Introduction
Gastric cancer is a common type of tumor with high
incidence and mortality rates which poses a significant threat to
human life. As the mechanism of gastric carcinogenesis is still
unknown, the study of gastric cancer initiation and progression and
the search for new therapeutic targets are currently hot research
topics. microRNA (miR) is a newly identified single-stranded
non-coded RNA containing 17–19 nucleotides that is a significant
regulator of gastric cancer initiation and progression. A series of
studies has reported that miR plays an essential regulatory role in
the proliferation, invasion and metastasis of gastric cancer cells
and in cell apoptosis (1–8). miR-196 was demonstrated to be associated
with involution of normal tissues, rib (9), tail (10)
and bone marrow (11), as well as
regulation of cancers of the rectum (12,13) and
liver (14), and appears to be a
potential target for cancer therapy (15). The PI3K/AKT/mTOR signaling pathway is
known to be essential to cell apoptosis. To determine the effect of
miR-196b in controlling apoptosis in gastric cancer cells, as well
as the underlying mechanism, here we examined the correlation
between miR-196b expression and apoptosis of MKN28 gastric cancer
cells and assessed its role in controlling the PI3K/AKT signaling
pathway.
Materials and methods
Lentivirus-based miR-196b vector
production and its delivery to gastric cancer MKN28 cells
The following study was approved by the Ethics
Committee of Wuhan University (Wuhan, China). Primers specific to
human miR-196b (gene ID 442920) were designed as follows:
5′-CGGTTAACCCCTTCCTTGACGCATTTG-'3 (sense) and
5′-CGACTCGAGAACCTAACCCTACCTGCTGTGA-3′ (anti-sense). The primers
were synchronized by Takara Biotechnology Co., Ltd. (Dalian, China)
by introducing HpaI and XhoI restriction enzyme sites
at the terminals. The normal male peripheral blood genomic DNA
template was used to amplify specific fragments by polymerase chain
reaction (PCR). Then the PCR product and pLL-3.7 plasmid (Takara
Biotechnology Co., Ltd.) were digested with HpaI and
XhoI and ligated overnight using T4 ligase at 16°C. The
ligated product was transferred to E.coli STBL-3 competent cells
(Takara Biotechnology Co., Ltd.) and selected for positive clones
by plating on an ampicillin+LB plate. pLL-miR-196 plasmid was
purified from the positive clones. Using Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA), 10 mg pLL-miR-196b and 5 mg pCMV-VSV-G were
delivered to HEK293FT cells (Cell Bank of the Chinese Academy of
Science, Beijing, China). Prior to transfection, the cells were
seeded on a six-well plate at a density of 2×106 cells
per well and co-cultured with modified RPMI-1640 medium containing
2 ml 10% fetal bovine serum (FBS; HyClone; GE Healthcare Life
Sciences, Logan, UT, USA) until confluence reached 70–90%.
Virus-containing cell culture fluid was collected at 48 h
post-transfection, centrifugated at 3000 × g for 15 min at 4°C,
filtered through a 0.45-µm filter, and maintained at −80°C. MKN28
gastric cancer cells (Cell Bank of the Chinese Academy of Science)
were incubated on a six-well plate at a density of 1×106
per well with modified RPMI-1640 medium supplemented with 2 ml 10%
FBS. After 24 h, virus-containing supernatant was delivered to the
cells to serve as the miR-196b group. For the control group, the
same procedure was carried out, with the exception that the
virus-containing supernatant was replaced with 100 µl
phosphate-buffered saline (PBS). The transduced cells demonstrated
green fluorescence under the fluorescence microscope. The
expression of miR-196b was measured using quantitative PCR
(qPCR).
qPCR
When cells were in the exponential phase of growth,
TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to isolate
total RNA. mRNA reverse transcription was generated using the
RevertAid First Strand cDNA synthesis kit (Thermo Fisher
Scientific, Inc.) following the instructions. The reaction system
was as follows: 2 µl 5X PrimeScript buffer, 2 µl total RNA (0.25
µg/µl), 1 µl primers (10 pM), 0.5 µl PrimerScript RT enzyme mix,
and 6.5 µl RNase-free dH2O. The reaction parameters were
37°C for 15 min, followed by 85°C for 5 sec and 4°C for the hold.
Quantitative analysis was performed using a Thunderbird SYBR® qPCR
mix kit (Toyobo Co., Ltd., Osaka, Japan). The reaction system was
as follows: 12.5 µl 2X qPCR mix, 2.0 µl of each 2.5 µM primer, 2.0
µl reverse transcription products, and 8.5 µl ddH2O. The
amplification parameters were 40 cycles of 95°C for 3 sec, 95°C for
5 sec, 60°C for 30 sec, and 68°C for 45 sec. For the qPCR primers,
see Table I.
 | Table I.Quantitative polymerase chain reaction
primers. |
Table I.
Quantitative polymerase chain reaction
primers.
| Gene | Primers (5′→3′) |
|---|
| miR-125a | Sense:
CGGTTAACCCCTTCCTTGACGCATTTG |
|
| Anti-sense:
CGACTCGAGAACCTAACCCTA |
|
| CCTGCTGTGA |
| PI3K | Sense:
GCCCAGGCTTACTACAGAG |
|
| Anti-sense:
AAGTAGGGAGGCATCTCG |
| AKT | Sense:
CTCATTCCAGACCCACGAC |
|
| Anti-sense:
ACAGCCCCGAAGTCCGTTA |
| mTOR | Sense:
ATGACGAGACCCAGGCTAA |
|
| Anti-sense:
GCCAGTCCTCTACAATACGC |
| β-actin | Sense:
ATCATGTTTGAGACCTTCAACA |
|
| Anti-sense:
CATCTCTTGGTCGAAGTCCA |
| U6 | Sense:
CTCGCTTCGGCAGCACA |
|
| Anti-sense:
AACGCTTCACGAATTTGCGT |
Western blot analysis
Cells were lysed for total protein extraction using
RIPA buffer (250 µl, Beyotime Institute of Biotechnology, Shanghai,
China) for 24 h after being incubated on a six-well plate. The
expression of miR-196a, PI3K, AKT and mTOR was measured using a
western blot kit (Biosdec, Wuhan, China). Rat anti-human miR-196a,
PI3K, AKT and mTOR antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Santa Cruz, Dallas, TX, USA).
Successively, 50 µg total protein was separated
using sodium dodecyl sulphate-polyacrylamide gel electrophoresis,
transferred to a 0.45 µm polyvinylidene fluoride membrane and
incubated overnight at 4°C with rat anti-human monoclonal antibody
(1:3000 dilution). Following decoloration, horseradish
peroxidase-conjugated goat anti-rabbit secondary antibody at a
1:3000 dilution was added for 30 min at room temperature and
unbound antibody was washed away. Proteins were visualized by
enhanced chemiluminescence detection reagents (Thermo Fisher
Scientific, Inc.) and exposed to X-ray film after raffinate
clearance. Exposure conditions and developing and fixing were based
on the light intensity. Developed films were processed with BanScan
software (Glyko Inc., Novato, CA, USA) to determine optical
densities.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
method for analysis of cell proliferative activity
Cells were seeded on a 96-well plate at a density of
2000 cells per well. When the cells reached 80% confluence, the
culture medium was removed before adding 20 µl MTT reaction
solution (5 mg/ml, Beyotime Institute of Biotechnology) to each
well. Once the cells had been incubated in the dark for 4 h at
37°C, the supernatant was removed and 150 µl DMSO (Beyotime
Institute of Biotechnology) was added to each well and agitated for
10 min at room temperature. Optical density was measured at 490 nm
using the enzyme-linked immunosorbent assay.
Flow cytometry for analysis of cell
cycle
Cells were seeded on a six-well plate at a density
of 1×106 per well and allowed to incubate until cell
adhesion using the conventional culture technique. The culture
medium was removed, then cells were suspended, centrifuged and then
fixed with precooled 75% ethanol overnight at −20°C. Cells were
centrifuged to remove supernatant and washed twice with PBS. A
total of 450 µl PBS was added to each well to resuspend cells.
Subsequently, 50 µl propidium iodide (0.5 mg/ml, Beyotime Institute
of Biotechnology) was added, agitated and the cells were maintained
in a water bath at 37°C for 30 min. Cells were again centrifuged to
remove the supernatant and PBS was used to resuspend the cells. The
cell cycle was determined by flow cytometer (BD-FACSCalibur, BD
Biosciences, San Jose, CA, USA).
Soft agar colony formation assay
Soft agar (5%) was added to the medium at a 1:9
ratio and mixed well before being placed on dishes and cooled at
room temperature. Exponentially growing cell suspension containing
1×103 cells/ml was prepared. Cell suspension (1.5 ml)
was then added to an equal volume of 0.5% soft agar, agitated and
incubated on a dish at 37°C under 5% CO2 for 2 to 3
weeks. The formation of colonies was calculated using the formula:
Colony formation rate (%) = (number of colonies/number of cells
incubated) × 100.
In vitro cell invasion assay
Cell invasion ability was tested using a Transwell
chamber model (Chemicon; EMD Millipore, Billerica, MA, USA). Cell
suspension was adjusted to a concentration of 1×105
cells/ml. Then, 50 µl cell suspension was placed in the upper
chamber. After 24 h, cells that had migrated to the lower chamber
were fixed with 10% formalin and stained with Giemsa to quantitate
the number of transmigrated cells under an inverted microscope. The
transmigration rate was the number of cells transmigrated over the
total number of cells.
Tumorigenecity test in nude mice
Six 5-week-old BALB/c nude mice purchased from Wuhan
University Center for Animal Experiments, China, were randomly
assigned to the miR-196a group and control group, with three mice
per group. In the miR-196a group, 1×105
miR-196a-transfected MKN28 cells were suspended in a serum-free
RPMI-1640 medium (0.1 ml), and administered by subcutaneous
injection into the back of the nude mice. Three days later, another
injection with the same number of cells at the same concentration
was administered. In the control group, the mice underwent the same
treatment as the miR-196a group with the exception that the MKN28
cells were not miR-196a-transfected. Animals were sacrificed 4
weeks after tumor formation and tumor weight was determined.
Statistical analysis
Data were expressed as the means ± standard
deviation and were processed with the paired t-test using SPSS 16.0
software (SPSS, Inc., Chicago, Illinois, USA). P<0.05 was
considered to indicate a statistically significant difference.
Results
Effect of miR-196b activation on
gastric cancer cell proliferation
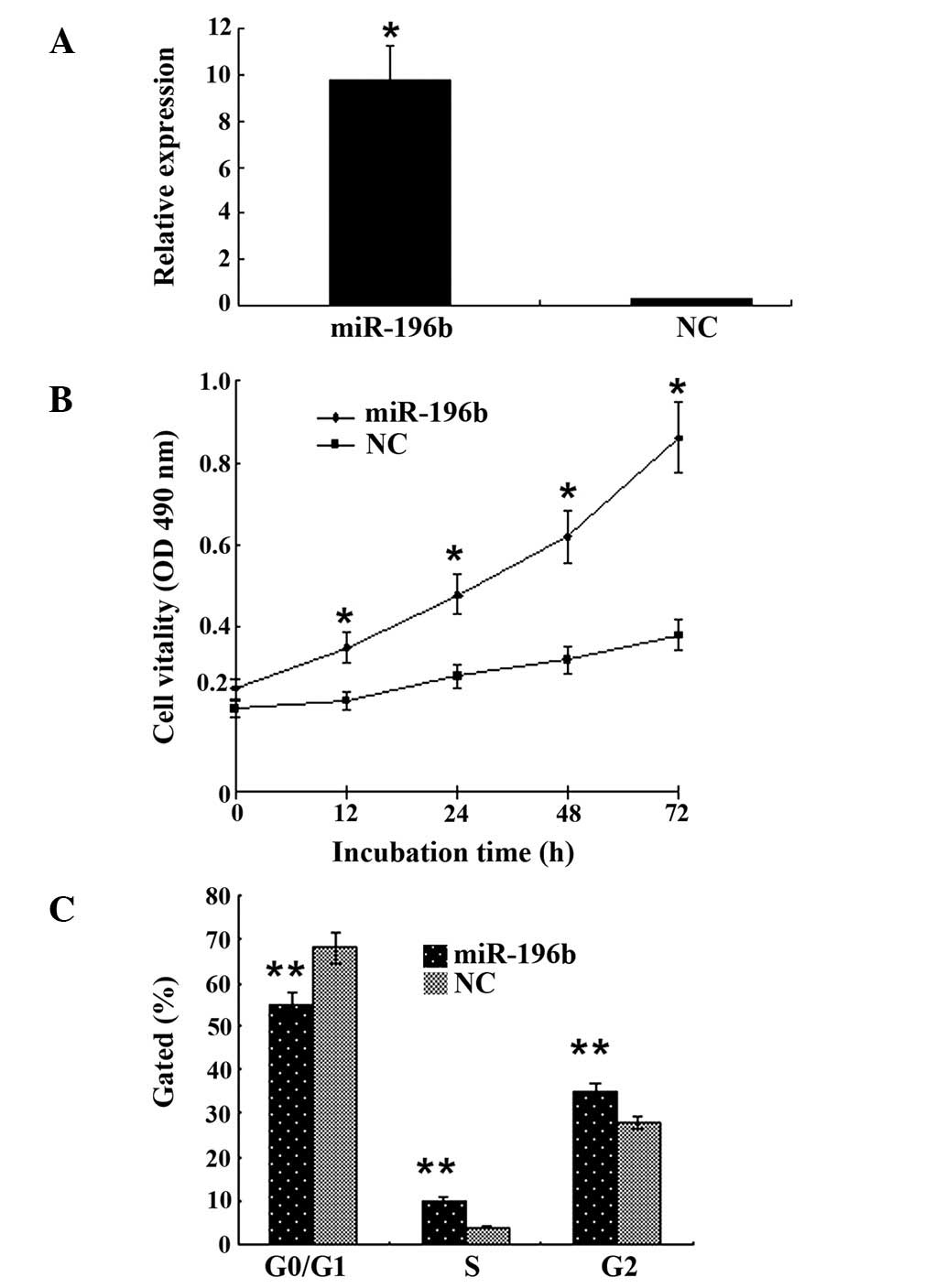
MKN28 cells exhibited green fluorescence when
transfected with lentivirus-mediated miR-196b. In addition, the
expression of miR-196b RNA was ~20-fold higher than that in the
control group (Fig. 1A). The results
of the MTT assay revealed that miR-196b activation enhanced the
proliferative ability of MKN28 cells compared with that of the
control group (P<0.01; Fig. 1B).
We assessed the effect of miR-196b on the cycle of gastric cancer
cells by flow cytometry and observed that gastric cancer cells in
the G0/G1 phase decreased, while those in the S and G2 phase of the
cell cycle increased compared with the control group (P<0.05;
Fig. 1C).
Effect of miR-196b on gastric cancer
cell cloning and invasion
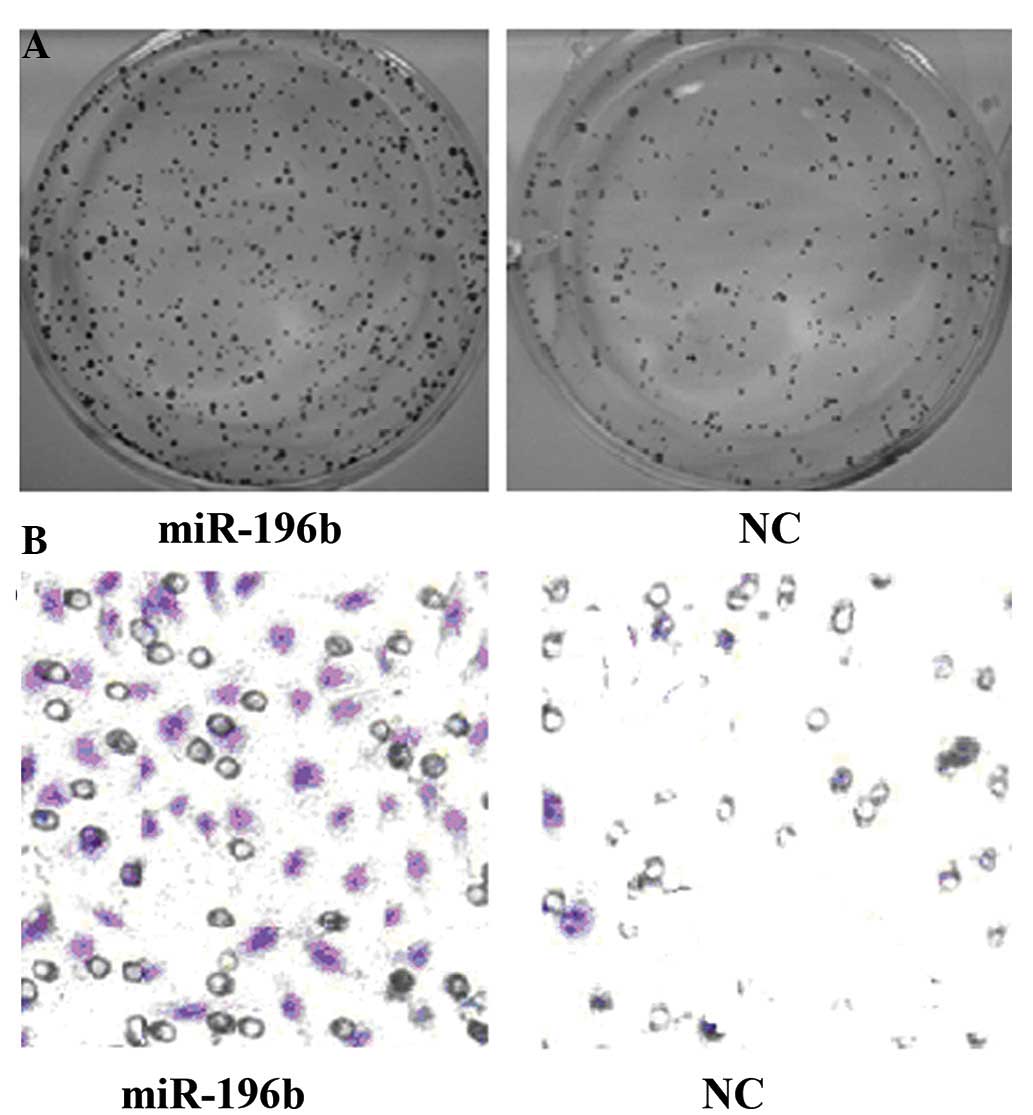
Colony-forming cell assay revealed that the cell
cloning efficiency increased to (78±12)% following miR-196b
activation compared with (32±8)% in the control group (P<0.01;
Fig. 2A). Transwell invasion assay
revealed (48±16)% cell migration following miR-196b activation,
which was far higher than the (12±4)% observed in the control group
(P<0.01; Fig. 2B).
In vitro tumor growth induced by
miR-196b transfection
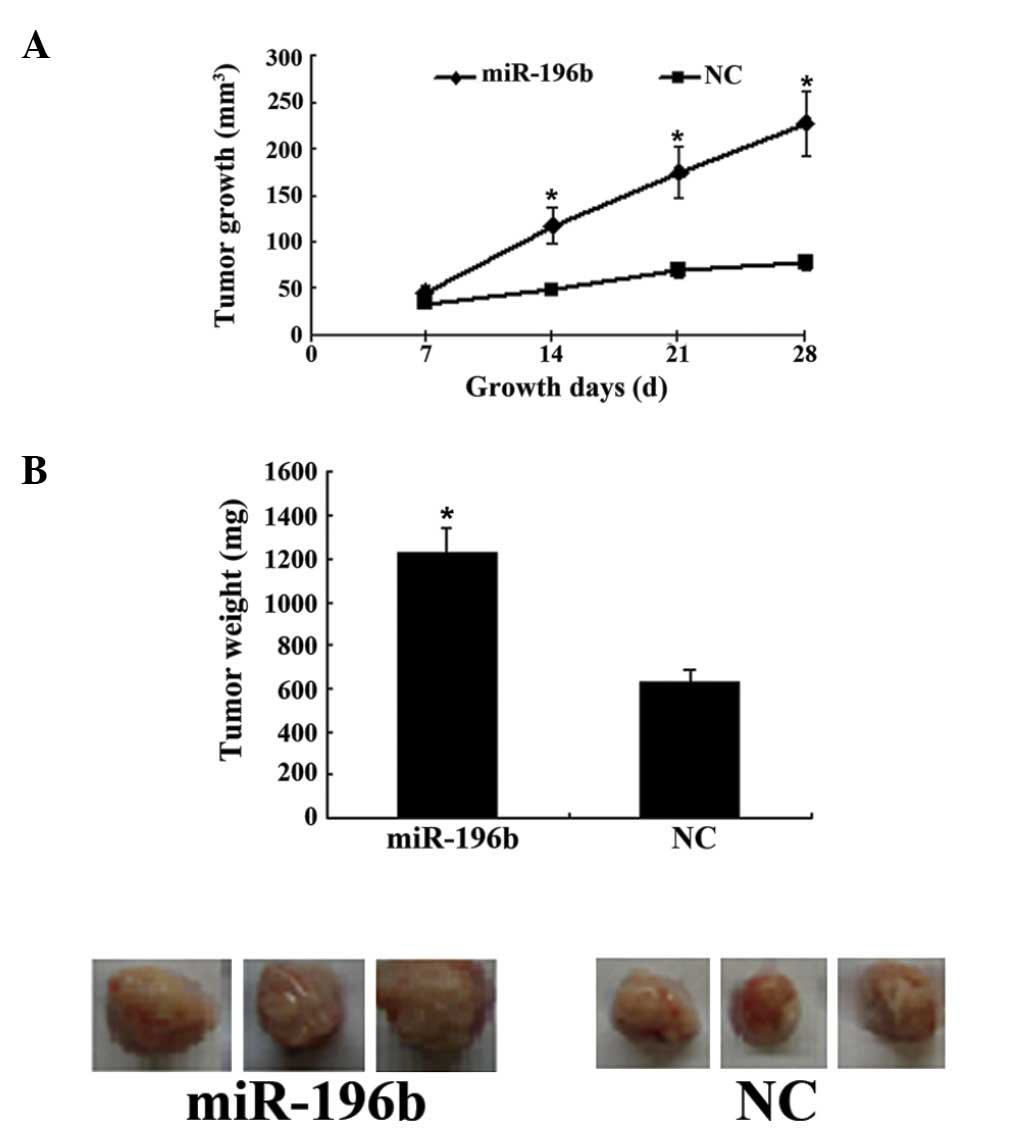
The results of the tumorigenecity test revealed
increased tumor growth following miR-196b transfection (Fig. 3A). Four weeks after injection of
miR-196b-transfected gastric cancer cells into nude mice, the tumor
weight was significantly higher than that of the control group
(P<0.01; Fig. 3B).
miR-196b regulates PI3K/AKT/mTOR
expression
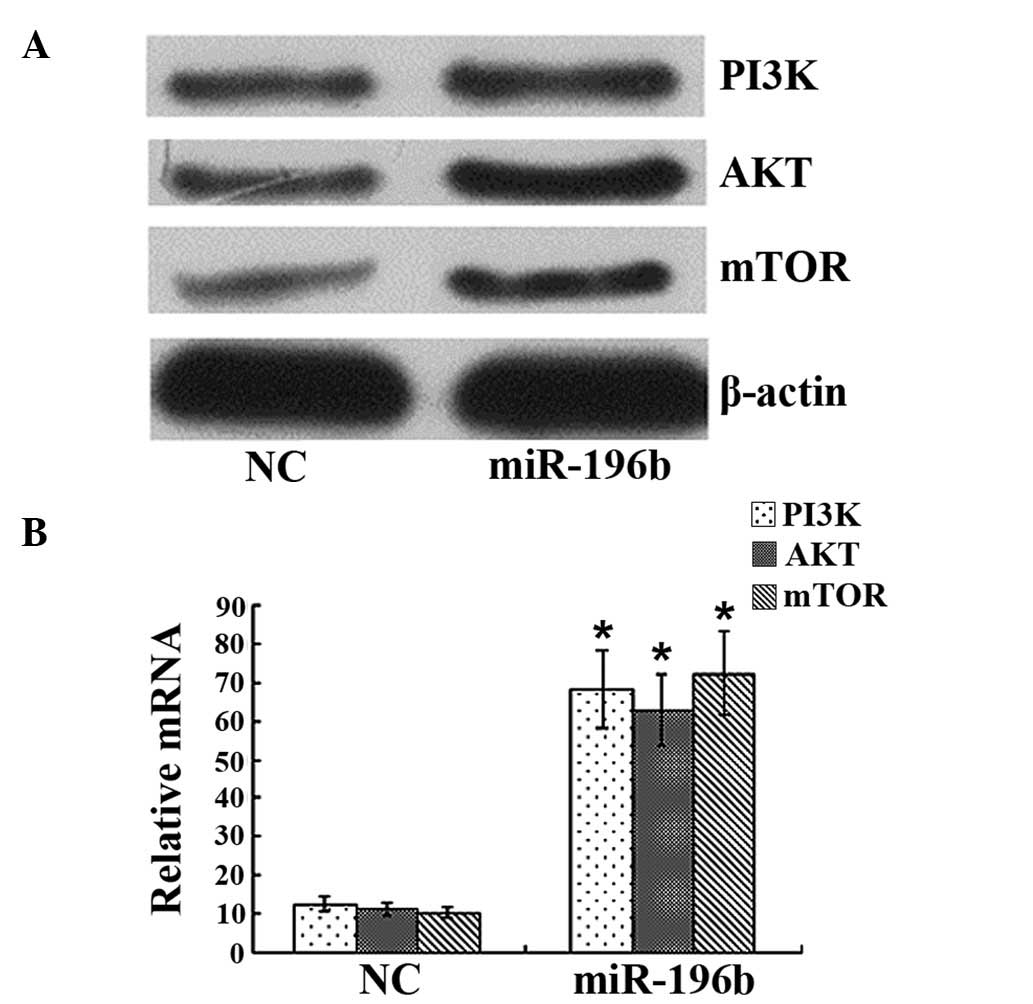
Western blot analysis revealed that the levels of
PI3K/AKT/mTOR protein in gastric cancer cells were 0.38±0.06,
0.46±0.08 and 0.22±0.06 in the miR-196b transfection group, which
was far higher than the levels 0.18±0.03, 0.16±0.04 and 0.09±0.03
observed in the negative control group (P<0.01; Fig. 4A). qPCR analysis demonstrated
significant upregulation of PI3K/AKT/mTOR mRNA in gastric cancer
cells following miR-196b transfection (Fig. 4B).
Discussion
A previous study reported that miR-196b played a
significant role in human tissue evolution and tumor growth with
particular emphasis on its role in the development and progression
of tumors (15). This provided a
promising targeted cancer therapy. miR-196b and miR-196a are each
members of the miR-196 family, despite miR-196b exhibiting one
basic group difference from miR-196a, they present similarities in
terms of their molecular structure and function. miR-196b was
demonstrated to have a positive effect on cancer proliferation, but
a negative effect on tumor cell apoptosis and differentiation was
implicated in the development and progression of leukemia and other
cancer types. The expression of miR-196b was upregulated in
short-term hematopoietic stem cells and downregulated in
highly-differentiated hematopoietic stem cells (16). In addition, in mixed-lineage leukemia
medullary cells, miR-196b demonstrated overexpression driven by the
pathogenically abnormal MLL fusion protein (16) and became a target of HOX genes
(17,18).
The regulatory role of miR-196b in gastric cancer
cells was investigated with lentivirus-mediated miR-196b
transfection into gastric cancer cells. To verify the role of
miR-196b in regulating gastric cancer cell proliferation, we
conducted an MTT assay to measure the proliferative ability of
gastric cancer cells following miR-196b activation, and observed
that miR-196b enhanced cell proliferation (Fig. 1B). In addition, changes in gastric
cancer cell cycle following miR-196b activation were detected using
flow cytometry. As a result, cells progressing from the G0/G1 phase
to the S phase were observed. Taken together, our results indicate
a significant regulatory role of miR-196b in gastric cancer cell
proliferation.
In our study, soft agar and Transwell assays were
performed to validate the role of miR-196b in regulating the
proliferation and migration of gastric cancer cells. Soft agar
colony formation assay is used to monitor tumor
anchorage-independent growth and tumor malignancy; i.e., a stronger
invasion ability of tumor cells is associated with a greater number
of cell colonies (19–21). The Transwell chamber model, which
imitates a cancer-associated microenvironment and extracellular
matrix, is known to be a reliable method for assaying cell invasion
ability (22). Our results identified
a marked increase in the cloning efficiency and migration rate of
gastric cancer cells following miR-196b activation, and implicated
a metastasis-promoting role of miR-196b in gastric cancer cells
(Fig. 2). In addition, to measure the
role of miR-196b in cancer genesis and growth in vitro, we
conducted tumorigenicity test in nude mice to assess cancer growth
following miR-196b activation. It was observed that miR-196b
induced tumor growth (Fig. 3).
The PI3K/AKT/mTOR signaling pathway is known to be
significant in the genesis, invasion and migration of gastric
cancer (23–26). Our study identified that PI3K/AKT/mTOR
mRNA and protein were upregulated in gastric cancer cells following
miR-196b activation, and implicated an active role of miR-196b in
gastric cancer cells via the PI3K/AKT/mTOR pathway.
High expression of miR-196b contributes to gastric
cancer proliferation and migration, and the mechanism is associated
with the interference of the PI3K/AKT/mTOR signaling pathway.
miR-196b may be a potential molecular target for gastric cancer
therapy.
References
|
1
|
Wu R, Li F, Zhu J, Tang R, Qi Q, Zhou X,
Li R, Wang W, Hua D and Chen W: A functional variant at mir-132-3p,
mir-212-3p and mir-361-5p binding site in cd80 gene alters
susceptibility to gastric cancer in a Chinese Han population. Med
Oncol. 31:602014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Xu YJ and Fan Y: Mir-215/192 participates
in gastric cancer progression. Clin Transl Oncol. 17:34–40. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Shen ZY, Zhang ZZ, Liu H, Zhao EH and Cao
H: Mir-375 inhibits the proliferation of gastric cancer cells by
repressing ERBB2 expression. Exp Ther Med. 7:1757–1761.
2014.PubMed/NCBI
|
|
4
|
Li H, Xie S, Liu M, Chen Z, Liu X, Wang L,
Li D and Zhou Y: The clinical significance of downregulation of
mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric
cancer tumorigenesis. Int J Oncol. 45:197–208. 2014.PubMed/NCBI
|
|
5
|
Xia J, Guo X, Yan J and Deng K: The role
of mir-148a in gastric cancer. J Cancer Res Clin Oncol.
140:1451–1456. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng Y, Li S, Ding Y, Wang Q, Luo H, Shi
Q, Hao Z, Xiao G and Tong S: The role of mir-18a in gastric cancer
angiogenesis. Hepatogastroenterology. 60:1809–1813. 2013.PubMed/NCBI
|
|
7
|
Duan Y, Hu L, Liu B, Yu B, Li J, Yan M, Yu
Y, Li C, Su L, Zhu Z, et al: Tumor suppressor miR-24 restrains
gastric cancer progression by downregulating regIV. Mol Cancer.
13:1272014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
He XJ, Ma YY, Yu S, Jiang XT, Lu YD, Tao
L, Wang HP, Hu ZM and Tao HQ: Up-regulated miR-199a-5p in gastric
cancer functions as an oncogene and targets klotho. BMC Cancer.
14:2182014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hornstein E, Mansfield JH, Yekta S, Hu JK,
Harfe BD, McManus MT, Baskerville S, Bartel DP and Tabin CJ: The
microRNA-miR-196 acts upstream of hoxb8 and shh in limb
development. Nature. 438:671–674. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sehm T, Sachse C, Frenzel C and Echeverri
K: miR-196 is an essential early-stage regulator of tail
regeneration, upstream of key spinal cord patterning events. Dev
Biol. 334:468–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kawasaki H and Taira K: MicroRNA-196
inhibits HOXB8 expression in myeloid differentiation of HL60 cells.
Nucleic Acids Symp Ser (Oxf). 48:211–212. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Brest P, Lapaquette P, Souidi M, Lebrigand
K, Cesaro A, Vouret-Craviari V, Mari B, Barbry P, Mosnier JF,
Hébuterne X, et al: A synonymous variant in IRGM alters a binding
site for miR-196 and causes deregulation of IRGM-dependent
xenophagy in crohn's disease. Nat Genet. 43:242–245. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hezova R, Kovarikova A, Bienertova-Vasku
J, Sachlova M, Redova M, Vasku A, Svoboda M, Radova L, Kiss I,
Vyzula R and Slaby O: Evaluation of SNPs in miR-196-a2, miR-27a and
miR-146a as risk factors of colorectal cancer. World J
Gastroenterol. 18:2827–2831. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hou W, Tian Q, Zheng J and Bonkovsky HL:
MicroRNA-196 represses Bach1 protein and hepatitis C virus gene
expression in human hepatoma cells expressing hepatitis C viral
proteins. Hepatology. 51:1494–1504. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chen C, Zhang Y, Zhang L, Weakley SM and
Yao Q: MicroRNA-196: Critical roles and clinical applications in
development and cancer. J Cell Mol Med. 15:14–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Popovic R, Riesbeck LE, Velu CS, Chaubey
A, Zhang J, Achille NJ, Erfurth FE, Eaton K, Lu J, Grimes HL, et
al: Regulation of mir-196b by MLL and its overexpression by MLL
fusions contributes to immortalization. Blood. 113:3314–3322. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schotte D, Lange-Turenhout EA, Stumpel DJ,
Stam RW, Buijs-Gladdines JG, Meijerink JP, Pieters R and Den Boer
ML: Expression of miR-196b is not exclusively MLL-driven but is
especially linked to activation of HOXA genes in pediatric acute
lymphoblastic leukemia. Haematologica. 95:1675–1682. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li Z, Huang H, Chen P, He M, Li Y,
Arnovitz S, Jiang X, He C, Hyjek E, Zhang J, et al: miR-196b
directly targets both HOXA9/MEIS1 oncogenes and FAS tumour
suppressor in MLL-rearranged leukaemia. Nat Commun. 3:6882012.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abbud-Antaki RA, Marhefka JN, DeLuca AL
and Zuromskis MP: The cancer biochip system: a functional genomic
assay for anchorage-independent three-dimensional breast cancer
cell growth. Horm Cancer. 3:261–270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guadamillas MC, Cerezo A and Del Pozo MA:
Overcoming anoikis-pathways to anchorage-independent growth in
cancer. J Cell Sci. 124:3189–3197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okabe H, Ishimoto T, Mima K, Nakagawa S,
Hayashi H, Kuroki H, Imai K, Nitta H, Saito S, Hashimoto D, et al:
CD44s signals the acquisition of the mesenchymal phenotype required
for anchorage-independent cell survival in hepatocellular
carcinoma. Br J Cancer. 110:958–966. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Marshall J: Transwell(®) invasion assays.
Methods Mol Biol. 769:97–110. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tapia O, Riquelme I, Leal P, Sandoval A,
Aedo S, Weber H, Letelier P, Bellolio E, Villaseca M, Garcia P and
Roa JC: The Pi3K/AKT/mTOR pathway is activated in gastric cancer
with potential prognostic and predictive significance. Virchows
Arch. 465:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sukawa Y, Yamamoto H, Nosho K, Ito M,
Igarashi H, Naito T, Mitsuhashi K, Matsunaga Y, Takahashi T, Mikami
M, et al: HER2 expression and Pi3K-Akt pathway alterations in
gastric cancer. Digestion. 89:12–17. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie X, Tang B, Zhou J, Gao Q and Zhang P:
Inhibition of the Pi3K/Akt pathway increases the chemosensitivity
of gastric cancer to vincristine. Oncol Rep. 30:773–782.
2013.PubMed/NCBI
|
|
26
|
Ye B, Jiang LL, Xu HT, Zhou DW and Li ZS:
Expression of Pi3K/AKT pathway in gastric cancer and its blockade
suppresses tumor growth and metastasis. Int J Immunopathol
Pharmacol. 25:627–636. 2012.PubMed/NCBI
|