Introduction
The Ras family small guanosine 5′-triphosphate
(GTP)-binding protein Rap2B is a member of the Ras oncogene family
(1). Ras proteins are known to be
promoters of tumorigenesis, and their expression has been observed
in a variety of human tumors (2). In
addition to the Rap family, Ras-related small GTPases include Ras,
Rho, adenosine diphosphate (ADP) ribosylation factor, Ras-related
nuclear protein and Rad, Rem, Rem2, Gem/Kir families (3–5). Ras is
important in the regulation of cell growth and differentiation
(6,7).
Furthermore, the Rap family presents 50–60% sequence homology with
the product of the Ras proto-oncogene (8). Rap2A, Rap2B and Rap2C belong to the Rap2
subfamily (8). Rap2B protein shares
~90% sequence homology with Rap2A, and 70% with RaplA and RaplB
(9). In addition, RaplB and Rap2B are
the only two members of the Rap family of GTPases that are
expressed at significant levels in circulating human platelets
(10–12). Additionally, Rap2B is
geranylgeranylated, and associates with the membranes of human
platelets and erythroleukemia cells (13,14).
The present review focusses on the possible
effectors and biological functions of Rap2B and summarizes current
progress in the field. Since an increasing number of studies
clearly supports the association between Rap2B and cancer, the
present review discusses the potential role of Rap2B as a target
for cancer therapy.
Identification and biological
characteristics of Rap2B
Identification of Rap2B
In 1990, Rap2B was first identified when a platelet
complementary (c)DNA library was screened (10,15).
Ohmstede et al (10) screened
the platelet cDNA expression library with the anti-H-Ras monoclonal
antibody M90, which is derived from human platelets. A specific
epitope on the Ras-encoded p21 protein (amino acids, 107–130) was
identified by the antibody, and an encoded partial amino acid
sequence of a protein that was closely associated with Rap2 was
identified by DNA sequence analysis of one clone (16). The authors revealed that this protein
had 90% homology with Rap2 at the amino acid level, with
variability at the carboxyl-terminus. The protein was named Rap2B.
Although Rap2B is 90% identical to Rap2 at the amino acid level
with variability at the carboxyl-terminus, the expression and
localization of different Rap2 family members remain
tissue-specific for the different isoforms (10). Thus, Rap1B and Rap2B are present in
the membrane of human platelets, while Rap2C localizes to the
plasma membrane of eukaryotic cells (17). Additional studies have demonstrated
that Rap2B is mainly expressed in human neutrophils, and the
expression of Rap1B in platelets is ~10-fold higher than the
expression of Rap2B (18). In
addition, Torti and Lapetina (17)
identified that the Rap2B protein, which is located at the cell
membrane, was subjected to post-translational modifications,
including isoprenylation, proteolysis and carboxymethylation. The
characteristic intracellular localization of Rap2B suggests that it
may exert a variety of cellular functions (19). Furthermore, an algorithm was developed
to predict amino acid positions that may be exchanged to create
switched functional mutants (20).
The algorithm was validated by rendering switch-of-function mutants
for Rap2B, which may be further investigated by combining
genome-wide experimental functional classification (20).
Basic structural features of
Rap2B
Since its identification in 1990, Rap2B has elicited
a considerable interest (1,10). The Rap2B gene, which has a
conservative Rap domain, is located at the 3q25.2 region of the
human chromosome (which has been extensively explored in previous
studies on cancer), and has four expressed sequence tags (21,22). The
cDNA of the Rap2B gene is composed of an open reading frame of 552
bp, which shares 84.2% nucleotide and 89.6% amino acid homology
with Rap2 (15). Using an integrative
genomic approach, Zhang et al (1) identified Rap2B as a conserved
p53-activated gene, which inhibited p53-mediated apoptosis
following DNA damage. Upon DNA damage, p53 directly binds to the
promoter of Rap2B and activates its transcription (1). Since the specificity of the gene
determines the structure and function of the protein, the Rap2B
protein is expected to exhibit a specific protein structure and
function (23).
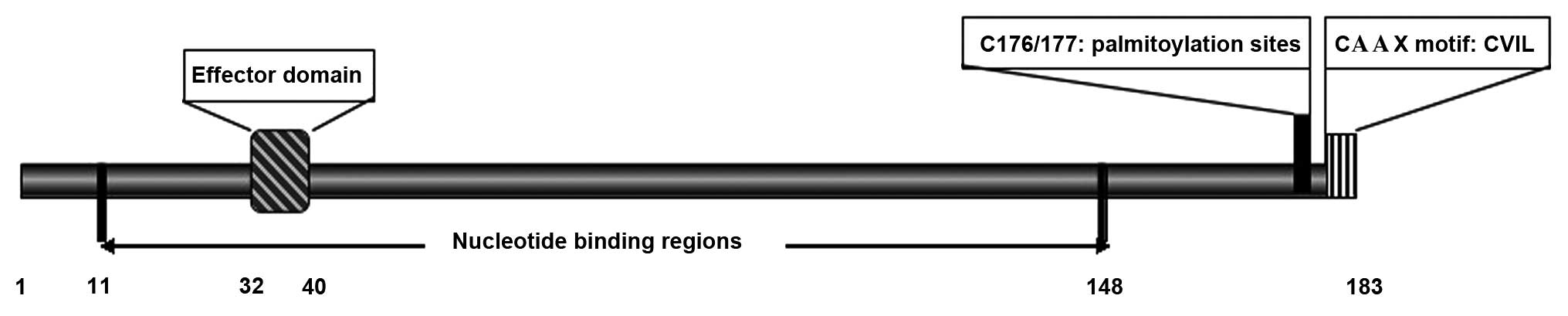
Rap2B encodes 183 amino acids, and is a Ras-related
GTP-binding protein of low molecular weight (24). The structure of the Rap2B protein is
similar to that of Ras proteins (8),
and consists of an effector domain (amino acids, 32–40) that
interacts with downstream effectors; nucleotide binding regions
(amino acids, 11–148) that mediate the interaction between
guanosine diphosphate (GDP) and GTP; and a carboxy-terminal CAAX
motif (consisting of a cysteine followed by two aliphatic residues
and one random amino acid) that targets proteins to the cell
membrane (Fig. 1) (5,25). All the
proteins of the Rap2 family present a cysteine residue (C180)
downstream of the cysteines C176 and C177 in the CAAX motif
(26). A previous study demonstrated
that palmitoylation occurs at the C176/177 sites, which requires
CAAX processing (27). The C-terminus
is also the site of sequential post-translational modifications
(26). Despite the fact that the open
reading frame of Rap2B shares 84.2% nucleotide and 89.6% amino acid
homology with Rap2, its C-terminal region is different (15). The Rap2B clone contains the amino acid
sequence CVIL, whereas Rap2 terminates with CNIQ (15). In addition, the insertion of a
polyisoprenic tail in the cysteine residue of the CAAX motif has
been demonstrated to be involved in post-translational
modifications (14).
Regulation of Rap2B activity
The regulation of Rap2B as a molecular switch of
signaling pathways is determined by its association with GDP (‘off’
position) or GTP (‘on’ position) (28). Similarly to other GTPases that serve
as molecular switches by cycling between GTP-bound active and
GDP-bound inactive forms (5), the
Rap2B protein is considered to be active when is associated with
GTP, and inactive when is associated with GDP (29). Therefore, the Rap2B protein functions
as a binary switch by cycling between two interconvertible states:
A GDP-bound inactive and a GTP-bound active form (28). A previous study comparing the kinetics
of nucleotide binding and release revealed that Rap2B bound GTP
more efficiently and possessed a faster rate of GDP release than
its highly homologous Rap2C (8).
Furthermore, in the presence of magnesium (Mg2+), the
relative affinity of Rap2B for GTP was ~7-fold higher than its
affinity for GDP (30). However,
under the same conditions, the relative affinity of Rap2C for GTP
was only ~2-fold higher, compared with its affinity for GDP
(8). In addition, the binding of GTP
to Rap2B was stronger and more rapid in the absence of
Mg2+ (30). Although the
specific reason was unclear, the present review hypothesizes that
this may be due to the concurrence of various intrinsic properties
of this protein (19). Rap2B encodes
intrinsic GTPase enzymatic activity, and the function of Rap2B is
regulated by guanine nucleotide exchange factors (GEFs) and
GTPase-activating proteins (GAPs). The rapid and sustained binding
of GTP to Rap2B has been previously observed to be induced by
thrombin, which stimulates heterotrimeric G protein-coupled
receptors (GPCRs), and the glycoprotein (GP) VI ligand convulxin,
which activates a tyrosine kinase-based signaling pathway (18). Thrombin- and convulxin-induced
activation of Rap2B were not observed to be dependent on
thromboxane A2 (18). However,
intracellular calcium (Ca2+) was observed to be capable
of regulating the activation of Rap2B. Furthermore, Rap2B
activation induced by thrombin was required for
phosphatidylinositol (PI) 3-kinase activity (18). In addition, Rap2B is also activated by
the GEF exchange protein directly activated by cyclic adenosine
monophosphate (cAMP) (Epac), which is regulated by cAMP (30–35).
Regulators of Rap2B activity
The process of cycling between GTP and GDP is
facilitated by cytosolic factors and generally induced by GEFs
(36). Inactivation occurs through
the intrinsic GTPase activity of Rap2B, which converts bound GTP
into GDP, and is stimulated by GAPs (28). Furthermore, in small GTP-binding
proteins, GAPs enhance the intrinsic GTPase activity to hydrolyze
GTP to GDP, whereas GEFs promote the release of bound GDP and the
capture of a new GTP molecule (25).
Rap2B and Rap have similar activation and deactivation regulatory
factors, including several GEFs and GAPs, which are capable of
regulating the activity of Rap proteins (37). GEFs that regulate the activity of
Rap2B include C3G (a GEF bound to the adaptor protein c-Crk), Epac,
Ras guanyl-releasing protein (GRP) 2 and Ras/Rap1A-associating
(RA)-GEF-1, while Rap GAPs include Rap1GAPII and suppressor of
phyA-105 1 (38–40).
Potential downstream effectors of Rap2B
Rap2B belongs to the Rap2 family of small
GTP-binding proteins, and shares 90% sequence homology with Rap2 at
the amino acid level, with marked variability at the
carboxyl-terminus (10). Although
specific downstream effectors of Rap2B are unknown, it may be
hypothesized that specific effectors of Rap2B are similar to those
of Rap2. There are various specific effectors of Rap2, including
mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4),
misshapen/nuclear factor kappa-light-chain-enhancer of activated B
cells (NF-κB)-inducing kinase-related kinase (MINK), tumor necrosis
factor receptor-associated factor 2 and noncatalytic region of
tyrosine kinase-interacting kinase (TNIK), protein tyrosine
phosphatase-like protein 1-associated RhoGAP 1 (PARG1) and Rap2
interacting protein 9 (RPIP9) (41–43). These
Rap2 specific effectors interact with Rap2 through the C-terminal
Citron homology domain (44).
MAP4K4 (also termed hepatocyte progenitor
kinase-like/germinal center kinase-like kinase), MINK and TNIK
belong to the human sterile 20 (STE20)/MAP4K family (45). MAP4K4 has been demonstrated to be
highly expressed in the majority of tumor cell lines. Machida et
al (44) revealed that MAP4K4
regulates c-Jun N-terminal kinase (JNK), and observed that
MAP4K4-induced activation of JNK was enhanced by Rap2. Collins
et al (46) demonstrated that
the effects of MAP4K4 on promoting cellular migration were mediated
through JNK, independently of activator protein 1 (AP-1) activation
and downstream transcription. MINK is highly expressed in the
brain, and its interaction with Rap2 is GTP-dependent and requires
the presence of phenylalanine at position 39 within the effector
region of Rap2 (43). TNIK was
observed to be activated by the palmitoylation-deficient mutant of
mouse Rap2B, and Rap2B promoted the growth and development of tumor
cells through the activation and interaction with TNIK (42). Furthermore, Rap2B does not induce a
TNIK-mediated cellular phenotype, but TNIK activation requires
palmitoylation-independent membrane-association of Rap2B (47). In addition, all Rap2 proteins,
including Rap2B, require palmitoylation for the induction of the
TNIK-mediated phenotype, which led to the suppression of
proliferation of human embryonic kidney (HEK)293T cells in a
previous study (47). Nonaka et
al (43) confirmed that MINK and
TNIK interact with a postsynaptic scaffold protein containing
tetratricopeptide repeats, ankyrin repeats and a coiled-coil
region, inducing its phosphorylation, which is enhanced by
Rap2.
PARG1 is a putative specific effector of Rap2 that
regulates Rho and exhibits RhoGAP activity in vitro
(25). PARG1 and ZK669.1a, a protein
that contains a RhoGAP domain, share a homology region, and the
Caenorhabditis elegans ortholog of Rap2 has been
demonstrated to interact with ZK669.1a (25). Rap2 suppresses the PARG1-induced
cytoskeletal alterations required for Rho inactivation in
vivo (25).
RPIP9 is coded by the multidrug resistance protein 1
gene, which is upregulated in numerous tumors, and overlaps with a
non-characterized gene transcribed from the opposite strand
(48). The predicted protein exhibits
high homology to human RPIP8, and has a RUN domain located near its
C-terminus (49). The activation of
RPIP9 occurs during malignant breast epithelial transformation, and
its expression increases with the progression of cancer toward an
invasive phenotype (48). However,
the role of Rap2 in breast cancer remains unknown. The specific
downstream effector of Rap2B is unclear. However, the open reading
frame of Rap2B shares 84.2% nucleotide and 89.6% amino acid
homology with Rap2. Therefore, the present study hypothesizes that
Rap2B regulates a variety of signaling pathways by interacting with
MAP4K4, MINK, TNIK, PARG1 and RPIP9 (Fig.
2).
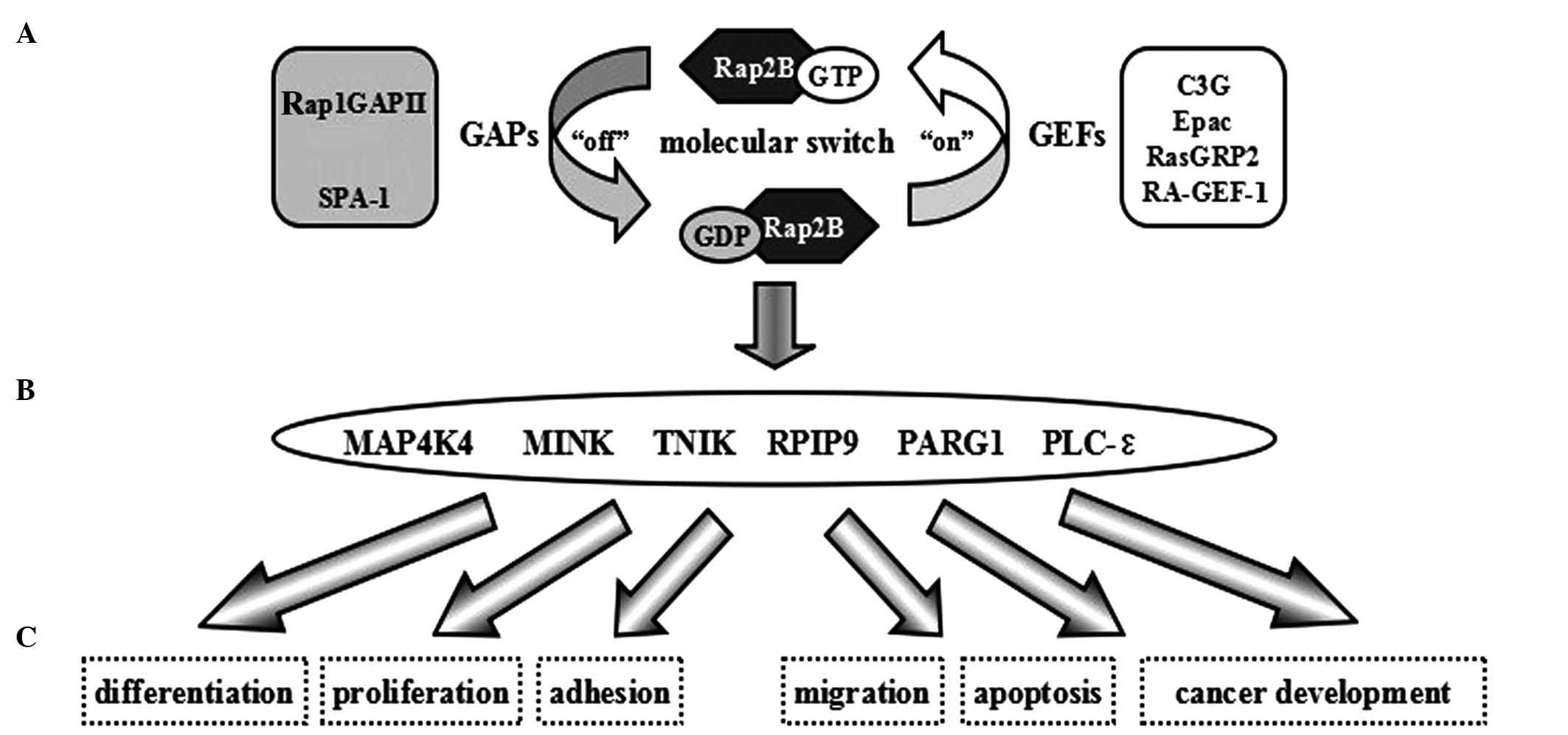 | Figure 2.Signaling pathway and biological
function of Rap2B, a Ras family small guanosine
5′-triphosphate-binding protein. (A) Rap2B functions as a binary
switch. (B) Specific effectors of Rap2B. (C) Rap2B regulates
various cellular biological functions by interacting with specific
effectors. GAP, GTPase-activating protein; SPA-1, suppressor of
phyA-105 1; GTP, guanosine 5′-triphosphate; GDP, guanosine
diphosphate; GEF, guanine nucleotide exchange factor; Epac,
exchange protein directly activated by cyclic adenosine
monophosphate; GRP, guanyl-releasing protein; RA,
Ras/Rap1A-associating; MAP4K4, mitogen-activated protein kinase
kinase kinase kinase 4; MINK, misshapen/NIK-related kinase; NIK,
NF-κB–inducing kinase; NF-κB, nuclear factor
kappa-light-chain-enhancer of activated B cells; TNIK, TRAF2 and
NCK-interacting kinase; TRAF2; TNF receptor-associated factor 2;
TNF, tumor necrosis factor; NCK, noncatalytic region of tyrosine
kinase; RPIP9, Rap2 interacting protein 9; PARG1, PTPL1-associated
RhoGAP 1; PLC, phospholipase C. |
Biological functions of Rap2B
Acting as molecular switches, small GTPases are able
to regulate several cellular processes, including adhesion,
proliferation, differentiation and apoptosis (27). In the Ras subfamily, the interaction
of the Ras family members depends on the potential for interactions
(which is dictated by their structure), the subcellular
localization of the particular protein and its potential regulators
and effectors (50). The affinities
of the different Ras members for their regulators or effectors and
their precise subcellular localization results in various
biological functions (51).
Rap2B, as a novel p53 target, regulates the
p53-mediated pro-survival function
The tumor suppressor p53 is a DNA sequence-specific
transcription factor and a stress sensor (52–54). Zhang
et al (1) identified Rap2B as
a novel p53 target that mediates cell survival following DNA
damage. Consistent with its pro-survival function, the authors also
revealed via analysis of cancer genomic data that Rap2B is
overexpressed in numerous types of tumors. Rap2B exhibits weak
transforming activity, which was observed using
anchorage-independent growth assays, suggesting that Rap2B is not
an oncogene by itself (1). This also
suggests that targeting Rap2B may sensitize tumor cells to
apoptosis induced by DNA damage (1).
Unpublished data by the authors of the present study also confirmed
that Rap2B is the direct target gene of p53, and has a
p53-dependent pro-survival function.
Translocation of Rap1B and Rap2B to the
cytoskeleton via von Willebrand factor (vWF) involves Fcγ receptor
II (FcγRII)-mediated protein tyrosine phosphorylation
Rap1B and Rap2B are the only members of the Rap
family of GTPases that are expressed at significant levels in
circulating human platelets (8,17).
Notably, Rap1B is ≥10 times more abundant than Rap2B in platelets
(17). Previous kinetic studies
demonstrated that the translocation of Rap1B to the cytoskeleton
preceded the translocation of Rap2B, which only occurs in the late
phase of platelet aggregation in thrombin-stimulated platelets
(55,56). Greco et al (18) revealed that the activation of Rap2B
may be stimulated by thrombin or convulxin in platelets. However,
the specific mechanisms of translocation of Rap1B and Rap2B to the
cytoskeleton remain unclear. The large GP vWF is synthesized by
megakaryocytes and endothelial cells, and is important in thrombus
formation and platelet adhesion (57). In platelets, the GPIb-IX–V complex, a
member of the leucine-rich GP gene family, is the main receptor for
vWF (58). In 1999, Torti et
al (59) revealed that the
translocation of Rap1B and Rap2B to the cytoskeleton was induced by
vWF, following binding of vWF to the GPIb-IX–V complex through a GP
IIb/IIIa-independent mechanism. In a previous study, Rap2B was
prevented from associating with the cytoskeleton by cytochalasin D,
which did not inhibit platelet aggregation (60). These results provide a novel role for
FcγRII in regulating the translocation of Rap proteins to the
cytoskeleton and mediating protein tyrosine phosphorylation.
Targeting of Rap2B to lipid rafts is
promoted by palmitoylation at C176 and C177, and is required for
efficient protein activation in blood platelets
Lipid rafts are dynamic membrane microdomains
abundant in cholesterol and glycosphingolipids (61), which appear to be important for human
platelet activation (62,63) and are also implicated in signal
transduction. Furthermore, there are specific differences in the
mechanisms for agonist-induced activation of Rap1B and Rap2B,
particularly in their dependence on secreted ADP in human platelets
(18). Previous studies have observed
that the thrombin-induced activation of Rap2B was significantly
reduced when secreted ADP was neutralized (64). In 2008, Canobbio et al
(27) observed that 20% of all the
membrane-bound Rap2B protein localizes to the membrane microdomain
lipid rafts, while the majority of membrane-associated Rap2B is
outside these microdomains. The authors also revealed that the
association of Rap2B to lipid rafts is promoted by palmitoylation
of C176 and C177, which are located at the C-terminal region of the
protein. These residues are required for the complete activation of
Rap2B, and are induced by stimulation of human platelets (27). Although additional studies are
required to fully elucidate the biochemical and functional
characterization of Rap2B in platelets, the above previous results
indicate a novel biochemical property of Rap2B, and demonstrate the
important role of Rap2B in regulating the activation and
aggregation of blood platelets.
Rap2B interacts with phospholipase C (PLC)-ε
and activates it
Inositol-specific mammalian PLC enzymes are
multidomain proteins whose functions are regulated by G proteins
(65). Heterotrimeric G proteins and
Ras-like GTPases directly activate the isozymes of PLC (66). In a wide variety of membrane
receptors, the hydrolysis of PI 4,5-bisphosphate via stimulation of
phosphoinositide-specific PLC, and the subsequent generation of
inositol 1,4,5-trisphosphate, is used as the main Ca2+
signaling pathway (67). In 2001,
Schmidt et al (68) identified
a novel PLC and Ca2+ signaling pathway that was mediated
by a small GTPase of the Rap family, and was triggered by cAMP. The
authors also provided evidence that these receptor responses,
mediated by Rap2B, activated Epac, which was regulated by cAMP
(31,32), and involved the PLC-ε isoform
(33–25). PLC-ε is a novel PLC that possesses a
cell division cycle 25 GEF domain and two Ras-associating domains,
of which RA2 is critical for Ras-mediated activation of the enzyme
(69,70). In 2003, Wing et al (71) reported that PLC-ε senses and mediates
the crosstalk between heterotrimeric and small GTPase signaling
pathways, acting as a multifunctional nexus protein. Additional
studies indicated that PLC-ε is important in promoting bladder cell
transitional proliferation, and small GTPases of the Ras and Rho
families have been observed to control the activity of PLC-ε
(72–74). The findings from the study by Lopez
et al (33) demonstrate that
PLC-ε is regulated by the heterotrimeric G protein Gα, and
activates the small G protein Ras/mitogen-activated protein kinase
(MAPK) signaling pathway. Stope et al (75) suggested that epidermal growth factor
receptor triggers the activation of Rap2B via PLCα1 activation and
tyrosine phosphorylation of the GEF RasGRP3 by the proto-oncogene
tyrosine-protein kinase c-Src, which results in the stimulation of
PLC-ε. In 2002, Evellin et al (76) demonstrated that PLC-ε appeared to be
stimulated by GPCRs through the formation of cAMP and activation of
Rap2B. Rap2B may promote cell adhesion, spread, migration and
polarity, in addition to integrin activation, axonal outgrowth and
phagocytosis, through interacting with R-Ras effectors such as
PLC-ε (76–83).
In addition, a diverse array of cellular functions
are coordinated by interferon (IFN)-γ through the transcriptional
regulation of immunologically relevant genes (84). In 2005, Gollob et al (85) revealed that the growth of melanoma
cells was affected by IFN-γ, and the anti-melanoma effect of IFN-γ
in the human melanoma DM6 cell line was associated with the
downregulation of multiple genes involved in G protein signaling
and PLC activation, including Rap2B and calpain 3, using DNA
microarray analysis. Therefore, the data provided novel insights
into the signaling events and gene expression alterations
associated with the growth inhibition and apoptosis of melanoma
cells, which may result in the identification of novel targets for
melanoma therapy (86). In summary,
following activation by heterotrimeric G protein signaling, Rap2B
interacts with PLC-ε, activating it, which affects cell growth by
activating the Ras-Raf-MAPK/extracellular signal-regulated kinase
(ERK) signaling pathway (71).
Rap2B is a novel candidate gene cloned from
lung cancer cells
Rap2B is one of the 50 novel candidate genes cloned
from differential expression cDNA libraries constructed in lung
cancer cells (87). In 2007, Liu
et al (88) used the
suppression subtractive hybridization method to identify
differentially expressed genes in lung squamous cell carcinoma
(SCC). The results revealed that the messenger RNA and protein
expression levels of Rap2B in lung cancer tissues was increased,
compared with normal tissues. A reporter gene assay demonstrated
that Rap2B activated the NF-κB pathway >3-fold, compared with
the mock vector (87). Although the
specific mechanism remains unclear, this observation implied that
Rap2B may play a potential role in the development and progression
of lung SCC.
Potential mechanism of Rap2B in tumor
development
Similarly to other Ras proteins, Rap2B is associated
with the occurrence and development of tumors (1,87). Certain
studies have reported an association between the function of Rap2B
and the development of malignant tumors, and increasing evidence
clearly confirms this association (1,87).
Although the mechanism of Rap2B in cancer is not clear, the
function of Rap2B relies on its gene homology and protein structure
(23). Therefore, it may be
hypothesized that Rap2B is involved in tumor development.
The formation of tumors results from abnormal
proliferation of normal cells, which usually present as abnormal
masses in the body (89). Numerous
studies have observed that the occurrence and development of cancer
are complex processes that are regulated by multiple genes and
factors (90). The abnormal
activation of oncogenes and inactivation of tumor suppressor genes
serve as a major contribution to tumor development (91). The open reading frame of Rap2B shares
84.2% nucleotide and 89.6% amino acid homology with Rap2 (15). Therefore, Rap2B may regulate a variety
of signaling pathways, in addition to cell spreading through its
interaction with MAP4K4, MINK, TNIK, PARG1 and RPIP9 (92,93). By
contrast, PLC-ε, which is activated by and interacts with Rap2B,
facilitates cell growth through activating the Ras-Raf-MAPK/ERK
pathway, which increases intracellular Ca+2 levels and
activates protein kinase C (75).
Therefore, Rap2B may promote the development of tumors through its
interaction with PLC-ε. In addition, it has been observed that ERK
activation by β2-adrenergic receptor and prostaglandin E1 receptor
in HEK293T cells and mouse neuroblastoma N1E-115 cells,
respectively, is mediated by cAMP-activated Epac proteins, which
leads to an increase in the intracellular levels of Ca+2
via Rap2B and PLC-ε stimulation (38,94). This
results in the activation of H-Ras, which triggers the MAPK cascade
(38), and may also regulate the
interaction between PLC-ε and Rap2B.
In vivo, the number and volume of Ras-induced
tumor nodules is increased significantly by JNK deficiency, and the
oncogenic effects of Ras are suppressed by the JNK signaling
pathway (95). Growth arrest and
apoptosis of certain tumor cells is caused by inhibition of JNK
(96–98). Phosphorylation of c-Jun and increase
of AP-1 transcriptional activity may be a result of activating
Rap2B by the JNK/MAPK signaling cascade (80). This interaction between JNK and Rap2B
is also a possible mechanism that explains tumor development.
Autophagy is considered a cell survival mechanism,
which is activated in response to various stress signals, including
high temperature, oxidative stress and accumulation of damaged
organelles (99). The pathogenesis of
clinically important disorders in a variety of organ systems
contribute to the dysregulation of autophagy (100). Staphylococcus aureus is a
pathogen that colonizes the lungs of patients with cystic fibrosis
(101) and causes serious infectious
diseases (102). In 2012, Mestre and
Colombo (103) demonstrated that
S. aureus induces an autophagic response to promote
bacterial growth. The authors also revealed that S.
aureus-induced autophagy may be regulated by RapGEF3 and Rap2B
through calpain activation, and activated RapGEF3 and Rap2B may
prevent the action of S. aureus by decreasing intracellular
cAMP levels. Therefore, Rap2B may regulate the autophagy and
survival of S. aureus to affect tumor development.
In 2004, McLeod et al (104) provided evidence for the regulation
of integrins by Rap2B in B cells. The authors described that cell
adhesion and spreading, in addition to actin polymerization and
integrin-mediated Pyk2 tyrosine phosphorylation, are regulated by
Rap GTPases in B cells. Consequently, the present review
hypothesizes that Rap2B may regulate the immune system by promoting
tumorigenesis. Although certain biochemical and functional roles of
Rap2B have been elucidated, additional studies are required to
fully understand the functional mechanism of Rap2B and its
association with tumor development.
Conclusion
Previous studies have suggested that targeting Rap2B
may sensitize tumor cells to undergo apoptosis in response to DNA
damage, since Rap2B is a conserved p53-activated gene (1). However, anticancer drugs that target
Rap2B remain a theory, since an improved understanding at a
molecular level of exactly how Rap2B functions as a tumor promoter
is required prior to the development of novel drugs targeting
Rap2B. Therefore, the identification of the functions of Rap2B may
result in novel avenues of research aimed to improve therapeutics
and prognosis of human malignancies.
Inhibiting the expression of Rap2B may potentially
be useful in cancer therapy, and has gained attention in the
treatment of various types of cancer that display increased
expression of Rap2B proteins (105).
This may offer novel therapeutic strategies for the treatment of
human carcinoma, and may eventually lead to the development of a
novel class of anticancer drugs that target Rap2B, and promote the
development of sensitive biomarkers for cancer diagnosis and
treatment. Future studies on Rap2B will provide evidence and
generate mechanistic hypotheses regarding the development of
cancer. Identifying and understanding the functionally important
Ras family of proteins may clarify the biology of cancer and lead
to novel therapeutic and diagnostic opportunities for patients
affected by this disease.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant nos.
81272207 and 81201637) and Natural Science Foundation for Colleges
and Universities in the Jiangsu Province (Beijing, China; grant no.
14KJB320023). Miss Jiehui Di was sponsored by the Qing Lan Project
of Jiangsu Province (Nanjing, China).
Glossary
Abbreviations
Abbreviations:
|
cAMP
|
cyclic adenosine monophosphate
|
|
Epac
|
exchange protein directly activated by
cAMP
|
|
GAP
|
GTPase-activating protein
|
|
GDP
|
guanosine diphosphate
|
|
GEF
|
guanine nucleotide exchange factor
|
|
GP
|
glycoprotein
|
|
GPCR
|
G protein-coupled receptor
|
|
GTP
|
guanosine 5′-triphosphate
|
|
JNK
|
c-Jun N-terminal kinase
|
|
MAP4K4
|
mitogen-activated protein kinase
kinase kinase kinase 4
|
|
MINK
|
misshapen/NIK-related kinase
|
|
NCK
|
noncatalytic region of tyrosine
kinase
|
|
NF-κB
|
nuclear factor
kappa-light-chain-enhancer of activated B cells
|
|
NIK
|
NF-κB-inducing kinase
|
|
PARG1
|
PTPL1-associated RhoGAP 1
|
|
PLC
|
phospholipase C
|
|
PI
|
phosphatidylinositol
|
|
PTPL1
|
protein tyrosine phosphatase-like
protein 1 RA, Ras/Rap1A-associating
|
|
RhoGAP
|
Rho GTPase-activating protein
|
|
RPIP9
|
Rap2 interacting protein 9
|
|
SCC
|
squamous cell carcinoma
|
|
TNF
|
tumor necrosis factor
|
|
TNIK
|
TRAF2 and NCK-interacting kinase
|
|
TRAF2
|
TNF receptor-associated factor 2
|
|
vWF
|
von Willebrand factor
|
References
|
1
|
Zhang X, He Y, Lee KH, Dubois W, Li Z, Wu
X, Kovalchuk A, Zhang W and Huang J: Rap2b, a novel p53 target,
regulates p53-mediated pro-survival function. Cell cycle.
12:1279–1291. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takashima A and Faller DV: Targeting the
RAS oncogene. Expert Opin Ther Targets. 17:507–531. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mackay DJ and Hall A: Rho GTPases. J Biol
Chem. 273:20685–20688. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vojtek AB and Der CJ: Increasing
complexity of the Ras signaling pathway. J Biol Chem.
273:19925–19928. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takai Y, Sasaki T and Matozaki T: Small
GTP-binding proteins. Physiol Rev. 81:153–208. 2001.PubMed/NCBI
|
|
6
|
Bourne HR, Sanders DA and McCormick F: The
GTPase superfamily: A conserved switch for diverse cell functions.
Nature. 348:125–132. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bourne HR, Sanders DA and McCormick F: The
GTPase superfamily: Conserved structure and molecular mechanism.
Nature. 349:117–127. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Paganini S, Guidetti GF, Catricalà S,
Trionfini P, Panelli S, Balduini C and Torti M: Identification and
biochemical characterization of Rap2C, a new member of the Rap
family of small GTP-binding proteins. Biochimie. 88:285–295. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Greco F, Ciana A, Pietra D, Balduini C,
Minetti G and Torti M: Rap2, but not Rap1 GTPase is expressed in
human red blood cells and is involved in vesiculation. Biochim
Biophys Acta. 1763:330–335. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ohmstede CA, Farrell FX, Reep BR,
Clemetson KJ and Lapetina EG: RAP2B: A RAS-related GTP-binding
protein from platelets. Proc Natl Acad Sci USA. 87:6527–6531. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lapetina EG, Lacal JC, Reep BR and Vedia
Molinay L: A ras-related protein is phosphorylated and translocated
by agonists that increase cAMP levels in human platelets. Proc Natl
Acad Sci USA. 86:3131–3134. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Klinz FJ, Seifert R, Schwaner I, Gausepohl
H, Frank R and Schultz G: Generation of specific antibodies against
the rap1A, rap1B and rap2 small GTP-binding proteins. Analysis of
rap and ras proteins in membranes from mammalian cells. Eur J
Biochem. 207:207–213. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Winegar DA, Molina y Vedia L and Lapetina
EG: Isoprenylation of rap2 proteins in platelets and human
erythroleukemia cells. J Biol Chem. 266:4381–4386. 1991.PubMed/NCBI
|
|
14
|
Farrell FX, Yamamoto K and Lapetina EG:
Prenyl group identification of rap2 proteins: A ras superfamily
member other than ras that is farnesylated. Biochem J. 289:349–355.
1993. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Farrell FX, Ohmstede CA, Reep BR and
Lapetina EG: cDNA sequence of a new ras-related gene (rap2b)
isolated from human platelets with sequence homology to rap2.
Nucleic Acids Res. 18:42811990. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lerosey I, Chardin P, de Gunzburg J and
Tavitian A: The product of the rap2 gene, member of the ras
superfamily. Biochemical characterization and site-directed
mutagenesis. J Biol Chem. 266:4315–4321. 1991.PubMed/NCBI
|
|
17
|
Torti M and Lapetina EG: Structure and
function of rap proteins in human platelets. Thromb Haemost.
71:533–543. 1994.PubMed/NCBI
|
|
18
|
Greco F, Sinigaglia F, Balduini C and
Torti M: Activation of the small GTPase Rap2B in agonist-stimulated
human platelets. J Thromb Haemost. 2:2223–2230. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen J, Liang H and Fernández A: Protein
structure protection commits gene expression patterns. Genome Biol.
9:R1072008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Heo WD and Meyer T: Switch-of-function
mutants based on morphology classification of Ras superfamily small
GTPases. Cell. 113:315–328. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Sun W, Zhang K, Zhang X, Lei W, Xiao T, Ma
J, Guo S, Shao S, Zhang H, Liu Y, et al: Identification of
differentially expressed genes in human lung squamous cell
carcinoma using suppression subtractive hybridization. Cancer Lett.
212:83–93. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
An Q, Pacyna-Gengelbach M, Schlüns K,
Deutschmann N, Guo S, Gao Y, Zhang J, Cheng S and Petersen I:
Identification of differentially expressed genes in immortalized
human bronchial epithelial cell line as a model for in vitro study
of lung carcinogenesis. Int J Cancer. 103:194–204. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schlicker A, Domingues FS, Rahnenführer J
and Lengauer T: A new measure for functional similarity of gene
products based on Gene Ontology. BMC Bioinformatics. 7:3022006.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Colicelli J: Human RAS superfamily
proteins and related GTPases. Sci STKE. 2004:RE132004.PubMed/NCBI
|
|
25
|
Myagmar BE, Umikawa M, Asato T, Taira K,
Oshiro M, Hino A, Takei K, Uezato H and Kariya K: PARG1, a
protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2
effector. Biochem Biophys Res Commun. 329:1046–1052. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Taguchi T and Misaki R: Palmitoylation
pilots ras to recycling endosomes. Small GTPases. 2:82–84. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Canobbio I, Trionfini P, Guidetti GF,
Balduini C and Torti M: Targeting of the small GTPase Rap2b, but
not Rap1b, to lipid rafts is promoted by palmitoylation at Cys176
and Cys177 and is required for efficient protein activation in
human platelets. Cell Signal. 20:1662–1670. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Raaijmakers JH and Bos JL: Specificity in
Ras and Rap signaling. J Biol Chem. 284:10995–10999. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hattori M and Minato N: Rap1 GTPase:
Functions, regulation, and malignancy. J Biochem. 134:479–484.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Molinay Vedia L, Ohmstede CA and Lapetina
EG: Properties of the exchange rate of guanine nucleotides to the
novel rap-2B protein. Biochem Biophys Res Commun. 171:319–324.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Rooij J, Zwartkruis FJ, Verheijen MH,
Cool RH, Nijman SM, Wittinghofer A and Bos JL: Epac is a Rap1
guanine-nucleotide-exchange factor directly activated by cyclic
AMP. Nature. 396:474–477. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
de Rooij J, Rehmann H, van Triest M, Cool
RH, Wittinghofer A and Bos JL: Mechanism of regulation of the Epac
family of cAMP-dependent RapGEFs. J Biol Chem. 275:20829–20836.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lopez I, Mak EC, Ding J, Hamm HE and
Lomasney JW: A novel bifunctional phospholipase c that is regulated
by Galpha 12 and stimulates the Ras/mitogen-activated protein
kinase pathway. J Biol Chem. 276:2758–2765. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kelley GG, Reks SE, Ondrako JM and Smrcka
AV: Phospholipase C(epsilon): A novel Ras effector. EMBO J.
20:743–754. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Song C, Hu CD, Masago M, Kariyai K,
Yamawaki-Kataoka Y, Shibatohge M, Wu D, Satoh T and Kataoka T:
Regulation of a novel human phospholipase C, PLCepsilon, through
membrane targeting by Ras. J Biol Chem. 276:2752–2757. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cherfils J and Chardin P: GEFs: Structural
basis for their activation of small GTP-binding proteins. Trends
Biochem Sci. 24:306–311. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Stork PJ: Does Rap1 deserve a bad Rap?
Trends Biochem Sci. 28:267–275. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Keiper M, Stope MB, Szatkowski D, Böhm A,
Tysack K, Dorp Vom F, Saur O, Weernink Oude PA, Evellin S, Jakobs
KH and Schmidt M: Epac- and Ca2+-controlled activation
of Ras and extracellular signal-regulated kinases by Gs-coupled
receptors. J Biol Chem. 279:46497–46508. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Rebhun JF, Castro AF and Quilliam LA:
Identification of guanine nucleotide exchange factors (GEFs) for
the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J
Biol Chem. 275:34901–34908. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gasper R, Sot B and Wittinghofer A: GTPase
activity of Di-Ras proteins is stimulated by Rap1GAP proteins.
Small GTPases. 1:133–141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ryu J, Futai K, Feliu M, Weinberg R and
Sheng M: Constitutively active Rap2 transgenic mice display fewer
dendritic spines, reduced extracellular signal-regulated kinase
signaling, enhanced long-term depression, and impaired spatial
learning and fear extinction. J Neurosci. 28:8178–8188. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Taira K, Umikawa M, Takei K, Myagmar BE,
Shinzato M, Machida N, Uezato H, Nonaka S and Kariya K: The Traf2-
and Nck-interacting kinase as a putative effector of Rap2 to
regulate actin cytoskeleton. J Biol Chem. 279:49488–49496. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Nonaka H, Takei K, Umikawa M, Oshiro M,
Kuninaka K, Bayarjargal M, Asato T, Yamashiro Y, Uechi Y, Endo S,
et al: MINK is a Rap2 effector for phosphorylation of the
postsynaptic scaffold protein TANC1. Biochem Biophys Res Commun.
377:573–578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Machida N, Umikawa M, Takei K, Sakima N,
Myagmar BE, Taira K, Uezato H, Ogawa Y and Kariya K:
Mitogen-activated protein kinase kinase kinase kinase 4 as a
putative effector of Rap2 to activate the c-Jun N-terminal kinase.
J Biol Chem. 279:15711–15714. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wright JH, Wang X, Manning G, LaMere BJ,
Le P, Zhu S, Khatry D, Flanagan PM, Buckley SD, Whyte DB, et al:
The STE20 kinase HGK is broadly expressed in human tumor cells and
can modulate cellular transformation, invasion, and adhesion. Mol
Cell Biol. 23:2068–2082. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Collins CS, Hong J, Sapinoso L, Zhou Y,
Liu Z, Micklash K, Schultz PG and Hampton GM: A small interfering
RNA screen for modulators of tumor cell motility identifies MAP4K4
as a promigratory kinase. Proc Natl Acad Sci USA. 103:3775–3780.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Uechi Y, Bayarjargal M, Umikawa M, Oshiro
M, Takei K, Yamashiro Y, Asato T, Endo S, Misaki R, Taguchi T and
Kariya K: Rap2 function requires palmitoylation and recycling
endosome localization. Biochem Biophys Res Commun. 378:732–737.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Raguz S, De Bella MT, Slade MJ, Higgins
CF, Coombes RC and Yagüe E: Expression of RPIP9 (Rap2 interacting
protein 9) is activated in breast carcinoma and correlates with a
poor prognosis. Int J Cancer. 117:934–941. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Wang S, Zhang Z, Ying K, Chen JZ, Meng XF,
Yang QS, Xie Y and Mao YM: Cloning, expression, and genomic
structure of a novel human Rap2 interacting gene (RPIP9). Biochem
Genet. 41:13–25. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Okamura SM, Oki-Idouchi CE and Lorenzo PS:
The exchange factor and diacylglycerol receptor RasGRP3 interacts
with dynein light chain 1 through its C-terminal domain. J Biol
Chem. 281:36132–36139. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Nomura K, Kanemura H, Satoh T and Kataoka
T: Identification of a novel domain of Ras and Rap1 that directs
their differential subcellular localizations. J Biol Chem.
279:22664–22673. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Vousden KH and Prives C: Blinded by the
light: The growing complexity of p53. Cell. 137:413–431. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Rozan LM and El-Deiry WS: p53 downstream
target genes and tumor suppression: A classical view in evolution.
Cell Death Differ. 14:3–9. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kruse JP and Gu W: Modes of p53
regulation. Cell. 137:609–622. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Fischer TH, Gatling MN, Lacal JC and White
GC II: rap1B, a cAMP-dependent protein kinase substrate, associates
with the platelet cytoskeleton. J Biol Chem. 265:19405–19408.
1990.PubMed/NCBI
|
|
56
|
Torti M, Ramaschi G, Sinigaglia F,
Lapetina EG and Balduini C: Association of the low molecular weight
GTP-binding protein rap2B with the cytoskeleton during platelet
aggregation. Proc Natl Acad Sci USA. 90:7553–7557. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Meyer D and Girma JP: von Willebrand
factor: Structure and function. Thromb Haemost. 70:99–104.
1993.PubMed/NCBI
|
|
58
|
Clemetson KJ: Platelet GPIb-V–IX complex.
Thromb Haemost. 78:266–270. 1997.PubMed/NCBI
|
|
59
|
Torti M, Bertoni A, Canobbio I, Sinigaglia
F, Lapetina EG and Balduini C: Rap1B and Rap2B translocation to the
cytoskeleton by von Willebrand factor involves FcgammaII
receptor-mediated protein tyrosine phosphorylation. J Biol Chem.
274:13690–13697. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Rosado JA and Sage SO: Farnesylcysteine
analogues inhibit store-regulated Ca2+ entry in human
platelets: Evidence for involvement of small GTP-binding proteins
and actin cytoskeleton. Biochem J. 347:183–192. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Pike LJ: Lipid rafts: Heterogeneity on the
high seas. Biochem J. 378:281–292. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Gousset K, Wolkers WF, Tsvetkova NM,
Oliver AE, Field CL, Walker NJ, Crowe JH and Tablin F: Evidence for
a physiological role for membrane rafts in human platelets. J Cell
Physiol. 190:117–128. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Bodin S, Tronchère H and Payrastre B:
Lipid rafts are critical membrane domains in blood platelet
activation processes. Biochim Biophys Acta. 1610:247–257. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Torti M, Ramaschi G, Sinigaglia F,
Lapetina EG and Balduini C: Glycoprotein IIb-IIIa and the
translocation of Rap2B to the platelet cytoskeleton. Proc Natl Acad
Sci USA. 91:4239–4243. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Drin G and Scarlata S: Stimulation of
phospholipase Cbeta by membrane interactions, interdomain movement,
and G protein binding - how many ways can you activate an enzyme?
Cell Signal. 19:1383–1392. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Hicks SN, Jezyk MR, Gershburg S, Seifert
JP, Harden TK and Sondek J: General and versatile autoinhibition of
PLC isozymes. Mol Cell. 31:383–394. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Ehrlich LS, Medina GN and Carter CA: ESCRT
machinery potentiates HIV-1 utilization of the
PI(4,5)P(2)-PLC-IP3R-Ca(2+) signaling cascade. J Mol Biol.
413:347–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Schmidt M, Evellin S, Weernink PA, von
Dorp F, Rehmann H, Lomasney JW and Jakobs KH: A new
phospholipase-C-calcium signalling pathway mediated by cyclic AMP
and a Rap GTPase. Nat Cell Biol. 3:1020–1024. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Kelley GG, Reks SE and Smrcka AV: Hormonal
regulation of phospholipase Cepsilon through distinct and
overlapping pathways involving G12 and Ras family G-proteins.
Biochem J. 378:129–139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Seifert JP, Zhou Y, Hicks SN, Sondek J and
Harden TK: Dual activation of phospholipase C-epsilon by Rho and
Ras GTPases. J Biol Chem. 283:29690–29698. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Wing MR, Bourdon DM and Harden TK:
PLC-epsilon: A shared effector protein in Ras-, Rho-, and G alpha
beta gamma-mediated signaling. Mol Interv. 3:273–280. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jin TG, Satoh T, Liao Y, Song C, Gao X,
Kariya K, Hu CD and Kataoka T: Role of the CDC25 homology domain of
phospholipase Cepsilon in amplification of Rap1-dependent
signaling. J Biol Chem. 276:30301–30307. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Song C, Satoh T, Edamatsu H, Wu D, Tadano
M, Gao X and Kataoka T: Differential roles of Ras and Rap1 in
growth factor-dependent activation of phospholipase C epsilon.
Oncogene. 21:8105–8113. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Wing MR, Snyder JT, Sondek J and Harden
TK: Direct activation of phospholipase C-epsilon by Rho. J Biol
Chem. 278:41253–41258. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Stope MB, Vom Dorp F, Szatkowski D, Böhm
A, Keiper M, Nolte J, Oude Weernink PA, Rosskopf D, Evellin S,
Jakobs KH and Schmidt M: Rap2B-dependent stimulation of
phospholipase C-epsilon by epidermal growth factor receptor
mediated by c-Src phosphorylation of RasGRP3. Mol Cell Biol.
24:4664–4676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Evellin S, Nolte J, Tysack K, vom Dorp F,
Thiel M, Weernink PA, Jakobs KH, Webb EJ, Lomasney JW and Schmidt
M: Stimulation of phospholipase C-epsilon by the M3 muscarinic
acetylcholine receptor mediated by cyclic AMP and the GTPase Rap2B.
J Biol Chem. 277:16805–16813. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Ivins JK, Yurchenco PD and Lander AD:
Regulation of neurite outgrowth by integrin activation. J Neurosci.
20:6551–6560. 2000.PubMed/NCBI
|
|
78
|
Jeong HW, Nam JO and Kim IS: The
COOH-terminal end of R-Ras alters the motility and morphology of
breast epithelial cells through Rho/Rho-kinase. Cancer Res.
65:507–515. 2005.PubMed/NCBI
|
|
79
|
Keely PJ, Rusyn EV, Cox AD and Parise LV:
R-Ras signals through specific integrin alpha cytoplasmic domains
to promote migration and invasion of breast epithelial cells. J
Cell Biol. 145:1077–1088. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Kwong L, Wozniak MA, Collins AS, Wilson SD
and Keely PJ: R-Ras promotes focal adhesion formation through focal
adhesion kinase and p130(Cas) by a novel mechanism that differs
from integrins. Mol Cell Biol. 23:933–949. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Self AJ, Caron E, Paterson HF and Hall A:
Analysis of R-Ras signalling pathways. J Cell Sci. 114:1357–1366.
2001.PubMed/NCBI
|
|
82
|
Sethi T, Ginsberg MH, Downward J and
Hughes PE: The small GTP-binding protein R-Ras can influence
integrin activation by antagonizing a Ras/Raf-initiated integrin
suppression pathway. Mol Biol Cell. 10:1799–1809. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Wozniak MA, Kwong L, Chodniewicz D, Klemke
RL and Keely PJ: R-Ras controls membrane protrusion and cell
migration through the spatial regulation of Rac and Rho. Mol Biol
Cell. 16:84–96. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Schroder K, Hertzog PJ, Ravasi T and Hume
DA: Interferon-gamma: An overview of signals, mechanisms and
functions. J Leukoc Biol. 75:163–189. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Gollob JA, Sciambi CJ, Huang Z and
Dressman HK: Gene expression changes and signaling events
associated with the direct antimelanoma effect of IFN-gamma. Cancer
Res. 65:8869–8877. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Avery-Kiejda KA, Bowden NA, Croft AJ,
Scurr LL, Kairupan CF, Ashton KA, Talseth-Palmer BA, Rizos H, Zhang
XD, Scott RJ and Hersey P: P53 in human melanoma fails to regulate
target genes associated with apoptosis and the cell cycle and may
contribute to proliferation. BMC Cancer. 11:2032011. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Fu G, Liu Y, Yuan J, Zheng H, Shi T, Lei
W, Xiao T, Gao Y and Cheng S: Identification and functional
analysis of a novel candidate oncogene RAP2B in lung cancer.
Zhongguo Fei Ai Za Zhi. 12:273–276. 2009.(In Chinese). PubMed/NCBI
|
|
88
|
Liu Y, Sun W, Zhang K, Zheng H, Ma Y, Lin
D, Zhang X, Feng L, Lei W, Zhang Z, et al: Identification of genes
differentially expressed in human primary lung squamous cell
carcinoma. Lung Cancer. 56:307–317. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tang X, Mo C, Wang Y, Wei D and Xiao H:
Anti-tumour strategies aiming to target tumour-associated
macrophages. Immunology. 138:93–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Boffetta P, Winn DM, Ioannidis JP, Thomas
DC, Little J, Smith GD, Cogliano VJ, Hecht SS, Seminara D, Vineis P
and Khoury MJ: Recommendations and proposed guidelines for
assessing the cumulative evidence on joint effects of genes and
environments on cancer occurrence in humans. Int J Epidemiol.
41:686–704. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Lee EY and Muller WJ: Oncogenes and tumor
suppressor genes. Cold Spring Harb Perspect Biol. 2:a0032362010.
View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tsygankova OM, Wang H and Meinkoth JL:
Tumor cell migration and invasion are enhanced by depletion of Rap1
GTPase-activating protein (Rap1GAP). J Biol Chem. 288:24636–24646.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Pannekoek WJ, Linnemann JR, Brouwer PM,
Bos JL and Rehmann H: Rap1 and Rap2 antagonistically control
endothelial barrier resistance. PLoS One. 8:e579032013. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Borland G, Smith BO and Yarwood SJ: EPAC
proteins transduce diverse cellular actions of cAMP. Br J
Pharmacol. 158:70–86. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Kennedy NJ, Sluss HK, Jones SN, Bar-Sagi
D, Flavell RA and Davis RJ: Suppression of Ras-stimulated
transformation by the JNK signal transduction pathway. Genes Dev.
17:629–637. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Potapova O, Gorospe M, Bost F, Dean NM,
Gaarde WA, Mercola D and Holbrook NJ: c-Jun N-terminal kinase is
essential for growth of human T98G glioblastoma cells. J Biol Chem.
275:24767–24775. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Bost F, McKay R, Bost M, Potapova O, Dean
NM and Mercola D: The Jun kinase 2 isoform is preferentially
required for epidermal growth factor-induced transformation of
human A549 lung carcinoma cells. Mol Cell Biol. 19:1938–1949. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Davis RJ: Signal transduction by the JNK
group of MAP kinases. Cell. 103:239–252. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Margaritopoulos GA, Tsitoura E, Tzanakis
N, Spandidos DA, Siafakas NM, Sourvinos G and Antoniou KM:
Self-eating: Friend or foe? The emerging role of autophagy in
idiopathic pulmonary fibrosis. BioMed Res Int. 2013:4204972013.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Haspel JA and Choi AM: Autophagy: A core
cellular process with emerging links to pulmonary disease. Am J
Respir Crit Care Med. 184:1237–1246. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Jarry TM, Memmi G and Cheung AL: The
expression of alpha-haemolysin is required for Staphylococcus
aureus phagosomal escape after internalization in CFT-1 cells.
Cell Microbiol. 10:1801–1814. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Mestre MB, Fader CM, Sola C and Colombo
MI: Alpha-hemolysin is required for the activation of the
autophagic pathway in Staphylococcus aureus-infected cells.
Autophagy. 6:110–125. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Mestre MB and Colombo MI:
Staphylococcus aureus promotes autophagy by decreasing
intracellular cAMP levels. Autophagy. 8:1865–1867. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
McLeod SJ, Shum AJ, Lee RL, Takei F and
Gold MR: The Rap GTPases regulate integrin-mediated adhesion, cell
spreading, actin polymerization, and Pyk2 tyrosine phosphorylation
in B lymphocytes. J Biol Chem. 279:12009–12019. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Di JH, Qu DB, Lu Z, Li LT, Cheng Q, Xin Y,
Zhang LZ, Zhang Y and Zheng JN: Rap2B promotes migration and
invasion of human suprarenal epithelioma. Tumour Biol.
35:9387–9394. 2014. View Article : Google Scholar : PubMed/NCBI
|