Introduction
More than 37,000 individuals develop pancreatic
adenocarcinoma annually in the United States, and the majority of
these succumb to the disease due to its aggressive characteristics
and the fact that a large number of patients present with
relatively advanced disease. The 5-year survival rate of patients
with pancreatic adenocarcinoma is <5%, therefore, improved
medical intervention is required (1,2). Surgical
resection offers the only option for a cure, however, resectable
disease is exhibited by only 15–20% of patients at the time of the
initial diagnosis; the majority of patients present with locally
advanced or metastatic cancer (1,2). Effective
systemic therapy is key for prolonging the survival of patients
exhibiting advanced pancreatic cancer.
Increased expression of the first member of the ErbB
family to be identified, epidermal growth factor receptor (EGFR),
and its ligand, epidermal growth factor (EGF), have been detected
in 40–60% of human pancreatic cancer cases. The co-expression of
EGFR and its ligand has been identified as a predictor of a poor
prognosis (3). The targeting of EGFR
with the tyrosine kinase inhibitor erlotinib demonstrated a marked
survival benefit when combined with gemcitabine treatment, compared
with gemcitabine treatment alone (4).
Human epidermal growth factor receptor 2 (HER2; ErbB2)-targeted
therapy has been demonstrated to significantly improve clinical
outcomes in breast and gastric cancer (5,6). A total
of 20% of pancreatic cancers demonstrate HER2 overexpression. When
monoclonal antibodies were utilized to target EGFR and HER2
synergistically in xenograft models, augmented inhibition of tumor
progression was observed, compared with single monoclonal antibody
treatment (P=0.006) or no treatment (P=0.0004), and a number of
complete remissions were evident (7).
Lapatinib is a tyrosine kinase inhibitor, which
binds EGFR and HER2 (8). In an
international phase III trial of HER2-positive breast cancers,
treatment with lapatinib and capecitabine [pro-drug of
5-fluorouracil (FU)] significantly improved the time to
progression, compared with capecitabine treatment alone (9). Therefore, in the present study, a
single-arm phase II study was conducted, in order to evaluate the
combination of lapatinib and capecitabine for the second-line
treatment of metastatic pancreatic cancer.
Biomarkers that predict responses to anticancer
therapy have been sought in order to identify effective treatments
and understand the mechanisms of resistance. MicroRNAs
(miRNAs/miRs) are small (~22-nt), non-coding RNAs that possess a
significant role in the control of a wide range of cellular
processes, including apoptosis, cell proliferation, the regulation
of embryonic stem cell development and cancer cell invasion
(10). A number of studies have
revealed that miRNA signatures may be used for distinguishing
between various cancers, and additionally for defining the
prognosis (11,12). A previous study revealed that, unlike
a number of other biomarker types, circulating miRNAs are stable,
making them reliable and robust biomarkers for cancer (13). Specific miRNAs (including miR-21,
miR-221, miR-210 and miR-7) have been implicated as downstream
effectors of the EGFR and HER2 signaling pathways (12–17). The
aim of the present study was to investigate whether the levels of
the aforementioned miRNA(s) in blood are able to predict the
clinical outcome for patients receiving lapatinib and capecitabine
treatment, and to evaluate how this group of miRNAs contribute to
the resistance to lapatinib and capecitabine treatment in
patients.
Materials and methods
Patients and clinical study
design
A total of 17 patients with metastatic,
gemcitabine-refractory pancreatic cancer were recruited at the
Lombardi Comphrensive Cancer Center (Washington, USA) between March
2009 and September 2013. The patient cohort included 13 males and 4
females, with a mean age of 61 years (range, 52–73 years). All
patients received continuous treatment with lapatinib (1,250 mg,
daily) and capecitabine (1,000 mg/m2, twice daily) on
days 1–14 of each 21-day cycle until disease progression occurred
or the patients were unable to tolerate chemotherapy. The primary
endpoint was median overall survival (OS). Serum samples were
collected at baseline (before treatment) and every 3 weeks during
the study for miRNA analysis. This study was approved by the
Institutional Review Board of Georgetown University (Washington,
USA) (IRB#CR00000441/2008-437) and written informed consent was
obtained from all patients.
Cell culture and pharmacological
agents
Human pancreatic cancer PANC-1, MIA PaCa-2 and
BXCP-3 cell lines were purchased from the American Type Culture
Collection (Manassas, VA, USA), and maintained in RPMI 1640 medium
supplemented with 10% fetal bovine serum (FBS) (Gibco; Thermo
Fisher Scientific Inc., Waltham, MA, USA). All cells were cultured
at 37°C in 5% CO2, with 100% humidity. The cells were
treated with 4 µM lapatinib and 16 µM 5-FU, or with anti-miRNA
oligonucleotides (AMOs) or a vehicle control at various doses at
37°C, and analyzed following 72 h of incubation. Lapatinib was
purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA) and
5-FU was obtained from Sigma-Aldrich (St. Louis, MO, USA), and
3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium
was obtained from Promega Corp. (Madison, WI, USA). The AMOs
hsa-miR-221-3p (MIMAT0000278) and hsa-miR-210 (MI0000286), and the
negative control (scrambled sequence) were purchased from Thermo
Fisher Scientific Inc.
Cell survival assay
The cells were seeded into 96-well plates at a
density of 1×104 cells/well in 50 µl RPMI 1640 medium
with 10% FBS (Gibco; Thermo Fisher Scientific Inc.), and incubated
for 24 h at 37°C. Subsequently, the cells were exposed to lapatinib
and/or 5-FU at increasing concentrations (0.25, 1, 4 and 16 µM) in
an additional 50 µl medium. Cell survival was assayed following 72
h of incubation using a CellTiter 96® Aqueous One Solution Cell
Proliferation Assay kit (Promega Corp.). Measurements were
performed in accordance with the manufacturer's protocols.
Assessment of cell survival rate was recorded as the relative
colorimetric change measured at 570 nm using a VICTOR2 Multilabel
Counter (PerkinElmer Finland, Turku, Finland).
Transfection with hsa-miR
Transfection of the cells with hsa-miR was performed
using Lipofectamine® RNAiMAX reagent (Invitrogen; Thermo Fisher
Scientific Inc.) according to the manufacturer's transfection
protocol. Briefly, the cells were seeded into 6-well
(2×105 cells/well) plates prior to transfection.
Following 24 h of incubation, hsa-miR or scrambled sequence were
diluted in serum-free medium, and incubated with Lipofectamine
RNAiMAX reagent for 10 min at room temperature. Complexes were
added dropwise onto cells. Cell survival was assayed 72 h after
transfection. Knockdown of miRNA levels was determined using
quantitative polymerase chain reaction (qPCR). Briefly, total RNA
enriched in miRNA was prepared from cell pellets (~106
cells) using the miRNAeasy Mini Kit (Qiagen, Inc., Valencia, CA,
USA). miRNA levels were determined following conversion of RNA to
cDNA using a RT2 miRNA first strand kit (Qiagen, Inc.).
cDNA was amplified using the Applied Biosystems 7900HT Fast
Real-Time PCR system (Thermo Fisher Scientific Inc.), SYBR qPCR
reaction mixture and miRNA specific primers (Qiagen, Inc.). PCR was
performed under the following conditions: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for
30 sec and 72°C for 30 sec (18,19). U6
spliceosomal RNA served as an internal control, and data was
quantified using the comparative Cq method (20).
In order to determine the effects of hsa-miR
pretreatment on cell sensitivity to lapatinib and 5-FU, the PANC-1
cells were seeded in 100-mm dishes at an initial density of
5×105 cells and transfected with hsa-miR or scramble
sequence for 24 h. The cells were subsequently collected and
transferred to a 96-well plate (1×104 cells/well).
Lapatinib (4 µM) and 5-FU (16 µM) were added in a combined
concentration following 24 h of incubation, with 5 replicate plate
columns per treatment. Following 72 h of treatment, cell survival
was determined using the CellTiter 96® Aqueous One Solution Cell
Proliferation Assay kit (Promega Corp.), as described above.
Reverse transcription-qPCR of
miRNA
Total RNA was isolated from the serum of patients or
cells using the QIAzol™ reagent (Qiagen, Inc.) as previously
described (18,19). The miRNA expression analysis was
performed using qPCR analysis as previously reported (18,19).
Briefly, serum samples were mixed at a ratio of 1:10 with
QIAzol™lysis reagent and vortexed for 1 min using a mini vortexer
(Thermo Fisher Scientific, Inc). Cell pellets (~106
cells) were mixed with 1 ml QIAzol™ reagent (Qiagen, Inc.). The
lysates were extracted using CHCl3 and the aqueous phase
was further processed, removing phenol and other contaminants, to
obtain total RNA enriched in miRNA using the miRNAeasy Mini Kit
(Qiagen, Inc.). miRNA levels were determined following conversion
of RNA to cDNA using the RT2 miRNA first strand kit
(Qiagen, Inc.) followed by amplification of cDNA in the Applied
Biosystems 7900HT Fast Real-Time PCR system (Thermo Fisher
Scientific, Inc.) using SYBR qPCR reaction mixture and miRNA
specific primers (Qiagen, Inc.). Amplification of cDNA was
performed under the following conditions: Initial denaturation at
95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for
30 sec and 72°C for 30 sec (18,19). U6
spliceosomal RNA served as an internal control, and data was
quantified using the comparative Cq method (20).
Statistical analysis
miRNA levels in the cells or patient specimens and
survival data were tested by a one-way analysis of variance test
using GraphPad Prism Software version 5.0 (GraphPad Software, Inc.,
La Jolla, CA, USA). Results are presented as the mean ± standard
deviation. OS and progression free survival curves were estimated
using the Kaplan-Meier method. OS and progression free survival are
presented as the median ± 95% confidence interval. Principal
component analysis was used to analyze the association between
serum miRNA levels and drug response. P﹤0.05 was considered to
indicate a statistically significant difference.
Results
Analysis of miRNA levels in patient
serum
A total of 17 patients presenting with advanced
pancreatic cancer, who demonstrated cancer progression following
first-line chemotherapy, were enrolled in an institutional review
board (IRB)-approved phase II clinical trial (IRB#
CR00000441/2008-437; Clinical Trials.gov
identifier, NCT00881621), and were administered 1,250 mg lapatinib
daily and 1,000 mg/m2 capecitabine twice daily, on days
1–14 of a 21-day cycle. A total of 8 patients, including 3
non-responders (NRS; defined as demonstrating disease progression
following 2 cycles of treatment) and 5 responders (RS; defined as
demonstrating stable disease following 2 cycles of treatment),
underwent serial serum sample collection at baseline, and at 3 and
6 weeks. The expression profile of a panel of miRNAs (miR-21,
miR-210, miR-221 and miR-7), which are associated with EGFR1 and
HER2 signaling pathways, was analyzed for fold-changes in
expression (compared with baseline).
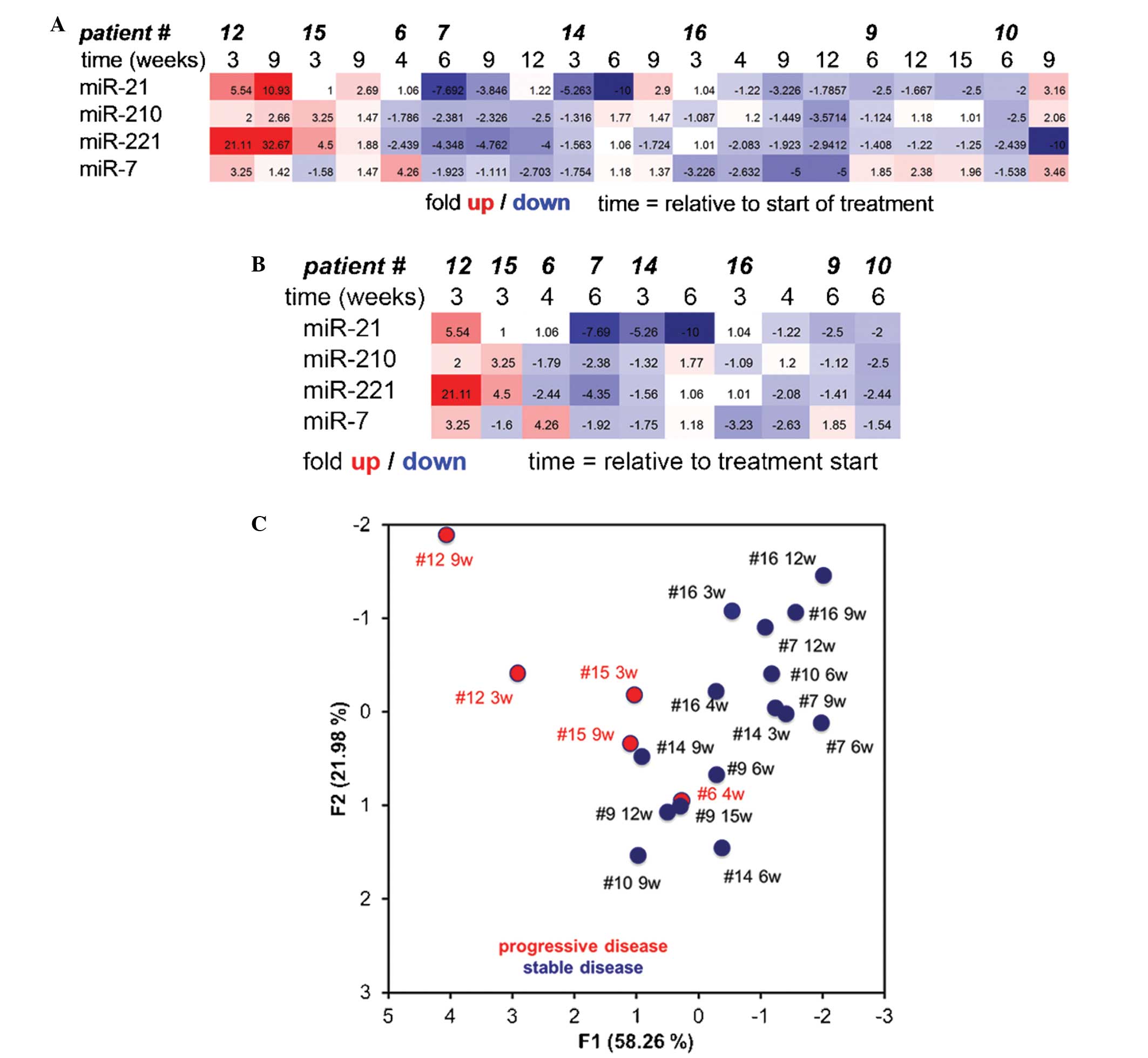
Heat chart analysis of the miRNA expression profiles
clearly demonstrated varying expression profiles between patient
numbers 6, 12 and 15 (NRS) and patient numbers 7, 9, 10, 14 and 16
(RS) (Fig. 1A). Most significantly,
heat chart analysis at early time points in treatment predicted the
subsequent prognosis of the patients as RS or NRS (Fig. 1B). Principal component analysis of the
data clearly separated RS from NRS utilizing all data, or data for
only early time points (Fig. 1C).
miR-221 and miR-210 levels increase in
chemoresistant pancreatic cancer cells treated with lapatinib and
5-FU in vitro, and suppression of miR-221 increases the sensitivity
of cancer cells to treatment
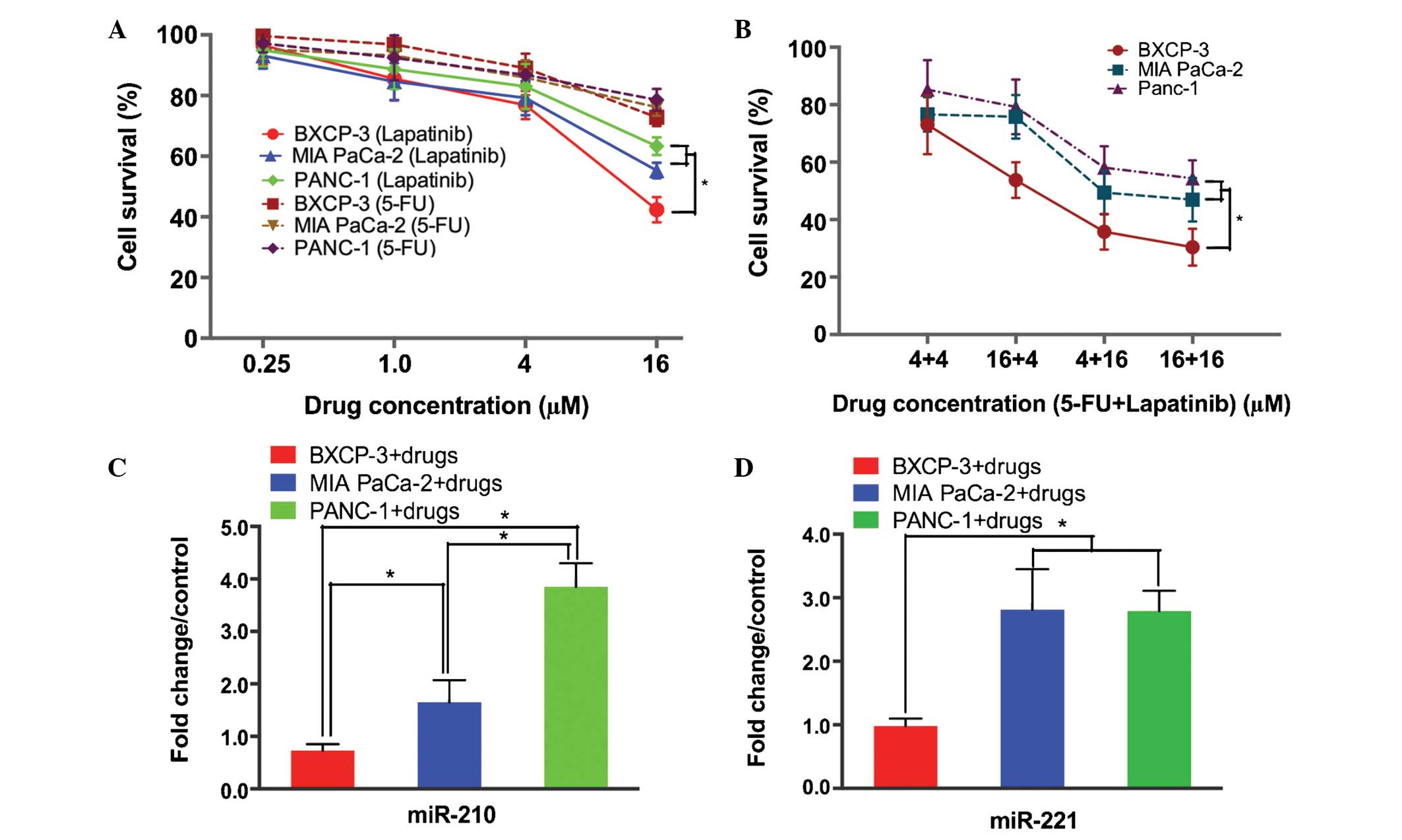
In order to confirm the observation that the panel
of miRNAs identified as being significant for the prediction of
patient responses to therapy were indeed associated with prognosis,
3 pancreatic cancer cell lines (PANC-1, MIA PaCa-2 and BXCP-3) with
varying levels of sensitivity to lapatinib and 5-FU were selected
in order to study the role of miRNA in treatment resistance. The
cell viability assay demonstrated that the PANC-1 and MIA PaCa-2
cells possessed increased resistance to lapatinib alone or in
combination with 5-FU treatment, compared with the BXCP-3 cells
(Fig. 2A and B). The miRNA analysis
revealed significant upregulation of miR-210 in the PANC-1
(1.65±0.42-fold; P<0.05) and MIA PaCa-2 (3.85±0.45-fold;
P<0.01) cells, however, no such upregulation was observed in the
BXCP-3 cells (0.73±0.12-fold; BXPC-3 + drug vs. PANC-1 + drug)
(Fig. 2C). Following treatment with
lapatinib and 5-FU, the levels of miR-221 were observed to be
increased in the PANC-1 and MIA PaCa-2 cells by 2.81±0.32-fold and
2.79±0.32-fold, respectively (P<0.01), however, no such increase
was observed in the BXCP-3 cells (0.98 ± 0.12 fold; BXPC-3 + drug
vs. PANC-1 + drug; P=0.0026) (Fig.
2D). There were no significant alterations observed in the
expression of miR-7 and miR-21 in all 3 pancreatic cancer cell
lines investigated (data not shown).
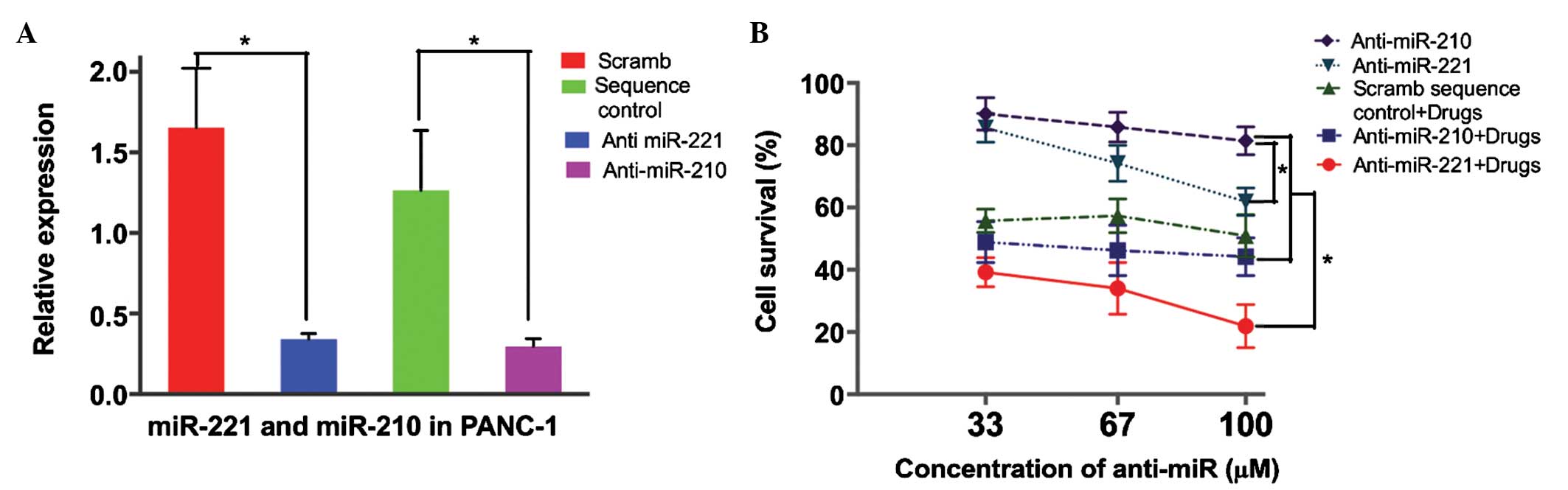
Based on observations from the cell lines and
patients, we hypothesized that an increase in miR-221 or miR-210
contributed to the resistance of cancer cells to lapatinib and
capecitabine treatment. In order to evaluate the effect of miR-221
or miR-210 inhibition on the response of pancreatic cancer cells to
lapatinib and 5-FU treatment, anti-miR-221 or anti-miR-210 were
transfected into the PANC-1 cells. This transfection resulted in a
4.9-fold decrease in miR-221 and a 4.2-fold decrease in miR-210
compared with a scramble sequence-transfected group (P=0.001;
Fig. 3A). The cell viability assay
demonstrated that anti-miR-221 transfection into the PANC-1 cells
induced the sensitivity of the cells to lapatinib and 5-FU
treatment in a dose-dependent manner, compared with no change in
sensitivity to treatment in the control cells transfected with
scramble sequence (scrambled sequence control + drug vs.
anti-miR-221 + drug; P=0.001; Fig.
3B). By contrast, decreasing the levels of miR-210 did not
alter the sensitivity of the PANC-1 cells to lapatinib and 5-FU
treatment (Fig. 3B).
Discussion
Chemoresistance is a significant cause of treatment
failure in pancreatic cancer (21,22). The
dual inhibition of EGFR and HER2 has been proposed as a potential
treatment for pancreatic adenocarcinoma based on the observed
increased levels of EGFR/HER2 heterodimers present in pancreatic
cancer cells (23). The present
investigation therefore consisted of a single-arm phase II study to
evaluate the combination of lapatinib and capecitabine for the
second-line treatment of metastatic pancreatic cancer. Notably, a
subset of patients existed (6/17) that responded to lapatinib and
capecitabine treatment with a mean overall survival time of 10.4
months (median, 8.3 months). In the search for a biomarker to
differentiate patients who responded to lapatinib and capecitabine
treatment from patients who were resistant to this treatment, the
present study identified that the increase in circulating miRNAs
from a targeted panel (associated with EGFR and HER2 signaling
pathways) that had been observed to be linked with a poor prognosis
and a lack of response to lapatinib and capecitabine treatment.
Similar pathway-specific patterns in circulating miRNAs between NRS
and RS have been observed in a previous study involving the
treatment of colon cancer patients with an antiangiogenic agent
(24).
In order to determine whether miRNAs serve purely as
a biomarker, or additionally contribute to the resistance of
pancreatic cancer cells to lapatinib and capecitabine treatment,
the present study performed additional experiments in 3 pancreatic
cancer cell lines that possessed various levels of sensitivity to
lapatinib and 5-FU (the active form of capecitabine) in
vitro. The present study identified that the levels of miR-210
and miR-221 were increased in response to drug treatment in the
resistant cells (PANC-1), compared with the levels in sensitive
cells (BXCP-3), which was in keeping with results obtained from the
patient serum samples of the NRS and RS groups. Unlike miR-210 or
miR-221, the expression of miR-7 and miR-21 in the pancreatic cell
lines did not alter in the same way as it did in patient serum
samples. This may be attributed to the differential response of
other cell types (including fibroblasts and lymphocytes) to
treatment with anticancer drugs. The potential significance of this
response with regard to patient outcomes may not be explained using
the present experimental model. The current study subsequently
demonstrated that blocking of the increase in miR-221 levels, but
not miR-210 levels, sensitized the pancreatic cancer cells to
lapatinib and 5-FU treatment. This observation supported the
hypothesis that miR-221 may possess a significant role in the
chemoresistance to lapatinib treatment. The results of the present
study support the idea that miR-221 may have potential as a
prognostic marker and potential target for therapeutic
interventions in pancreatic cancer (25,26). It is
notable to consider the reported ability of certain natural
compounds to downregulate miR-221 in pancreatic cancer cells in
preclinical studies (27,28). If proven safe to administer to
patients, these agents require evaluation for their ability to
downregulate miR-221 in clinical studies.
The present study demonstrated that a subset of
pancreatic cancer patients received benefits from lapatinib, a
treatment that induces the combined inhibition of the EGFR and HER2
signaling pathways. An increase in miR-221 levels in the blood,
detected 3 weeks after the beginning of lapatinib and capecitabine
treatment, may predict treatment failure and a lack of clinical
benefit in patients exhibiting pancreatic cancer. The results of
the present study require the performance of future studies in
order to evaluate the role of miR-221 in the prediction of
lapatinib treatment failure, as well as the effect of a combined
lapatinib and anti-miR-221 agent on the patient response to
treatment.
Acknowledgements
The present study was supported by the American
Cancer Society (grant no. 118525-MRSG-10-068-01-TBE), as well as by
the Ruesch Center for the Cure of Gastrointestinal Cancer.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Valsecchi ME, Díaz-Cantón E, de la Vega M
and Littman SJ: Recent treatment advances and novel therapies in
pancreas cancer: A review. J Gastrointest Cancer. 45:190–201.
2014.PubMed/NCBI
|
|
3
|
Vaccaro V, Gelibter A, Bria E, Iapicca P,
Cappello P, Di Modugno F, Pino MS, Nuzzo C, Cognetti F, Novelli F,
et al: Molecular and genetic bases of pancreatic cancer. Curr Drug
Targets. 13:731–743. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Moore MJ, Goldstein D, Hamm J, Figer A,
Hecht JR, Gallinger S, Au HJ, Murawa P, Walde D, Wolff RA, et al:
National Cancer Institute of Canada Clinical Trials Group:
Erlotinib plus gemcitabine compared with gemcitabine alone in
patients with advanced pancreatic cancer: A phase III trial of the
National Cancer Institute of Canada Clinical Trials Group. J Clin
Oncol. 25:1960–1966. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Won E, Janjigian YJ and Ilson DH: HER2
directed therapy for gastric/esophageal cancers. Curr Treat Options
Oncol. 15:395–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rimawi MF, Schiff R and Osborne CK:
Targeting HER2 for the treatment of breast cancer. Annu Rev Med.
66:111–128. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Larbouret C, Robert B, Navarro-Teulon I,
Thèzenas S, Ladjemi MZ, Morisseau S, Campigna E, Bibeau F, Mach JP,
Pèlegrin A and Azria D: In vivo therapeutic synergism of
anti-epidermal growth factor receptor and anti-HER2 monoclonal
antibodies against pancreatic carcinomas. Clin Cancer Res.
13:3356–3362. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xia W, Mullin RJ, Keith BR, Liu LH, Ma H,
Rusnak DW, Owens G, Alligood KJ and Spector NL: Anti-tumor activity
of GW572016: A dual tyrosine kinase inhibitor blocks EGF activation
of EGFR/erbB2 and downstream Erk1/2 and AKT pathways. Oncogene.
21:6255–6263. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cameron D, Casey M, Press M, Lindquist D,
Pienkowski T, Romieu CG, Chan S, Jagiello-Gruszfeld A, Kaufman B,
Crown J, et al: A phase III randomized comparison of lapatinib plus
capecitabine versus capecitabine alone in women with advanced
breast cancer that has progressed on trastuzumab: Updated efficacy
and biomarker analyses. Breast Cancer Res Treat. 112:533–543. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Walther A, Johnstone E, Swanton C, Midgley
R, Tomlinson I and Kerr D: Genetic prognostic and predictive
markers in colorectal cancer. Nat Rev Cancer. 9:489–499. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Drakaki A and Iliopoulos D: MicroRNA-gene
signaling pathways in pancreatic cancer. Biomed J. 36:200–208.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garofalo M, Romano G, Di Leva G, Nuovo G,
Jeon YJ, Ngankeu A, Sun J, Lovat F, Alder H, Condorelli G, et al:
EGFR and MET receptor tyrosine kinase-altered microRNA expression
induces tumorigenesis and gefitinib resistance in lung cancers. Nat
Med. 18:74–82. 2012.
|
|
16
|
Seike M, Goto A, Okano T, Bowman ED,
Schetter AJ, Horikawa I, Mathe EA, Jen J, Yang P, Sugimura H, et
al: MiR-21 is an EGFR-regulated anti-apoptotic factor in lung
cancer in never-smokers. Proc Natl Acad Sci USA. 106:12085–12090.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chou YT, Lin HH, Lien YC, Wang YH, Hong
CF, Kao YR, Lin SC, Chang YC, Lin SY, Chen SJ, et al: EGFR promotes
lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc
pathway that targets the Ets2 transcriptional repressor ERF. Cancer
Res. 70:8822–8831. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Larbouret C, Gaborit N, Chardès T, Coelho
M, Campigna E, Bascoul-Mollevi C, Mach JP, Azria D, Robert B and
Pèlegrin A: In pancreatic carcinoma, dual EGFR/HER2 targeting with
cetuximab/trastuzumab is more effective than treatment with
trastuzumab/erlotinib or lapatinib alone: Implication of receptors'
down-regulation and dimers' disruption. Neoplasia. 14:121–130.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
LaConti JJ, Shivapurkar N, Preet A, Mays
Deslattes A, Peran I, Kim SE, Marshall JL, Riegel AT and Wellstein
A: Tissue and serum microRNAs in the Kras(G12D) transgenic animal
model and in patients with pancreatic cancer. PLoS One.
6:e206872011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Shivapurkar N, Weiner LM, Marshall JL,
Madhavan S, Mays Deslattes A, Juhl H and Wellstein A: Recurrence of
early stage colon cancer predicted by expression pattern of
circulating microRNAs. PLoS One. 9:e846862014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Andersson R, Aho U, Nilsson BI, Peters GJ,
Pastor-Anglada M, Rasch W and Sandvold ML: Gemcitabine
chemoresistance in pancreatic cancer: Molecular mechanisms and
potential solutions. Scand J Gastroenterol. 44:782–786. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ghaneh P, Kawesha A, Evans JD and
Neoptolemos JP: Molecular prognostic markers in pancreatic cancer.
J Hepatobiliary Pancreat Surg. 9:1–11. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sarkar S, Dubaybo H, Ali S, Goncalves P,
Kollepara SL, Sethi S, Philip PA and Li Y: Down-regulation of
miR-221 inhibits proliferation of pancreatic cancer cells through
up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer
Res. 3:465–477. 2013.PubMed/NCBI
|
|
25
|
Shivapurkar N, Mikhail S, Navarro R, Bai
W, Marshall J, Hwang J, Pishvaian M, Wellstein A and He AR:
Decrease in blood miR-296 predicts chemotherapy resistance and poor
clinical outcome in patients receiving systemic chemotherapy for
metastatic colon cancer. Int J Colorectal Dis. 28:8872013.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kawaguchi T, Komatsu S, Ichikawa D,
Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T,
Hirajima S, et al: Clinical impact of circulating miR-221 in plasma
of patients with pancreatic cancer. Br J Cancer. 108:361–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Su A, He S, Tian B, Hu W and Zhang Z:
MicroRNA-221 mediates the effects of PDGF-BB on migration,
proliferation, and the epithelial-mesenchymal transition in
pancreatic cancer cells. PLoS One. 8:e713092013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Basu A, Alder H, Khiyami A, Leahy P, Croce
CM and Haldar S: MicroRNA-375 and MicroRNA-221: Potential noncoding
RNAs associated with antiproliferative activity of benzyl
isothiocyanate in pancreatic cancer. Genes Cancer. 2:108–119. 2011.
View Article : Google Scholar : PubMed/NCBI
|