Introduction
MicroRNAs (miRNAs) are small non-coding RNAs of
21–25 nucleotides in length that control gene expression via
post-transcriptional regulation (1).
Contrasting with the maturation process of normal coding RNAs, the
miRNA genes are initially transcribed by RNA polymerase II into a
primary transcript in the nucleus, where the hairpin structure is
processed into precursor miRNA by a microprocessing complex that
includes Drosha and DiGeorge syndrome chromosomal (or critical)
region 8 (DGCR8) (1). Subsequently,
the ~70 nucleotide-long precursor miRNA is exported into the
cytoplasm, where it undergoes secondary processing by Dicer, and
one strand of the hairpin is then incorporated into a
ribonucleoprotein complex known as miRNA-induced silencing complex
(1). Once matured, a single miRNA may
target miscellaneous messenger RNAs (mRNAs), and an individual mRNA
may be regulated by various miRNAs (1).
miRNAs have significant roles during stem cell
maintenance and differentiation (2).
In addition, they have frequently been used as markers for certain
types of cancer cells, by comparing the measured levels of known
miRNAs with those present in their wild-type counterparts (2,3). Screens
investigating differential expression of miRNAs in various cancer
cell populations have been previously performed. The present review
will focus on the results of five screens conducted in the past
recent years, which investigated prostate cancer (3), neuroblastoma (4), pancreatic cancer (5), chronic myeloid leukemia (CML) (6) and osteosarcoma cells (7). Specifically, Liu et al (3) used three prostate cancer metastatic cell
lines, namely LAPC9, LAPC4 and Du145 [bone cluster of
differentiation (CD)44+/CD44− subpopulation
sorting based on LAPC9, LAPC4 and Du145 cell lines; side population
(SP)/non-SP sorting based on LAPC9 cell line; and
CD133+/CD133− subpopulation sorting based on
LAPC4 cell line], and compared the expression levels of
miscellaneous miRNAs (with vs. without markers) in these cells.
Following testing for 324 DGCR8 miRNAs by reverse
transcription-quantitative polymerase chain reaction, 137 miRNAs
were identified to be expressed in the three cell types, of which,
4 were downregulated, confirming that miRNAs may be differentially
expressed in various cancer cell populations (3). Samaraweera et al (4) compared the miRNA expression levels of
two types of tumorigenic neuroblastic cells, namely stem cells
(I-cells) and neuronal cells (N-cells), with those of
non-tumorigenic non-neuronal cells (S-cells) by microarray
analysis. A total of 313 distinct miRNAs were assayed, among which,
17 displayed reduced expression levels and 3 displayed increased
expression levels in neuronal cells, compared with non-neuronal
cells. A total of 11 miRNAs were observed to be downregulated in
I-cells, compared with N-cells (4).
Consistently, the differential expression profile of miRNAs in
distinct cell compartments applies also to pancreatic cancer, CML
and osteosarcoma, as demonstrated by the following studies: Bao
et al (5) isolated triple
positive CD44+/CD133+/epithelial cell
adhesion molecule+ cells from two pancreatic cancer cell
lines termed MiaPaCa-2 and L3.6pl, and identified >400 miRNAs
that were differentially expressed in these cancer stem cell-like
populations by microarray analysis. Using this type of analysis,
Zhu et al (6) studied the
differences in expression levels of 275 known human miRNAs in bone
marrow stromal cells derived from patients with CML and normal
human subjects, and identified 19 downregulated and 6 upregulated
miRNAs. Furthermore, Feng et al (7) compared the gene expression profile of
the osteosarcoma stem cell line 3AB-OS with that of its parental
cell line MG63 by microarray analysis, and identified 189
differentially expressed miRNAs in addition to chromosome copy
number variations and dysregulated genes.
In the present review, those miRNAs that were
identified to be differentially expressed in ≥2 of the
aforementioned screens are discussed, and their interaction with
signaling networks is emphasized. A notable point is that, although
certain miRNAs were observed to be commonly downregulated or
differentially regulated in the above screens, no miRNA was
identified to be commonly upregulated in those studies (Table I).
 | Table I.Summary of miRs reviewed in the
present study. |
Table I.
Summary of miRs reviewed in the
present study.
| miR | Cancer screen where
miR was identified | Signaling pathway
involved | Expression
levels |
|---|
| let-7 | Prostate
cancer/osteosarcoma | Development and
LIN-41 | Downregulated |
| miR-34a | Prostate
cancer/osteosarcoma | Snail, p53 and
Wnt | Downregulated |
| miR-183 | Prostate
cancer/osteosarcoma | DKK3 and Smad4 | Downregulated |
| miR-15 | Prostate
cancer/osteosarcoma | BCL2, MCL1, CCND1
and WNT3A | Downregulated |
| miR-130a | Pancreatic
cancer/chronic myeloid leukemia | C/EBP, HOXA5, RUNX3
and Wnt | Downregulated |
| miR-21 | Pancreatic cancer
cells/neuroblastoma | PTEN and SPRY1 | Differentially
regulated |
| miR-335 | Pancreatic cancer
cells/neuroblastoma | TGF, RTK, ATM and
Oct4-pRB | Differentially
regulated |
| miR-133a | Prostate
cancer/osteosarcoma | Snail and
others | Differentially
regulated |
Commonly downregulated microRNAs
let-7
The let-7 family of miRNAs was initially identified
in Caenorhabditis elegans, where it participates in the
transition of later larval to adult stage (8). The let-7 family has 13 members in
humans, namely let-7a-1, a-2, a-3, 7b, 7c, 7d, 7e, 7f-1, 7f-2, 7g,
7i, miR-98 and miR-202 (9,10). In the aforementioned screens, the
let-7 family members involved were all identified to be
downregulated. Thus, in prostate cancer and osteosarcoma, 4 members
of the let-7 family were downregulated (let-7a, b, e and f),
whereas in pancreatic cancer and CML let-7a and let-7f were
observed to be downregulated (3,5–7). Let-7 is highly conserved across
bilaterians, and is involved in developmental switches in C.
elegans and mammals (11). It has
been reported that let-7 may be involved in neural cell
proliferation and differentiation in C. elegans,
Drosophila and mammals (12–14). In
addition to its developmental roles, let-7 has also been identified
to be involved in neuronal cell aging by suppressing the expression
of abnormal cell lineage protein 41 (LIN-41). In old
neurons, let-7 binds to the 3′ untranslated region of the
LIN-41 gene and inhibits its expression, which results in
the downregulation of the axonal regeneration abilities of the aged
anterior ventral microtubule axon in C. elegans (15). In cancer cells, the majority of the
let-7 family members were observed to be downregulated (16). It has been reported that let-7
manifests tumor suppressive effects in prostate cancer cells
(17). Furthermore, differential
mechanisms between let-7 and miR-34a on the regulation of the cell
cycle in prostate cancer cells have been previously reported. Thus,
miR-34a primarily induced G1 phase cell cycle arrest followed by
cell senescence, while let-7 mainly induced G2/M arrest (17).
miR-34a and miR-183
miR-34a and miR-183 were identified to be
downregulated in CD44+ prostate cancer and osteosarcoma
cells (3,7). Enforced expression of miR-34a in bulk or
purified CD44+ prostate cancer cells was demonstrated to
inhibit clonogenic expansion, tumor regeneration and metastasis
(3). In addition, CD44 has been
identified as a direct and functional target of miR-34a, which may
induce G1 phase cell cycle arrest (17). The miR-34 family members may be
induced by tumor protein p53, and may be able to directly suppress
epithelial-mesenchymal transition (EMT) by inhibiting the
expression of Snail, a transcription factor required for EMT
(18). In addition, miR-34 has been
observed to associate the tumor suppressor p53 to the
wingless-related integration site (Wnt) signaling pathway by
directly binding to the 3′ untranslated region of wingless-type
mouse mammary tumor virus integration site family, member 1 (WNT1),
WNT3, low density lipoprotein receptor-related protein 6 (LRP6),
axis inhibition protein 2 (AXIN2), β-catenin and lymphoid
enhancer-binding factor 1 (LEF1) (19). Therefore, the expression of miR-34 may
be capable of inhibiting metastasis (20). The miR-183 family of genes, including
miR-96, −182 and −183, are expressed coordinately in the sensory
cells of zebrafish, chicken and mouse (21–26), where
they are required for sensory cell differentiation and functioning.
Ueno et al (27) observed that
miR-183 was able to suppress the expression of Dickkopf-related
protein 3 and mothers against decapentaplegic homolog 4 (Smad4) in
prostate cancer, resulting in tumor growth inhibition.
Additionally, it has been demonstrated that miR-183 is able to
suppress the metastasis of osteosarcoma cells via downregulation of
the expression of Ezrin (28).
miR-15
miR-15 was identified to be downregulated in screens
of CD133+ subpopulation and SP of prostate cancer and
osteosarcoma cells (3,7). The miR-15 gene is located on the
chromosome 13q14, and was previously identified to be downregulated
in chronic lymphocytic leukemia and pituitary adenomas (29,30).
miR-15 downregulates oncogenes, including B-cell lymphoma 2,
myeloid cell leukemia 1, cyclin D1 and WNT3A (31). The repression of miR-15 and miR-16 in
hypoxia was identified to be caused by hypoxia inducible factor-2α
via c-myc signaling (32).
miR-130a
miR-130a was observed to be downregulated in
pancreatic cancer and CML (5,6). miR-130a is a c-myc-responsive
gene, which has been identified to regulate CCAAT/enhancer-binding
protein β, homeobox A5, runt-related transcription factor 3 and Wnt
signaling (33–35). Previous studies have demonstrated that
the expression of miR-130a is increased in non-small cell lung
cancer (36), whereas in glioblastoma
patients, increased levels of miR-130a are associated with
long-term survival (37).
Differentially regulated microRNAs in
various types of cancer
miR-21
In CD133+ prostate cancer cells, miR-21
was identified to be upregulated, whereas in neuroblastoma it was
observed to be downregulated (3,4). In
pancreatic cancer, miR-21-3p was observed to be upregulated,
whereas miR-21-5p was observed to be downregulated (5). The miR-21 gene is located on the
chromosome 17q23.2, and its potential targets include phosphatase
and tensin homolog (PTEN) and sprouty homolog 1 (SPRY1) (38). The expression levels of miR-21 have
been revealed to correlate with cell proliferation in
hepatocellular cancer (39). It has
additionally been reported that miR-21 is able to downregulate the
tumor suppressor programmed cell death 4, and therefore promote the
metastatic activities of colorectal and gastric cancer (40,41).
miR-335
miR-335 is upregulated in pancreatic cancer, but
downregulated in tumorigenic neuroblastoma cells (5,7). Lynch
et al (42) reported that
miR-335 was able to suppress metastasis of neuroblastoma cells by
directly repressing the downstream effector proteins of the
transforming growth factor (TGF)-β pathway, namely Rho-associated,
coiled-coil containing protein kinase 1 (ROCK1) and
mitogen-activated protein kinase 1 (MAPK1), which consequently
reduced the phosphorylation levels of myosin light chain and
inhibited the invasiveness of neuroblastoma cells. In addition to
its regulatory function in signaling pathways, miR-335 has roles in
at least two pathways involved in cell cycle control, namely ataxia
telangiectasia mutated (ATM)-dependent DNA damage control (43) and octamer-binding transcription factor
4-retinoblastoma protein (Oct4-pRB)-dependent stem cell
renewal/cell cycle control (44). In
the ATM pathway, irradiation-activated ATM downregulates miR-335
via cyclic adenosine monophosphate response element binding
protein, which subsequently activates C terminal binding protein
interaction protein, which in turn recruits breast cancer 1, early
onset to double strand breaks (43).
During stem cell renewal, miR-335 is able to regulate the cell
cycle by controlling the activities of the Oct4-pRb signaling
pathway. When differentiation begins, miR-335 is upregulated, and
thus downregulates the expression of Oct4, which impairs the rapid
division cycle of stem cell renewal (44). Previous studies have demonstrated that
the expression levels of miR-335 are cancer type-specific. Thus,
downregulation of miR-335 is observed in certain types of
metastatic breast cancer (45),
whereas upregulation is observed in the breast cancer cell line
MCF7 (43). Regarding its role in
ATM-dependent DNA damage control, Martin et al (43) suggested that differential expression
levels of miR-335 may explain the variations in terms of resistance
to chemotherapy or radiotherapy observed among different
patients.
miR-133a
miR-133a was identified to be downregulated in
CD133+ prostate cancer cells but upregulated in
osteosarcoma cells (3,7). The location and expression of miR-133a,
as well as its target genes, have been previously reviewed
elsewhere (46), and therefore will
not be discussed in the present review. A notable point is that in
the majority of the cancer stem cells listed in the present review,
miR-133a was observed to be downregulated (46). Numerous overexpression studies have
previously demonstrated that miR-133a is able to effectively
suppress tumor growth (17,46). Accordingly, the observation that
miR-133a was upregulated in osteosarcoma was unexpected (7). miR-133a was also observed to be
expressed in cultured embryonic rat cardiomyocytes (7). Furthermore, a recent study demonstrated
that miR-133a was able to promote cardiac reprogramming via
suppression of the Snail1 gene (47).
Based on the differences between osteosarcoma and prostate or other
types of cancer such as lung cancer, it is possible to conclude
that, since osteosarcoma is a mesodermal tissue-derived tumor, this
may explain the upregulation of miR-133a observed in 3AB-OS
osteosarcoma cells (7,17). Nevertheless, additional studies are
required to validate this hypothesis.
Signalling cascades with microRNA
interaction
Prior to miRNAs becoming a relevant topic in cancer
research, signaling networks were considered the key regulators of
tumor development, and this vision currently remains (1). The miRNAs listed in the preceding
sections of the present review have been previously associated with
Wnt, receptor tyrosine kinase (RTK) and TGF-β/bone morphogenetic
protein (BMP) signaling pathways, which involve genes encoding cell
cycle checkpoint proteins and transcriptional regulatory proteins
(1,20,48).
Wnt
Wnt signaling is dysregulated in various types of
cancer, as discussed elsewhere (48).
The crosstalk between miR-30 family members and the canonical Wnt
signaling pathway has been discussed in detail previously (49). Previous studies have implicated
miR-34a, miR-15 and miR-130 in the regulation of Wnt signaling, and
these miRNAs, in addition to miR-183, were observed to be
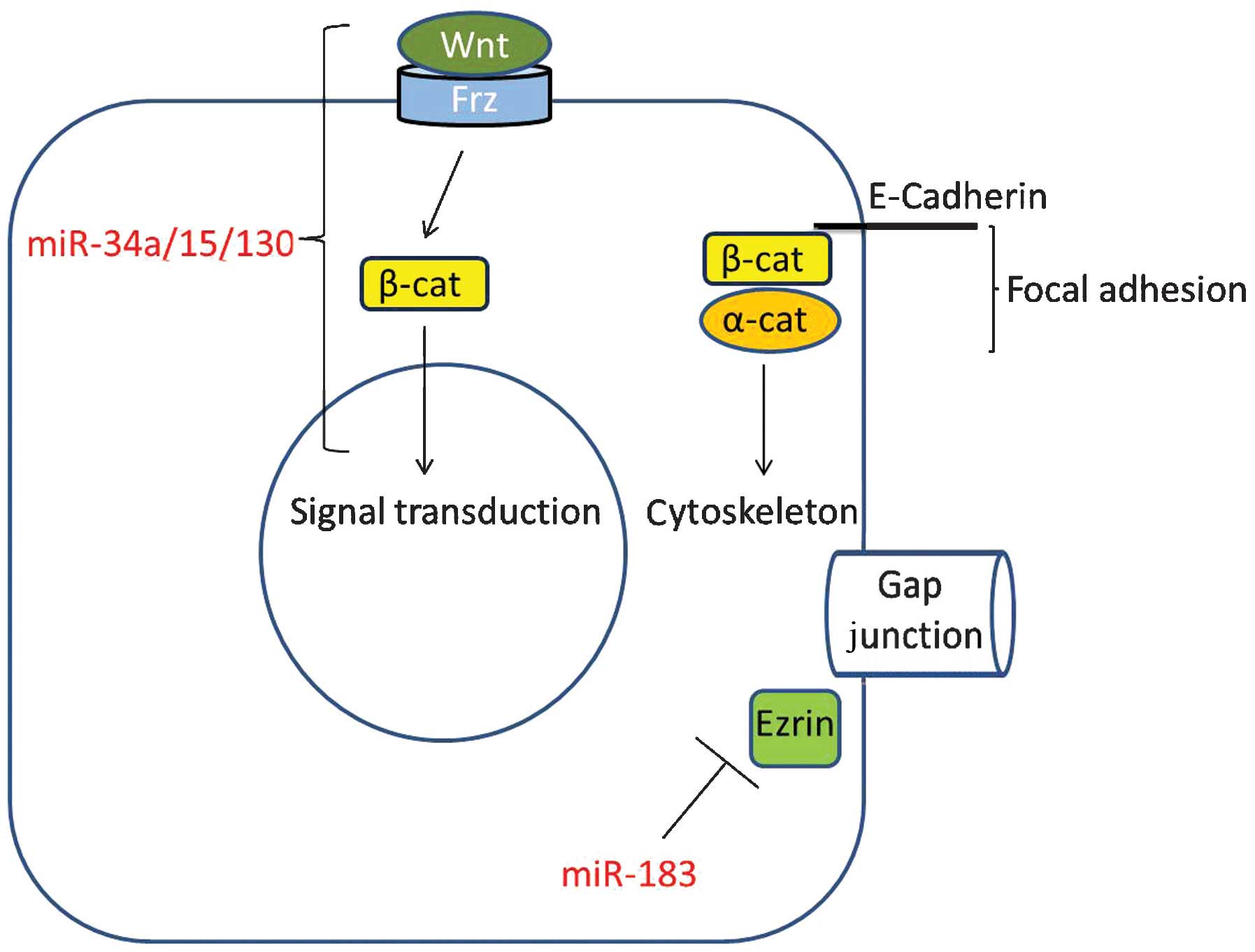
downregulated in prostate cancer and osteosarcoma cells (3,5,7). Wnt signaling is relevant in terms of one
of its key downstream components, namely β-catenin. This protein is
a signaling molecule and a component of focal adhesions, which
forms a complex with E-cadherin and α-catenin (50). Therefore, miR-183 may be involved in
the regulatory network of intercellular junctions (Fig. 1), since miR-183 downregulates the
expression of Ezrin (a key component of the gap junction of
adjacent epithelial cells), thus potentially inhibiting the
invasion and metastasis of osteosarcoma cells (28).
TGF-β/BMP
BMP signaling is a type of TGF-β signaling that is
considered to be the ‘master regulator’ of EMT and cancer
metastasis (51). Accordingly, it is
not surprising that miRNA-335, which functions to repress ROCK1
(the downstream target of TGF-β), is downregulated in tumorigenic
neuroblastoma cells (42). In
addition, miR-335 represses MAPK1, which is a common downstream
factor in RTK signaling, while miR-183 represses the expression of
Smad4, thus inhibiting BMP signaling (52). Therefore, miRNAs appear to directly
regulate the expression of key signaling molecules and provide an
additional regulatory layer for the signaling network.
RTK
SPRY1 and PTEN (targeted by miR-21), as well as
MAPK1 (targeted by miR-335), are downstream factors of RTK
signaling, which includes signal transduction induced by epidermal
growth factor, fibroblast growth factor, vascular endothelial
growth factor and insulin-like growth factor signals (53). All these signaling pathways have been
implicated in cancer initiation and development (53,54).
miR-21 regulates RAS1, a key signaling molecule that is a common
downstream target of RTK signals, which is located downstream of
SPRY1 and PTEN and upstream of MAPK1, and participates in the
growth of diverse types of cancer (39–41).
Cell cycle checkpoints
It has been previously reported that miR-34 may be
induced by p53 and subsequently cause G1 phase cell cycle arrest
(3,17), while miR-335 may be downregulated by
activated ATM (43). Therefore, it
appears that the genes involved in cell cycle checkpoint control
are primarily upstream of miRNA expression. The loss of checkpoint
control is required for cancer initiation and development (43,44). Thus,
there is a clear direct connection between miRNAs and cancer
formation, which will not be discussed further in the present
review. As previously indicated, miR-34 may be capable of binding
to the 3′ untranslated region of WNT1, WNT3, LRP6, AXIN2, β-catenin
and LEF1, thus suppressing the Wnt signaling pathway (19). These scenarios suggest that miRNAs
possess a bridging role, connecting Wnt signaling with cell cycle
control. Future studies may confirm additional miRNAs to act by
bridging the gap between signal transduction processes and cell
cycle control.
Conclusion
The majority of miRNAs observed to be downregulated
during tumor progression are considered to be tumor suppressive,
and may be under the control of tumor suppressors such as p53.
miRNAs regulate signal transduction by modulating the expression
levels of key regulatory proteins, which may be involved in
identical or differing signaling pathways to those of their
corresponding miRNA. In addition, miRNAs may be modulated by cell
cycle checkpoint proteins, thus connecting signaling networks with
cell cycle control. Compared with previous studies on
protein-encoding genes, the current research on miRNAs is at its
initial stages, and it is predicted that the functions exhibited by
different miRNAs may be as complex as those displayed by
protein-encoding genes. In conclusion, the functional regulatory
mechanisms of a biological entity are composed of protein-encoding
and non-encoding genes, including miRNAs, which together fine-tune
the processes that form a living organism.
Acknowledgements
The present study was financially supported by the
National Natural Science Foundation of China (Beijing, China; grant
no's. 81372750 and 81572518), the Shanghai Natural Science
Foundation of China (Shanghai, China; grant no. 12ZR1425200), the
Science & Technology Development Fund of Shanghai Pudong
(Shanghai, China; grant no. PKJ2014-Y10) and the Scientific
Research Foundation for the Returned Overseas Chinese Scholars from
the State Education Ministry of China (Beijing, China; grant no.
2013-1792).
References
|
1
|
Bartel DP: MicroRNAs: Genomics biogenesis,
mechanism, and function. Cell. 116:281–297. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gangaraju VK and Lin H: MicroRNAs: Key
regulators of stem cells. Nat Rev Mol Cell Biol. 10:116–125. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu C, Kelnar K, Liu B, Chen X,
Calhoun-Davis T, Li H, Patrawala L, Yan H, Jeter C, Honorio S, et
al: The microRNA miR-34a inhibits prostate cancer stem cells and
metastasis by directly repressing CD44. Nat Med. 17:211–215. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Samaraweera L, Grandinetti KB, Huang R,
Spengler BA and Ross RA: MicroRNAs define distinct human
neuroblastoma cell phenotypes and regulate their differentiation
and tumorigenicity. BMC Cancer. 14:3092014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bao B, Ali S, Ahmad A, Li Y, Banerjee S,
Kong D, Aboukameel A, Mohammad R, Van Buren E, Azmi AS and Sarkar
FH: Differentially expressed miRNAs in cancer-stem-like cells,
Markers for tumor cell aggressiveness of pancreatic cancer. Stem
Cells Dev. 23:1947–1958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhu X, Lin Z, Du J, Zhou X, Yang L and Liu
G: Studies on microRNAs that are correlated with the cancer stem
cells in chronic myeloid leukemia. Mol Cell Biochem. 390:75–84.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Feng Z, Takahashi R, Nakamura T, Sato D,
Shirasawa N, Nakayama A, Kurashige S, Kosawada T, Kitajima T and
Umezu M: Expression of microRNA-1, microRNA-133a and Hand2 protein
in cultured embryonic rat cardiomyocytes. In Vitro Cell Dev
Biol Anim. 50:700–706. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Thornton JE and Gregory RI: How does Lin28
let-7 control development and disease? Trends Cell Biol.
22:474–482. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ruby JG, Jan C, Player C, Axtell MJ, Lee
W, Nusbaum C, Ge H and Bartel DP: Large-scale sequencing reveals
21U-RNAs and additional microRNAs and endogenous siRNAs in C.
elegans. Cell. 127:1193–1207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: MicroRNAs and developmental
timing. Curr Opin Genet Dev. 21:511–517. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Abrahante JE, Daul AL, Li M, Volk ML,
Tennessen JM, Miller EA and Rougvie AE: The Caenorhabditis
elegans hunchback-like gene lin-57/hbl-1 controls developmental
time and is regulated by microRNAs. Dev Cell. 4:625–637. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lin SY, Johnson SM, Abraham M, Vella MC,
Pasquinelli A, Gamberi C, Gottlieb E and Slack FJ: The C
elegans hunchback homolog, hbl-1, controls temporal patterning
and is a probable microRNA target. Dev Cell. 4:639–650. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao C, Sun G, Li S, Lang MF, Yang S, Li W
and Shi Y: MicroRNA let-7b regulates neural stem cell proliferation
and differentiation by targeting nuclear receptor TLX signaling.
Proc Natl Acad Sci USA. 107:1876–1881. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zou Y, Chiu H, Zinovyeva A, Ambros V,
Chuang CF and Chang C: Developmental decline in neuronal
regeneration by the progressive change of two intrinsic timers.
Science. 340:372–376. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Boyerinas B, Park SM, Hau A, Murmann AE
and Peter ME: The role of let-7 in cell differentiation and cancer.
Endocr Relat Cancer. 17:F19–F36. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Liu C, Kelnar K, Vlassov AV, Brown D, Wang
J and Tang DG: Distinct microRNA expression profiles in prostate
cancer stem/progenitor cells and tumor-suppressive functions of
let-7. Cancer Res. 72:3393–3404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kim NH, Kim HS, Li XY, Lee I, Choi HS,
Kang SE, Cha SY, Ryu JK, Yoon D, Fearon ER, et al: A p53/miRNA-34
axis regulates Snail1-dependent cancer cell epithelial-mesenchymal
transition. J Cell Biol. 195:417–433. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cha YH, Kim NH, Park C, Lee I, Kim HS and
Yook JI: MiRNA-34 intrinsically links p53 tumor suppressor and Wnt
signaling. Cell Cycle. 11:1273–1281. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Rokavec M, Öner MG, Li H, Jackstadt R,
Jiang L, Lodygin D, Kaller M, Horst D, Ziegler PK, Schwitalla S, et
al: IL-6R/STAT3/miR-34a feedback loop promotes EMT-mediated
colorectal cancer invasion and metastasis. J Clin Invest.
124:1853–1867. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wienholds E, Kloosterman WP, Miska E,
Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen
S and Plasterk RH: MicroRNA expression in zebrafish embryonic
development. Science. 309:310–311. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Shao P, Zhou H, Xiao ZD, He JH, Huang MB,
Chen YQ and Qu LH: Identification of novel chicken microRNAs and
analysis of their genomic organization. Gene. 418:34–40. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kloosterman WP, Wienholds E, de Bruijn E,
Kauppinen S and Plasterk RH: In situ detection of miRNAs in
animal embryos using LNA-modified oligonucleotide probes. Nat
Methods. 3:27–29. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
24
|
Weston MD, Pierce ML, Rocha-Sanchez S,
Beisel KW and Soukup GA: MicroRNA gene expression in the mouse
inner ear. Brain Res. 1111:95–104. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xu S, Witmer PD, Lumayag S, Kovacs B and
Valle D: MicroRNA (miRNA) transcriptome of mouse retina and
identification of a sensory organ-specific miRNA cluster. J Biol
Chem. 282:25053–25066. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weston MD, Pierce ML, Jensen-Smith HC,
Fritzsch B, Rocha-Sanchez S, Beisel KW and Soukup GA: MicroRNA-183
family expression in hair cell development and requirement of
microRNAs for hair cell maintenance and survival. Dev Dyn.
240:808–819. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ueno K, Hirata H, Shahryari V, Deng G,
Tanaka Y, Tabatabai ZL, Hinoda Y and Dahiya R: MicroRNA-183 is an
oncogene targeting Dkk-3 and SMAD4 in prostate cancer. Br J Cancer.
108:1659–1667. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhu J, Feng Y, Ke Z, Yang Z, Zhou J, Huang
X and Wang L: Down-regulation of miR-183 promotes migration and
invasion of osteosarcoma by targeting Ezrin. Am J Pathol.
180:2440–2451. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Calin GA, Dumitru CD, Shimizu M, Bichi R,
Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bottoni A, Piccin D, Tagliati F, Luchin A,
Zatelli MC and Uberti Degli EC: miR-15a and miR-16-1
down-regulation in pituitary adenomas. J Cell Physiol. 204:280–285.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Aqeilan RI, Calin GA and Croce CM: miR-15a
and miR-16-1 in cancer, Discovery, function and future
perspectives. Cell Death Differ. 17:215–220. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xue G, Yan HL, Zhang Y, Hao LQ, Zhu XT,
Mei Q and Sun SH: c-Myc-mediated repression of miR-15-16 in hypoxia
is induced by increased HIF-2α and promotes tumor angiogenesis and
metastasis by upregulating FGF2. Oncogene. 34:1393–1406. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yang F, Miao L, Mei Y and Wu M: Retinoic
acid-induced HOXA5 expression is co-regulated by HuR and miR-130a.
Cell Signal. 25:1476–1485. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q
and Zhang J: Upregulated miR-130a increases drug resistance by
regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell.
Biochem Biophys Res Commun. 425:468–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Larsen MT, Häger M, Glenthøj A, Asmar F,
Clemmensen SN, Mora-Jensen H, Borregaard N and Cowland JB:
miRNA-130a regulates C/EBP-ε expression during granulopoiesis.
Blood. 123:1079–1089. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang XC, Tian LL, Wu HL, Jiang XY, Du LQ,
Zhang H, Wang YY, Wu HY, Li DG, She Y, et al: Expression of
miRNA-130a in nonsmall cell lung cancer. Am J Med Sci. 340:385–388.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qiu S, Lin S, Hu D, Feng Y, Tan Y and Peng
Y: Interactions of miR-323/miR-326/miR-329 and
miR-130a/miR-155/miR-210 as prognostic indicators for clinical
outcome of glioblastoma patients. J Transl Med. 11:102013.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Haghikia A and Hilfiker-Kleiner D:
MiRNA-21: A key to controlling the cardiac fibroblast compartment?
Cardiovasc Res. 82:1–3. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Asangani IA, Rasheed SA, Nikolova DA,
Leupold JH, Colburn NH, Post S and Allgayer H: MicroRNA-21 (miR-21)
post-transcriptionally downregulates tumor suppressor Pdcd4 and
stimulates invasion, intravasation and metastasis in colorectal
cancer. Oncogene. 27:2128–2136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li L, Zhou L, Li Y, Lin S and Tomuleasa C:
MicroRNA-21 stimulates gastric cancer growth and invasion by
inhibiting the tumor suppressor effects of programmed cell death
protein 4 and phosphatase and tensin homolog. J BUON. 19:228–236.
2014.PubMed/NCBI
|
|
42
|
Lynch J, Fay J, Meehan M, Bryan K, Watters
KM, Murphy DM and Stallings RL: MiRNA-335 suppresses neuroblastoma
cell invasiveness by direct targeting of multiple genes from the
non-canonical TGF-β signalling pathway. Carcinogenesis. 33:976–985.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Martin NT, Nakamura K, Davies R, Nahas SA,
Brown C, Tunuguntla R, Gatti RA and Hu H: ATM-dependent MiR-335
targets CtIP and modulates the DNA damage response. PLoS Genet.
9:e10035052013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Schoeftner S, Scarola M, Comisso E,
Schneider C and Benetti R: An Oct4-pRb axis, controlled by MiR-335,
integrates stem cell self-renewal and cell cycle control. Stem
Cells. 31:717–728. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tavazoie SF, Alarcón C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massagué J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Nohata N, Hanazawa T, Enokida H and Seki
N: MicroRNA-1/133a and microRNA-206/133b clusters, Dysregulation
and functional roles in human cancers. Oncotarget. 3:9–21. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Muraoka N, Yamakawa H, Miyamoto K,
Sadahiro T, Umei T, Isomi M, Nakashima H, Akiyama M, Wada R,
Inagawa K, et al: MiR-133 promotes cardiac reprogramming by
directly repressing Snai1 and silencing fibroblast signatures. EMBO
J. 33:1565–1581. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Atlasi Y, Looijenga L and Fodde R: Cancer
stem cells, pluripotency, and cellular heterogeneity, A WNTer
perspective. Curr Top Dev Biol. 107:373–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhao JJ and Carrasco RD: Crosstalk between
microRNA30a/b/c/d/e-5p and the canonical Wnt pathway: Implications
for multiple myeloma therapy. Cancer Res. 74:5351–5358. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Fagotto F: Looking beyond the Wnt pathway
for the deep nature of β-catenin. EMBO Rep. 14:422–433. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fazilaty H, Gardaneh M, Bahrami T,
Salmaninejad A and Behnam B: Crosstalk between breast cancer stem
cells and metastatic niche, Emerging molecular metastasis pathway?
Tumour Biol. 34:2019–2030. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Miyazono K: Signal transduction by bone
morphogenetic protein receptors: F unctional roles of Smad
proteins. Bone. 25:91–93. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Shibuya M: VEGFR and type-V RTK activation
and signaling. Cold Spring Harb Perspect Biol. 5:a0090922013.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Wang D, Du L, Liu Q, Liu X and Wang Z:
Receptor tyrosine kinase alterations and therapeutic opportunities
in squamous cell carcinoma of the lung. Cancer Chemother Pharmacol.
72:725–731. 2013. View Article : Google Scholar : PubMed/NCBI
|