.969 | 82 | 30 | 0.003 | 85 | 24 | 0.006 | 45 | 68 | 0.152 | 80 | 37 | 0.248 |
| + | – | – | – | 18 | 15 |
| 21 | 8 |
| 15 | 18 |
| 17 | 15 |
| 8 | 23 |
| 18 | 14 |
|
| Cathepsin G |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | – | – | – | 75 | 21 | 0.023 | 81 | 30 | 0.035 | 84 | 25 | 0.025 | 41 | 69 | 0.925 | 78 | 31 | 0.073 |
| + | – | – | – | – | – | – | 21 | 15 |
| 25 | 20 |
| 26 | 18 |
| 16 | 26 |
| 22 | 21 |
|
| Cathepsin K |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | – | – | – | – | – | – | 68 | 26 | 0.124 | 69 | 23 | 0.747 | 36 | 55 | 0.180 | 66 | 33 | 0.259 |
| + | – | – | – | – | – | – | – | – | – | 21 | 15 |
| 26 | 10 |
| 10 | 27 |
| 22 | 17 |
|
| Cathepsin L |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | – | – | – | – | – | – | – | – | – | 78 | 27 | 0.292 | 44 | 58 | 0.032 | 72 | 37 | 0.111 |
| + | – | – | – | – | – | – | – | – | – | – | – | – | 31 | 16 |
| 12 | 36 |
| 27 | 24 |
|
| Cathepsin V |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 45 | 62 | 0.059 | 70 | 45 | 0.673 |
| + | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 11 | 32 |
| 24 | 18 |
|
| VEGF |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 47 | 12 | 0.001 |
| + | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 54 | 46 |
|
Association of cathepsins B, D, G, K,
L and V as well as RECK and VEGF expression with
clinicopathological parameters
The expression of cathepsins B, D, G, K, L and V as
well as RECK and VEGF were subsequently evaluated for their
associations with the clinicopathological parameters of breast
cancer patients. The results demonstrated that cathepsin L
expression was associated with advanced tumor stage, while the
expression of cathepsins B (P=0.033) and K (P=0.038) was associated
with ER expression (P=0.022) (Table
II). In addition, cathepsin K expression was found to be
associated with PR expression (P=0.022) and it was revealed that
RECK expression was associated with ErbB2 expression (P=0.023) as
well as lymph node metastasis (P=0.023) (Table II). Furthermore, cathepsin V
expression was associated with distant metastasis (P=0.035), while
cathepsin D expression was associated with breast cancer metastasis
to the chest (P=0.041) (Table II).
However, there was no association identified between protein
expression and patient age, tumor size, histological grade,
menopausal status, bone metastasis, lung metastasis, hepatic
metastasis, brain metastasis or opposite breast metastasis
(Table II).
 | Table II.Association of cathepsins B, D, C, K,
L and V as well as RECK and VEGF expression with
clinicopathological parameters from breast cancer patients. |
Table II.
Association of cathepsins B, D, C, K,
L and V as well as RECK and VEGF expression with
clinicopathological parameters from breast cancer patients.
|
| Cathepsin B | Cathepsin D | Cathepsin G | Cathepsin K | Cathepsin L | Cathepsin V | RECK | VEGF |
|---|
|
|
|
|
|
|
|
|
|
|
|---|
| Characteristic | Low | High | Low | High | Low | High | Low | High | Low | High | Low | High | Low | High | Low | High |
|---|
| Age |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
<50 | 41 | 37 | 66 | 21 | 68 | 26 | 59 | 19 | 60 | 31 | 63 | 31 | 62 | 35 | 33 | 59 |
|
>50 | 36 | 28 | 56 | 12 | 55 | 21 | 44 | 21 | 55 | 20 | 55 | 15 | 46 | 31 | 29 | 44 |
|
P-value |
| 0.661 |
| 0.327 |
| 0.997 |
| 0.292 |
| 0.304 |
| 0.103 |
| 0.573 |
| 0.611 |
| Tumor size |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| <2.0
cm | 21 | 14 | 26 | 5 | 25 | 12 | 26 | 7 | 33 | 6 | 24 | 12 | 12 | 26 | 19 | 18 |
| 2.0–5.0
cm | 42 | 40 | 76 | 22 | 79 | 27 | 56 | 27 | 66 | 36 | 75 | 24 | 38 | 60 | 71 | 38 |
| >5.0
cm | 14 | 11 | 20 | 6 | 19 | 8 | 21 | 6 | 17 | 9 | 19 | 10 | 12 | 17 | 18 | 11 |
|
P-value |
| 0.670 |
| 0.733 |
| 0.695 |
| 0.359 |
| 0.075 |
| 0.406 |
| 0.663 |
| 0.298 |
| Histological
grade |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| Grade
1 | 18 | 19 | 31 | 10 | 32 | 12 | 26 | 7 | 32 | 13 | 28 | 15 | 33 | 12 | 18 | 27 |
| Grade
2 | 46 | 38 | 71 | 18 | 69 | 29 | 58 | 30 | 66 | 30 | 68 | 25 | 55 | 46 | 33 | 61 |
| Grade
3 | 13 | 8 | 20 | 5 | 22 | 6 | 19 | 3 | 17 | 8 | 22 | 6 | 20 | 8 | 11 | 15 |
|
P-value |
| 0.615 |
| 0.852 |
| 0.694 |
| 0.099 |
| 0.950 |
| 0.435 |
| 0.051 |
| 0.739 |
| Tumor stage |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
0/I | 11 | 6 | 14 | 1 | 15 | 4 | 14 | 3 | 18 | 0 | 12 | 6 | 7 | 12 |
9 | 9 |
|
IIA/IIB | 42 | 41 | 72 | 23 | 77 | 29 | 59 | 23 | 61 | 39 | 69 | 30 | 35 | 64 | 69 | 37 |
|
III/IV | 23 | 16 | 33 | 8 | 29 | 13 | 28 | 13 | 32 | 12 | 34 | 10 | 20 | 25 | 27 | 19 |
|
P-value |
| 0.464 |
| 0.289 |
| 0.723 |
| 0.553 |
| 0.004 |
| 0.582 |
| 0.578 |
| 0.420 |
| ER |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 29 | 14 | 43 | 9 | 44 | 13 | 39 | 8 | 35 | 15 | 42 | 10 | 40 | 17 | 24 | 28 |
| + | 47 | 51 | 78 | 24 | 78 | 34 | 63 | 32 | 79 | 36 | 75 | 36 | 68 | 48 | 37 | 75 |
|
P-value |
| 0.033 |
| 0.374 |
| 0.300 |
| 0.038 |
| 0.868 |
| 0.081 |
| 0.140 |
| 0.106 |
| PR |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 22 | 17 | 46 | 8 | 45 | 15 | 44 | 9 | 36 | 17 | 43 | 13 | 41 | 18 | 23 | 31 |
| + | 54 | 48 | 75 | 25 | 77 | 32 | 58 | 31 | 78 | 34 | 74 | 33 | 67 | 47 | 38 | 72 |
|
P-value |
| 0.712 |
| 0.142 |
| 0.545 |
| 0.022 |
| 0.824 |
| 0.304 |
| 0.168 |
| 0.316 |
| ErbB2 |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 15 | 21 | 35 | 11 | 37 | 14 | 31 | 13 | 37 | 21 | 34 | 14 | 37 | 12 | 21 | 27 |
| + | 61 | 43 | 86 | 21 | 84 | 33 | 72 | 27 | 76 | 39 | 82 | 32 | 70 | 53 | 40 | 75 |
|
P-value |
| 0.078 |
| 0.550 |
| 0.920 |
| 0.835 |
| 0.233 |
| 0.888 |
| 0.023 |
| 0.281 |
| Lymph node |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Negative | 35 | 25 | 49 | 13 | 58 | 15 | 44 | 14 | 50 | 19 | 46 | 24 | 39 | 35 | 24 | 45 |
|
Positive | 39 | 39 | 70 | 20 | 63 | 31 | 55 | 26 | 62 | 31 | 70 | 21 | 67 | 29 | 36 | 56 |
|
P-value |
| 0.330 |
| 0.854 |
| 0.074 |
| 0.307 |
| 0.430 |
| 0.116 |
| 0.023 |
| 0.572 |
| Menopausal
status |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Pre-menopausal | 43 | 40 | 67 | 23 | 74 | 27 | 61 | 21 | 63 | 34 | 66 | 33 | 66 | 36 | 33 | 63 |
|
Post-menopausal | 33 | 25 | 55 | 10 | 49 | 20 | 42 | 19 | 52 | 17 | 52 | 13 | 41 | 30 | 29 | 39 |
|
P-value |
| 0.551 |
| 0.127 |
| 0.747 |
| 0.466 |
| 0.152 |
| 0.063 |
| 0.354 |
|
0.282 |
| Distant
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 56 | 39 | 71 | 23 | 79 | 28 | 58 | 25 | 73 | 33 | 69 | 35 | 39 | 68 | 69 | 42 |
| + | 21 | 26 | 51 | 10 | 44 | 19 | 45 | 15 | 42 | 18 | 49 | 11 | 23 | 35 | 39 | 24 |
|
P-value |
| 0.108 |
| 0.230 |
| 0.574 |
| 0.501 |
| 0.829 |
| 0.035 |
| 0.685 | | 0.973 |
| Bone
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 62 | 50 | 94 | 27 | 96 | 37 | 75 | 32 | 87 | 44 | 89 | 39 | 50 | 82 | 88 | 50 |
| + | 15 | 15 | 28 | 6 | 27 | 10 | 28 | 8 | 28 | 7 | 29 | 7 | 12 | 21 | 20 | 16 |
|
P-value |
| 0.601 |
| 0.557 |
| 0.924 |
| 0.374 |
| 0.122 |
| 0.193 |
| 0.872 |
| 0.366 |
| Lung
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 69 | 52 | 99 | 28 | 103 | 39 | 82 | 34 | 97 | 42 | 95 | 41 | 51 | 89 | 91 | 54 |
| + | 8 | 13 | 23 | 5 | 20 | 8 | 21 | 6 | 18 | 9 | 23 | 5 | 11 | 14 | 17 | 12 |
|
P-value |
| 0.108 |
| 0.624 |
| 0.905 |
| 0.460 |
| 0.748 |
| 0.817 |
| 0.472 |
| 0.675 |
| Hepatic
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 71 | 55 | 106 | 31 | 109 | 42 | 88 | 36 | 104 | 45 | 103 | 43 | 57 | 91 | 100 | 56 |
| + | 6 | 10 | 16 | 2 | 14 | 5 | 15 | 4 | 11 | 6 | 15 | 3 | 5 | 12 |
8 | 10 |
|
P-value |
| 0.154 |
| 0.262 |
| 0.891 |
| 0.471 |
| 0.666 |
| 0.255 |
| 0.463 |
| 0.104 |
| Brain
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 77 | 65 | 120 | 33 | 120 | 47 | 100 | 40 | 113 | 51 | 116 | 46 | 62 | 101 | 109 | 64 |
| + | 1 | 0 |
2 | 0 |
3 | 0 |
3 | 0 |
2 | 0 |
2 | 0 | 0 | 2 |
0 | 2 |
|
P-value |
| 0.357 |
| 0.459 |
| 0.280 |
| 0.275 |
| 0.343 |
| 0.374 |
| 0.270 |
| 0.069 |
| Chest
metastasis |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 74 | 58 | 108 | 141 | 111 | 44 | 91 | 37 | 104 | 48 | 107 | 43 | 56 | 94 | 98 | 61 |
| + | 3 | 7 | 14 | 0 | 12 | 3 | 12 | 3 | 11 | 3 | 11 | 3 | 6 | 9 | 10 | 5 |
|
P-value |
| 0.111 |
| 0.041 |
| 0.488 |
| 0.467 |
| 0.431 |
| 0.564 |
| 0.839 | | 0.701 |
| Opposite
breast |
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
| − | 77 | 63 | 117 | 32 | 119 | 45 | 99 | 39 | 111 | 49 | 113 | 46 | 60 | 99 | 103 | 65 |
| + | 0 | 2 |
5 | 1 |
4 | 2 |
4 | 1 |
4 | 2 |
5 | 01 | 2 | 4 |
5 | 1 |
|
P-value |
| 0.121 |
| 0.778 |
| 0.751 |
| 0.686 |
| 0.888 |
| 0.156 |
| 0.827 |
|
0.275 |
Association of cathepsins B, D, G, K,
L and V as well as RECK and VEGF expression with the prognosis of
breast cancer patients
Associations of the expression of cathepsins B, D,
G, K, L and V as well as RECK and VEGF proteins with breast cancer
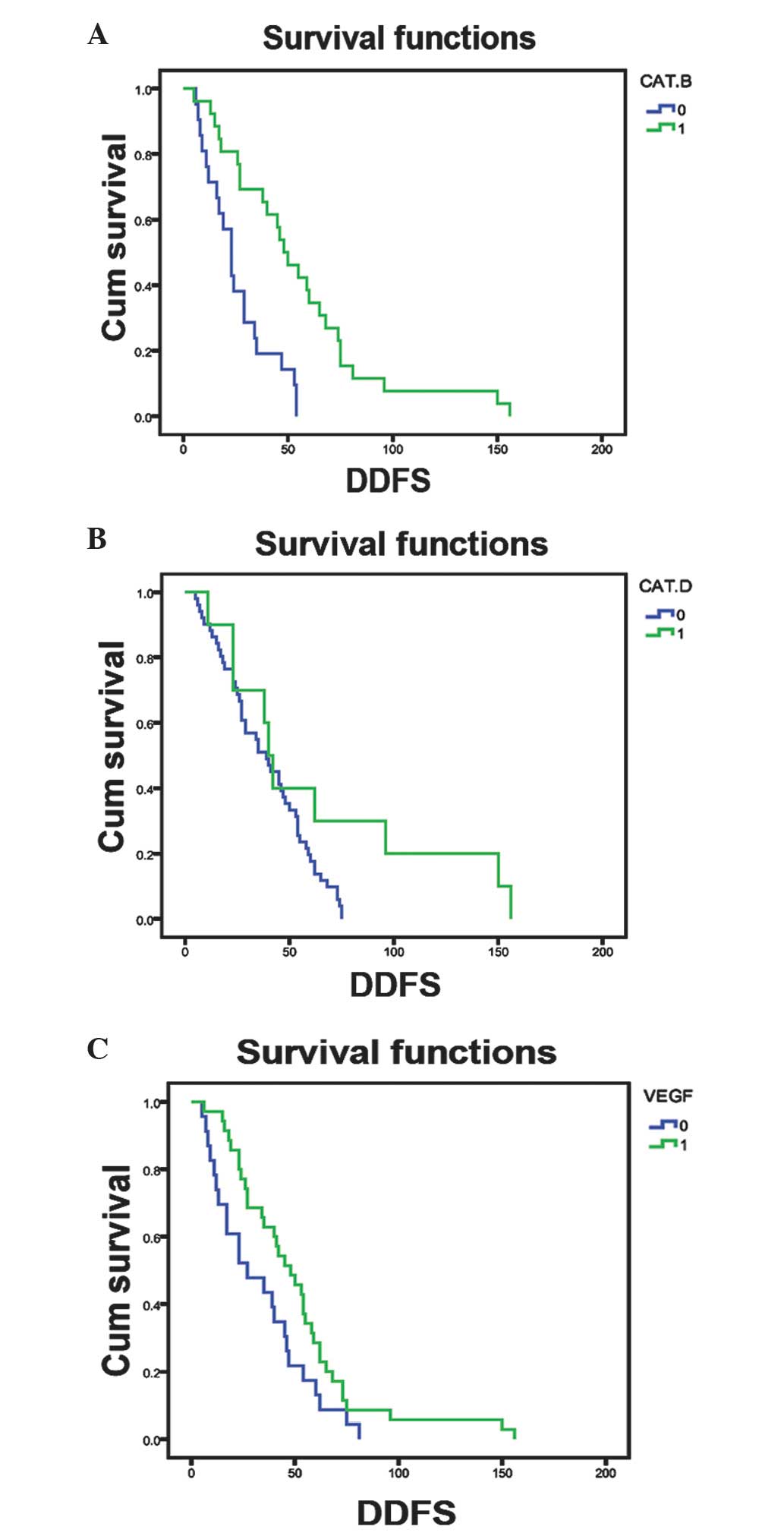
patient prognosis were determined. The results revealed that high
expression levels of cathepsin D and B and VEGF were associated
with poor disease-free survival in breast cancer patients (Fig. 2). However, the expression of other
proteins did not show any association with disease-free survival of
these patients (data not shown).
Furthermore, univariate analysis data indicated that
breast cancer metastasis to the bone and the expression of
cathepsin B proteins were associated with poor disease-free
survival. In addition, multivariate analysis demonstrated that PR
expression was associated with poor disease-free survival and
cathepsin B expression was only marginally associated with poor
disease-free survival (P=0.058) (Table
III).
 | Table III.Cox regression analysis of
disease-free survival according to clinicopathological parameters
and protein expression. |
Table III.
Cox regression analysis of
disease-free survival according to clinicopathological parameters
and protein expression.
|
| Univariate
analysis | Multivariate
analysis |
|---|
|
|
|
|
|---|
|
| Exp (B) | 95% CI | P-value | Exp (B) | 95% CI | P-value |
|---|
| pTNM | 1.496 | 0.939–2.384 | 0.090 | 1.652 | 0.816–3.341 | 0.163 |
| Bone
metastasis | 1.780 | 1.067–2.970 | 0.027 | 2.069 | 0.930–4.606 | 0.75 |
| Cathepsin B | 0.280 | 0.140–0.561 | 0.000 | 0.471 | 0.216–1.026 | 0.058 |
| ER | 0.625 | 0.370–1.055 | 0.078 | 0.613 | 0.246–1.529 | 0.294 |
| PR | 0.641 | 0.384–1.070 | 0.089 | 0.470 | 0.224–0.985 | 0.046 |
| Cathepsin D | 0.461 | 0.205–1.035 | 0.061 | 0.770 | 0.187–3.165 | 0.717 |
| VEGF | 0.580 | 0.338–0.995 | 0.048 | 0.429 | 0.209–0.882 | 0.021 |
Discussion
The development of breast cancer involves a
progressive multistage process, from premalignant lesion, to ductal
carcinoma in situ, to invasive carcinoma and then to
metastatic disease (19). Given the
variability in clinical progression and its high incidence and
mortality rates, the identification of biomarkers that are able to
predict tumor behavior may be particularly important in breast
cancer (19). Therefore, the current
study aimed to detected the expression of different cathepsins as
well as RECK and VEGF proteins in breast cancer tissue specimens.
The results demonstrated that cathepsin B was expressed in 109/142
(76.76%), cathepsin D in 91/155 (58.71%), cathepsin G in 99/171
(57.89%), cathepsin K in 97/143 (67.83%), cathepsin L in 95/166
(57.23%), cathepsin V in 45/164 (27.44%), RECK in 67/175 (38.29%)
and VEGF in 100/164 (60.98%) samples of breast cancer tissues. It
was also determined that the expression of cathepsin L was
associated with advanced tumor stage, the expression of cathepsins
B and K was associated with positive ER expression and that of
cathepsin K was associated with PR expression. In addition, the
expression of cathepsins V and D was associated with breast cancer
metastasis. Furthermore, the expression of cathepsins B and D as
well as that of VEGF was associated with poor disease-free survival
in breast cancer patients. Univariate analysis revealed that breast
cancer metastasis to the bone as well as the expression of
cathepsin B and VEGF proteins were associated with poor
disease-free survival. In addition, multivariate analysis
demonstrated that PR and VEGF expression were significantly
associated with poor disease-free survival of the patients, while
cathepsin B expression was only marginally associated with poor
disease-free survival. Therefore, the present results indicated
that targeting cathepsin B and D may have a potential therapeutic
benefit for breast cancer patients.
Cathepsins are a superfamily of proteins that are
highly expressed in various types of human cancers and have been
associated with cancer metastasis (6). Each cathepsin member has relatively
different functions and thus, has a different role under normal as
well as disease conditions. In cancer, cathepsins have been
reported to have various functions, including the regulation of
tissue remodeling, cell proliferation and angiogenesis, as well as
cancer progression and metastasis (6). Several members of the cathepsin family
have previously been studied in breast cancer (31). The results of the present study
indicated that the expression of cathepsin V was associated with
distant breast cancer metastasis and expression of cathepsin D was
associated with breast cancer metastasis to the chest. These
results were consistent with those of previously published studies
suggesting that members of the cathepsin family were highly
expressed in metastatic tumors (10,11,14,19).
Cathepsin D protein has been reported to be overexpressed in breast
cancer and hyper-secreted by epithelial breast cancer cells
(21,32,33). Other
previous studies have suggested that cathepsin D expression may be
an independent predictor for poor prognosis of breast cancer
patients as cathepsin D expression was associated with a high
incidence of cancer metastasis (20,34).
Consistent with these previous findings, the current study also
demonstrated that high expression of cathepsin D protein was
associated with poor prognosis of breast cancer patients; however,
multivariate analysis failed to indicate that this was an
independent predictor of disease-free survival. In addition,
numerous studies have suggested that cathepsin B overexpression was
associated with the invasive and metastatic phenotypes of various
cancers (15,35). However, the role of cathepsin B in
breast cancer remains to be fully elucidated (23,36). In
the current study, expression of cathepsin B showed a trend
(P=0.058) to independently predict breast cancer disease-free
survival; however, this results was not statistically significant.
Thus, further studies are required with a larger sample size in
order to verify the role of cathepsins in breast cancer and their
potential use as biomarkers for disease progression.
Numerous previous efforts have been made to develop
specific inhibitors to target the activity of each cathepsin family
member (26,37,38). For
example, intraperitoneal administration of a highly selective
cathepsin B inhibitor, such as CA-074, was reported to reduce the
metastatic potential of breast cancer cells in nude mice (39). In addition, inhibitors of the other
cysteine cathepsins, including S, L, C and K, are also available
(40).
In conclusion, the present study demonstrated that
the altered expression of cathepsins, in particular cathepsins B
and D, was associated with breast cancer progression and poor
disease-free survival. However, future studies are required in
order to investigate whether the targeting of these two cathepsins,
B and D, may have potential therapeutic use for attenuating the
progression of breast cancer.
Acknowledgements
This study was supported by the Cooperation Project
of China and Slovenia (grant no. 10–15 to Tao Sun).
Glossary
Abbreviations
Abbreviations:
|
ER
|
estrogen receptor
|
|
PR
|
progesterone-receptor
|
|
RECK
|
reversion-inducing cysteine-rich
protein with Kazal motifs
|
|
VEGF
|
vascular endothelial growth factor
|
References
|
1
|
Figueroa-Magalhaes MC, Jelovac D, Connolly
RM and Wolff AC: Treatment of HER2-positive breast cancer. Breast.
23:128–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herold CI and Anders CK: New targets for
triple-negative breast cancer. Oncology (Williston Park).
27:846–854. 2013.PubMed/NCBI
|
|
3
|
DeSantis C, Ma J, Bryan L and Jemal A:
Breast cancer statistics, 2013. CA Cancer J Clin. 64:52–62. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Turk B, Turk D and Turk V: Lysosomal
cysteine proteases: More than scavengers. Biochim Biophys Acta.
1477:98–111. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
López-Otín C and Overall CM: Protease
degradomics: A new challenge for proteomics. Nat Rev Mol Cell Biol.
3:509–519. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gocheva V and Joyce JA: Cysteine
cathepsins and the cutting edge of cancer invasion. Cell Cycle.
6:60–64. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen JM, Dando PM, Rawlings ND, et al:
Cloning, isolation, and characterization of mammalian legumain, an
asparaginyl endopeptidase. J Biol Chem. 272:8090–8098. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen JM, Rawlings ND, Stevens RA and
Barrett AJ: Identification of the active site of legumain links it
to caspases, clostripain and gingipains in a new clan of cysteine
endopeptidases. FEBS Lett. 441:361–365. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Im E and Kazlauskas A: The role of
cathepsins in ocular physiology and pathology. Exp Eye Res.
84:383–388. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tan GJ, Peng ZK, Lu JP and Tang FQ:
Cathepsins mediate tumor metastasis. World J Biol Chem. 4:91–101.
2013.PubMed/NCBI
|
|
11
|
Mohamed MM and Sloane BF: Cysteine
cathepsins: Multifunctional enzymes in cancer. Nat Rev Cancer.
6:764–775. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Jedeszko C and Sloane BF: Cysteine
cathepsins in human cancer. Biol Chem. 385:1017–1027. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krepela E: Cysteine proteinases in tumor
cell growth and apoptosis. Neoplasma. 48:332–349. 2001.PubMed/NCBI
|
|
14
|
Joyce JA and Hanahan D: Multiple roles for
cysteine cathepsins in cancer. Cell Cycle. 3:1516–1619. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gondi CS and Rao JS: Cathepsin B as a
cancer target. Expert Opin Ther Targets. 17:281–291. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Frlan R and Gobec S: Inhibitors of
cathepsin B. Curr Med Chem. 13:2309–2327. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Masson O, Bach AS, Derocq D, et al:
Pathophysiological functions of cathepsin D: Targeting its
catalytic activity versus its protein binding activity? Biochimie.
92:1635–1643. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Benes P, Vetvicka V and Fusek M: Cathepsin
D-many functions of one aspartic protease. Crit Rev Oncol Hematol.
68:12–28. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clezardin P: Therapeutic targets for bone
metastases in breast cancer. Breast Cancer Res. 13:2072011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Huang L, Liu Z, Chen S, Liu Y and Shao Z:
A prognostic model for triple-negative breast cancer patients based
on node status, cathepsin-D and Ki-67 index. PLoS One.
8:e830812013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dian D, Heublein S, Wiest I, et al:
Significance of the tumor protease cathepsin D for the biology of
breast cancer. Histol Histopathol. 29:433–438. 2014.PubMed/NCBI
|
|
22
|
Kawakubo T, Yasukochi A, Toyama T, et al:
Repression of cathepsin E expression increases the risk of mammary
carcinogenesis and links to poor prognosis in breast cancer.
Carcinogenesis. 35:714–726. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bengsch F, Buck A, Günther SC, et al: Cell
type-dependent pathogenic functions of overexpressed human
cathepsin B in murine breast cancer progression. Oncogene.
33:4474–4484. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Esteva FJ and Hortobagyi GN: Prognostic
molecular markers in early breast cancer. Breast Cancer Res.
6:109–118. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Rochefort H, Garcia M, Glondu M, et al:
Cathepsin D in breast cancer: Mechanisms and clinical applications,
a 1999 overview. Clin Chim Acta. 291:157–170. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tomoo K: Development of cathepsin
inhibitors and structure-based design of cathepsin B-specific
inhibitor. Curr Top Med Chem. 10:696–707. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Turk V, Stoka V, Vasiljeva O, Renko M, Sun
T, Turk B and Turk D: Cysteine cathepsins: From structure, function
and regulation to new frontiers. Biochim Biophys Acta. 1824:68–88.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hashida S, Kominami E and Katunuma N:
Inhibitions of cathepsin B and cathepsin L by E-64 in vivo. II.
Incorporation of [3H] E-64 into rat liver lysosomes in vivo.
J Biochem. 91:1373–1380. 1982.PubMed/NCBI
|
|
29
|
Hara K, Kominami E and Katunuma N: Effect
of proteinase inhibitors on intracellular processing of cathepsin
B, H and L in rat macrophages. FEBS Lett. 231:229–231. 1988.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Tsukuba T, Okamoto K, Yasuda Y, et al: New
functional aspects of cathepsin D and cathepsin E. Mol Cells.
10:601–611. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Garcia M, Platet N, Liaudet E, et al:
Biological and clinical significance of cathepsin D in breast
cancer metastasis. Stem Cells. 14:642–650. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vetvicka V and Fusek M: Procathepsin D as
a tumor marker, anti-cancer drug or screening agent. Anticancer
Agents Med Chem. 12:172–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nicotra G, Castino R, Follo C, Peracchio
C, Valente G and Isidoro C: The dilemma: Does tissue expression of
cathepsin D reflect tumor malignancy? The question: Does the assay
truly mirror cathepsin D mis-function in the tumor? Cancer Biomark.
7:47–64. 2010.
|
|
34
|
Markićević M, Kanjer K, Mandušić V, et al:
Cathepsin D as an indicator of clinical outcome in early breast
carcinoma during the first 3 years of follow-up. Biomark Med.
7:747–758. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong YJ, Shao LH and Li Y: Cathepsin
B-cleavable doxorubicin prodrugs for targeted cancer therapy
(Review). Int J Oncol. 42:373–383. 2013.PubMed/NCBI
|
|
36
|
Nouh MA, Mohamed MM, El-Shinawi M, et al:
A potential prognostic marker for inflammatory breast cancer. J
Transl Med. 9:12011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Deaton DN and Tavares FX: Design of
cathepsin K inhibitors for osteoporosis. Curr Top Med Chem.
5:1639–1675. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Hoegl L, Korting HC and Klebe G:
Inhibitors of aspartic proteases in human diseases: Molecular
modeling comes of age. Pharmazie. 54:319–329. 1999.PubMed/NCBI
|
|
39
|
Withana NP, Blum G, Sameni M, et al:
Cathepsin B inhibition limits bone metastasis in breast cancer.
Cancer Res. 72:1199–1209. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Frizler M, Stirnberg M, Sisay MT and
Gutschow M: Development of nitrile-based peptidic inhibitors of
cysteine cathepsins. Curr Top Med Chem. 10:294–322. 2010.
View Article : Google Scholar : PubMed/NCBI
|