Introduction
Prostate cancer (PCa) is one of the most prevalent
malignant tumours and is the second leading cause of
cancer-associated mortality among men in Western countries
(1). For men with localised PCa,
radical prostatectomy (RP) is considered to be the ideal therapy;
however, determining the optimal management strategy for locally
advanced PCa remains a challenging issue (2). Traditionally, the malignant properties
of PCa are characterised based predominantly on clinical stage,
biopsy Gleason score (GS) and serum prostate-specific antigen (PSA)
level, individually or collectively. These factors are helpful for
guiding treatment decisions; however, they have limited predictive
ability (3). Therefore, there is a
critical need to develop novel prognostic predictors to improve
clinical strategies for the treatment of PCa.
Extra-prostatic extension (EPE) is defined as the
presence of cancer extending beyond the prostate gland, and it has
long been considered an unfavourable prognostic factor in terms of
cancer progression and survival (4–7).
Identifying the presence of EPE is likely to reduce the chance of
positive surgical margins; furthermore, it may be helpful for
identifying patients who require postoperative adjuvant treatment.
Traditionally, it has been difficult to assess patients who are at
high risk for EPE based on a single preoperative
clinicopathological variable or imaging information due to limited
sensitivity (8–12). Hence, it is of practical significance
to develop a new approach for predicting EPE.
Transmembrane protease serine 2
(TMPRSS2)-ETS-related gene (ERG) is the most common
gene fusion in PCa; however, its prognostic value remains largely
elusive (13–15). Our group has previously reported an
initial scoring system for assessing ERG rearrangements in
biopsy samples, based on the use of fluorescence in situ
hybridisation (FISH), for the diagnosis of PCa and the risk
assessment of lymph node metastasis. This proposed system has
demonstrated excellent sensitivity and specificity (16,17). In
clinical practice, an increase in ERG rearrangements was
observed to be associated with more aggressive characteristics.
Thus, the aim of the current study was to explore the utility of
ERG rearrangements and other preoperative parameters for
predicting EPE in patients with clinically localised PCa.
Materials and methods
Patients and samples
This study included 409 consecutive patients who
underwent RP at the Third Affiliated Hospital of Sun Yat-Sen
University (Guangzhou, China) between January 2008 and June 2013.
Of these patients, 103 without complete information prior to biopsy
or with suspected metastasis by bone scan, computed tomography scan
or magnetic resonance imaging (MRI) were excluded from the study.
Finally, 306 cases with clinically localised PCa were enrolled in
this retrospective analysis. The diagnosis of PCa was confirmed via
transrectal ultrasound (TRUS)-guided needle biopsy preoperatively
(median total biopsy cores, 12; range, 10–16). This study was
approved by the Institutional Ethics Committee of the Third
Affiliated Hospital of Sun Yat-Sen University, and all patients
signed informed consent forms prior to the intervention.
Biopsies and corresponding prostatectomy specimens
were retrospectively collected from 306 PCa patients for analysis.
The selection of slides for FISH analysis was performed by the
pathologist conducting the diagnosis. The ERG rearrangement
was calibrated with a dual-colour break-apart FISH assay (Beijing
GP Medical Technologies, Ltd., Beijing, China) as previously
described (16–18).
Pathological analysis
Morphological diagnoses were conducted according to
the International Union Against Cancer 2009 staging classification
guidelines for PCa (19), and
histological analyses were performed according to the Gleason
grading system (20). EPE was defined
as the presence of any malignant cell beyond the prostatic capsule
(≥pT3a) according to the criteria described by Epstein (21), or the presence of pathologically
confirmed positive lymph node metastasis.
Assessment of ERG rearrangements via
FISH
FISH analysis was conducted according to the
manufacturer's protocols (Beijing GP Medical Technologies, Ltd.)
with certain modifications. Briefly, 3-mm tissue sections were
obtained from tissue blocks and mounted on poly-L-lysine-coated
slides (Beijing GP Medical Technologies, Ltd.). Following
deparaffinisation, the tissue sections were dehydrated in 100, 85
and 70% ethanol for 2 min each. Subsequent to washing in deionised
water for 5 min, the sections were boiled in deionised water at
100°C for 27 min and then digested with Proteinase K (Beijing GP
Medical Technologies, Ltd.) at 37°C for 10 min. The sections on the
slides were then dried, and hybridisation was performed as
described previously (16,17). Next, the slides were counterstained
and mounted with DAPI, examined under an oil objective at 100x
magnification using an Olympus fluorescence microscope (BX51;
Olympus Corp., Tokyo, Japan) and imaged with a CCD camera (DP70;
Olympus Corp.) using the PathFinder CellScan software system
(IMSTAR S.A., Paris, France).
According to our scoring system, two yellow
(red/green fusion) signals in a cell indicate a normal signal
pattern, whereas the presence of one yellow/one green or one
yellow/one green/one red signal in a cell commonly represents an
abnormal signal pattern indicative of a partial deletion or
translocation, respectively, of ERG.
During the evaluation of the FISH results, each
slide was reviewed and ≥400 epithelial cells were scored, with the
strongest abnormal signals in the ‘z’ axis. ERG
rearrangement rate in the patient was calculated using the
following formula: ERG rearrangement rate (%) = number of cells
exhibiting an abnormal signal pattern/number of cancer cells
(16,17). All slices were independently assessed
by three experienced researchers (Dr Li Lu, Dr Hao Zhang and Dr
Guo-Liang Hou) who were blinded to the clinicopathological
parameters, and any discordant results were reassessed until a
consensus was achieved (Fig.
1A–F).
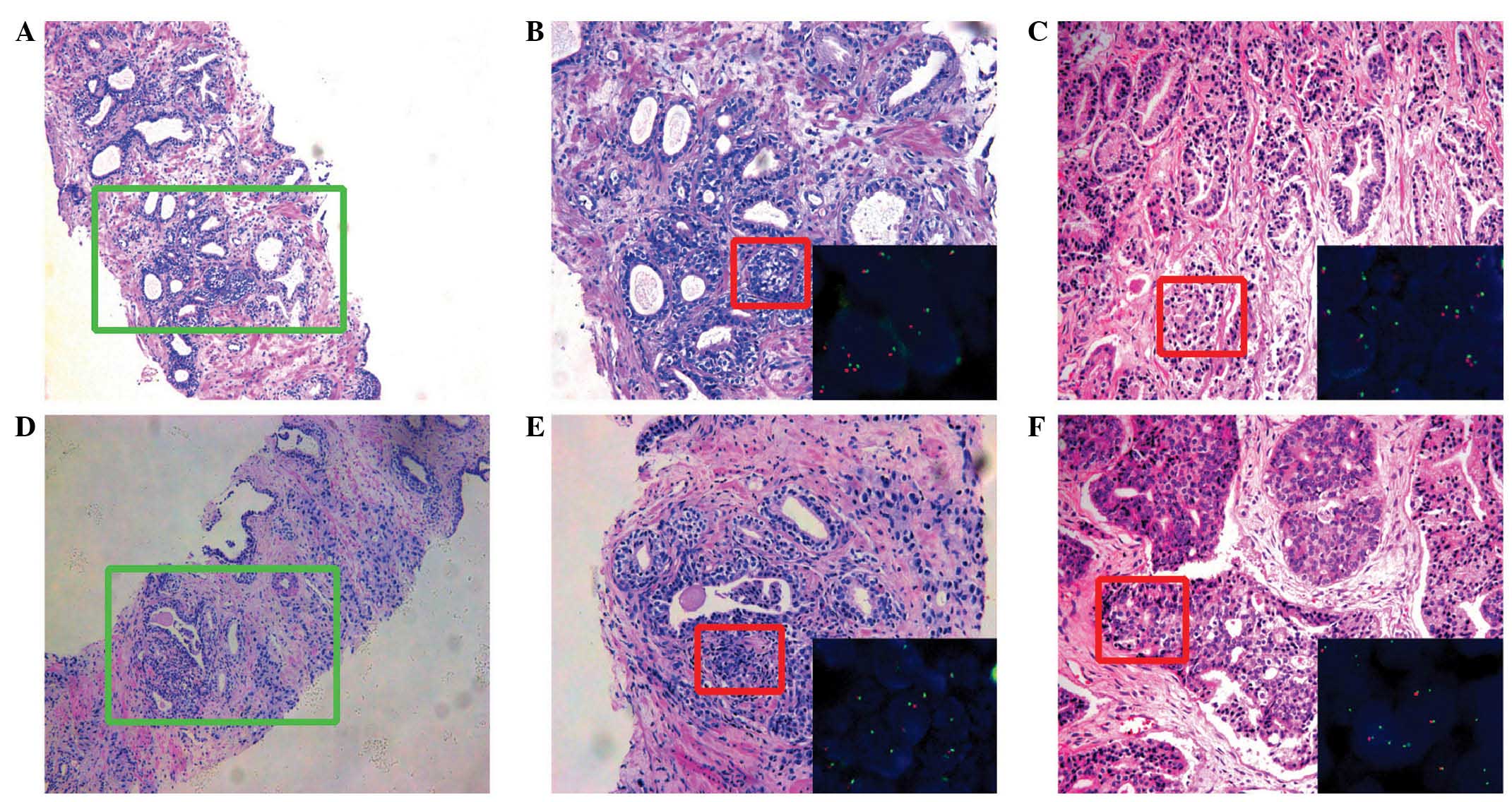 | Figure 1.Hematoxylin and eosin staining and
the corresponding fluorescence in situ hybridization images
demonstrating ERG rearrangements. (A) Prostatic biopsy
tissue with prostate cancer glands (GS 3+4) (magnification, x100).
(B) Image of the green boxed area in part A (magnification, x200);
inset (lower right) shows ERG probe image of the red boxed
area in part B (magnification, x1,000). (C) The corresponding
prostatectomy tissue from part A with organ-confined disease (GS
3+4) (magnification, x200); inset (lower right) shows ERG
probe image of the red boxed area in part C (magnification,
x1,000). (D) Prostatic biopsy tissue with prostate cancer glands
(GS 3+4) (magnification, x100). (E) Image of the green boxed area
in part D (magnification, x200); inset (lower right) shows
ERG probe image of the red boxed area in part E
(magnification, x1,000). (F) The corresponding prostatectomy sample
shown in part D with extra-prostatic extension, GS 4+4
(magnification, x200); inset (lower right) shows ERG probe
image of the red boxed area in part F, demonstrating an ERG
rearrangement (magnification, x1,000). ERG, ETS-related
gene; GS, Gleason score. |
Statistical analysis
Clinicopathological parameters were analysed using
the χ2 test or independent samples t-test (paired
samples t-test for comparison between preoperative and
postoperative ERG rearrangement rate, and independent samples
t-test for comparison between EPE group and localised PCa
group). Spearman's rank correlation coefficients were calculated to
explore the relationship between ERG rearrangement and
clinicopathological outcome. Receiver operating characteristic
(ROC) analysis and binary logistic regression were used to evaluate
the predictive values of ERG rearrangement and other
variables for EPE. The 10-fold leave-one-out cross-validation
(LOOCV) approach was used to validate the predictive performance of
ERG rearrangement. Statistical analyses were performed using
SPSS software version 13.0 (SPSS, Inc., Chicago, IL, USA) and
MedCalc software version 12.7 (Medcalc Software, Ostend, Belgium).
LOOCV analysis was performed in R 2.5.1 (http://cran.r-project.org). All tests were two-tailed,
and P<0.05 was considered to indicate statistical
significance.
Results
Patient characteristics
The characteristics of the 306 patients are
summarised in Table I. Pathological
examination indicated that a total of 220 patients (71.9%) had
organ-confined disease, and 86 (28.1%) showed evidence of EPE in
the prostatectomy specimen (EPE group). No significant differences
were observed in the comparison of mean ERG rearrangements
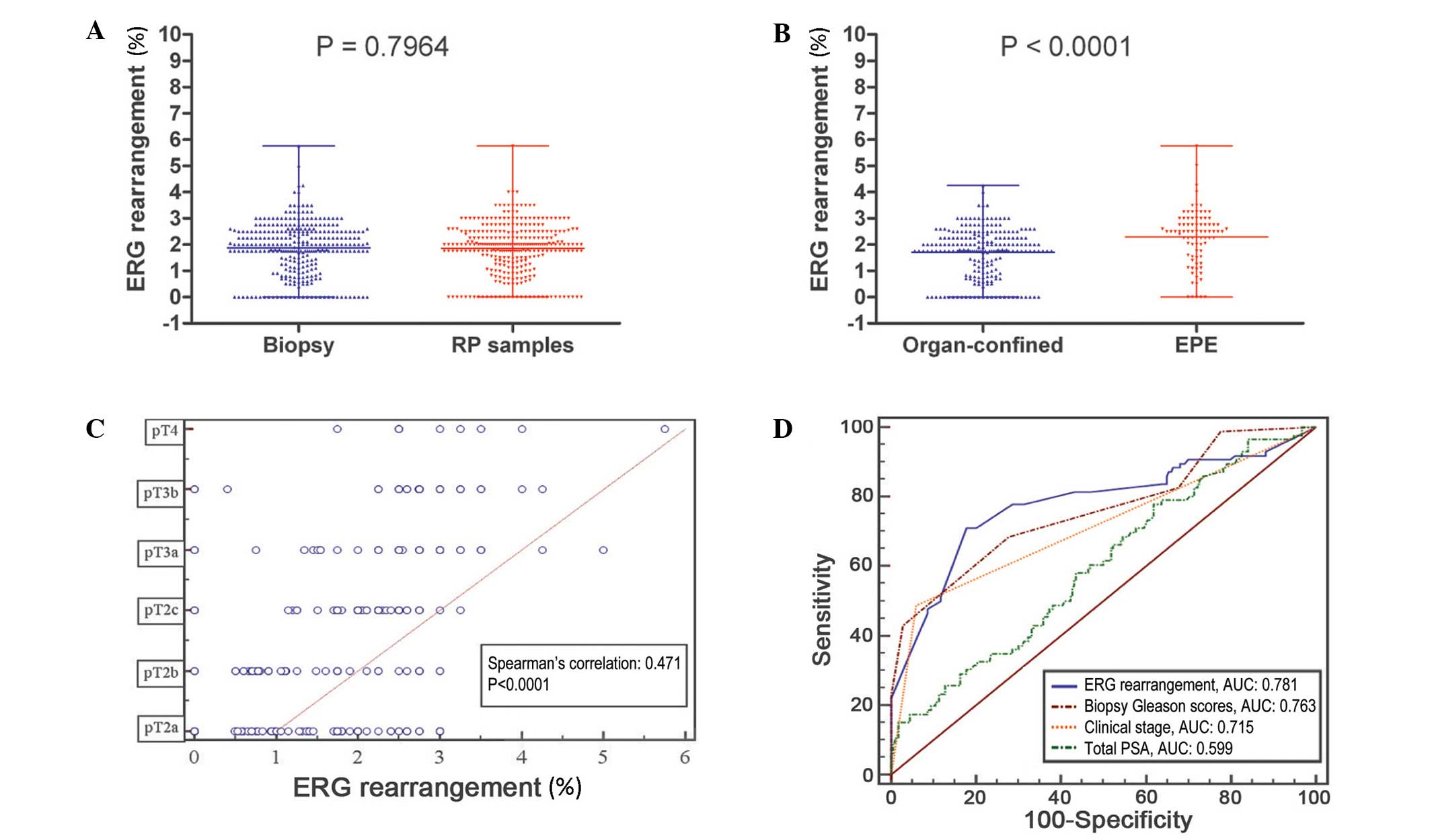
between biopsies and prostatectomy specimens (P=0.796) (Fig. 2A); however, the differences in
ERG rearrangements in the biopsy specimens were significant
between the EPE group and the group with organ-confined disease
(P<0.0001) (Fig. 2B).
 | Table I.Clinicopathological characteristics
of the study sample (n=306). |
Table I.
Clinicopathological characteristics
of the study sample (n=306).
| Characteristic | Value |
|---|
| Age, years; median
(range) | 69 (43–89) |
| tPSA level, ng/ml;
median (range) | 12.25
(2.75–45.79) |
| PV, ml; median
(range) | 49 (13–105) |
| TBC, n; median
(range) | 12 (10–16) |
| PBC, n; median
(range) | 3 (1–8) |
| Clinical T
classification, n (%) |
|
|
T1b-c | 63
(20.6) |
|
T2a | 112 (36.6) |
|
T2b | 76
(24.8) |
|
T2c | 55
(18.0) |
| Biopsy GS, n
(%) |
|
| ≤6 | 75
(57.2) |
| 7 | 81
(26.5) |
| ≥8 | 50
(16.3) |
| Pathological TNM
stage, n (%) |
|
|
T2a | 56
(18.3) |
|
T2b | 40
(13.0) |
|
T2c | 132 (43.1) |
|
T3a | 39
(12.7) |
|
T3b | 30 (9.8) |
| T4 | 9
(2.9) |
| Pathological GS, n
(%) |
|
| ≤6 | 163 (53.3) |
| 7 | 79
(25.8) |
| ≥8 | 64
(20.9) |
| Seminal vesicle
invasion, n (%) |
|
|
Negative | 265 (86.8) |
|
Positive | 41
(13.4) |
| Lymph node
metastasis, n (%) |
|
|
Negative | 280 (91.5) |
|
Positive | 26 (8.5) |
| Capsule state, n
(%) |
|
|
Organ-confined | 220 (71.9) |
|
Extra-prostatic extension | 86
(28.1) |
Table II summarises
the clinicopathological characteristics of the patients in the EPE
group and the group with organ-confined disease. There was a
significantly higher total PSA (tPSA) level (P=0.001) and a higher
percentage of positive biopsy cores (PBCs) in the EPE group
(P<0.0001), and significant differences were also identified
between the stratified biopsy GSs (≤6, 7 and ≥8) and clinical T
classifications (T1b-T1c, T2a-T2b and T2c) (P<0.0001). However,
there were no differences with regard to age (P=0.558) or prostate
volume (P=0.604).
 | Table II.Comparison of clinicopathological
features between the patients with and without EPE. |
Table II.
Comparison of clinicopathological
features between the patients with and without EPE.
| Feature | Organ-confined | EPE | P-value |
|---|
| Cases, n (%) | 220 (71.9) | 86 (28.1) |
|
| Age, years; mean ±
SD | 68.64±7.668 | 69.22±8.031 | 0.558 |
| tPSA level, ng/ml;
mean ± SD | 14.00±8.590 | 18.05±11.774 | 0.001 |
| Prostate volume,
ml; mean ± SD | 50.49±14.339 | 51.43±13.938 | 0.604 |
| Percentage of PBC,
%; mean ± SD | 21.56±10.462 | 36.41±10.680 | <0.0001 |
| Biopsy Gleason
scores, n (%) |
|
| <0.0001 |
| ≤6 | 151 (86.3) | 24 (13.7) |
|
| 7 | 56
(69.1) | 25 (30.9) |
|
| ≥8 | 13
(26.0) | 37 (74.0) |
|
| Clinical TNM stage,
n (%) |
|
| <0.0001 |
|
Tb-1c | 47
(74.6) | 16 (25.4) |
|
|
T2a-T2b | 160 (85.1) | 28 (14.9) |
|
|
T2c | 13
(23.6) | 42 (76.4) |
|
ERG rearrangement for predicting
EPE
A significant positive association was identified
between the ERG rearrangement rate and pathological T
classification [r=0.471; 95% confidence interval (CI), 0.379–0.554;
P<0.0001; Fig. 2C].
ROC analysis was used to explore the performance of
ERG rearrangement rates in the biopsy specimens for
assessing the risk of EPE. ERG rearrangement was expressed
as a continuous variable, and the results revealed that the area
under the curve (AUC) was 0.781 (95% CI, 0.730–0.826). An optimal
cut-off value of 2.25% was established, with a sensitivity of
70.24% (95% CI, 62.6–78.9%) and a specificity of 80.43% (95% CI,
75.4–85.1%) (Fig. 2D).
To investigate the independent risk factors for the
EPE of PCa, ERG rearrangement and other preoperative
variables, including tPSA, biopsy GS and clinical T classification,
were included in a logistic regression analysis, and the AUC of
each parameter was compared. The results indicated that ERG
rearrangement had a better predictive value for EPE compared with
tPSA (AUC, 0.599; 95% CI, 0.541–0.654; P<0.0001); a slight,
non-significant difference existed compared to clinical T
classification (AUC, 0.715; 95% CI, 0.660–0.765; P=0.094), and
ERG rearrangement had a similar predictive value to biopsy
GS (AUC, 0.763; 95% CI, 0.711–0.810; P=0.695) (Fig. 2D).
Multivariate logistic regression models revealed
that ERG rearrangement in the biopsy sample was an
independent predictor of EPE [odds ratio (OR), 1.997; 95% CI,
1.277–3.124; P=0.002]. In addition, biopsy GSs of 7 (OR, 2.669; 95%
CI, 1.116–6.383; P=0.027) and ≥8 (OR, 39.032; 95% CI,
10.397–146.527; P<0.0001), and a clinical T classification of
T2c (OR, 9.103; 95% CI, 3.338–24.824; P<0.0001) were independent
predictors of EPE of clinically localised PCa. However, age, tPSA,
prostate volume, number of PBCs, percentage of PBCs, biopsy GSs of
≤6 and clinical T1b-1c and T2a-T2b staging were not independent
risk factors for EPE (Table
III).
 | Table III.Multivariate logistic regression
analysis of extra-prostatic extension prediction. |
Table III.
Multivariate logistic regression
analysis of extra-prostatic extension prediction.
| Variable | Odds ratio | 95% CI | P-value |
|---|
| Age |
0.992 | 0.945–1.041 | 0.750 |
| tPSA level |
0.971 | 0.927–1.016 | 0.203 |
| ERG
rearrangement |
1.997 | 1.277–3.124 | 0.002 |
| Prostate
volume |
0.997 | 0.968–1.026 | 0.966 |
| Number of PBCs |
1.956 | 0.449–8.525 | 0.372 |
| Percentage of
PBCs | 70.338 | – | 0.961 |
| Biopsy Gleason
scores |
|
|
|
| ≤6 | – | – | – |
| 7 |
2.669 | 1.116–6.383 | 0.027 |
| ≥8 | 39.032 | 10.397–146.527 | <0.001 |
| Clinical T
classification |
|
|
|
|
T1b-T2b | – | – | – |
|
T2c |
9.103 | 3.338–24.824 | <0.001 |
Validation of the ability of ERG
rearrangement to predict EPE
Ten-fold LOOCV was used to validate the power of
ERG rearrangement for predicting EPE. For all cases that
were excluded from the model, a cut-off was assigned using 10-fold
cross-validation, and a predictive value was determined for the
excluded cases. In this LOOCV model, ERG rearrangement
performed well for predicting EPE, with a sensitivity of 76.923%
and a specificity of 71.429%. The 95% CI for the AUC was
0.724–0.958. Thus, the null hypothesis of ROC <0.5 (random
prediction) could not be rejected (Table
IV).
 | Table IV.10-fold leave-one-out cross
validation of ERG rearrangement for prediction of
extra-prostatic extension. |
Table IV.
10-fold leave-one-out cross
validation of ERG rearrangement for prediction of
extra-prostatic extension.
| No. | Sensitivity, % | Specificity, % | AUC | Cut-off (%) |
|---|
| 1 | 54.545 | 80.000 | 0.905 | 2.175 |
| 2 | 62.750 | 73.684 | 0.780 | 2.450 |
| 3 | 64.500 | 70.000 | 0.876 | 2.175 |
| 4 | 63.636 | 68.421 | 0.854 | 2.450 |
| 5 | 70.000 | 64.286 | 0.818 | 2.450 |
| 6 | 76.923 | 71.429 | 0.776 | 2.450 |
| 7 | 77.778 | 75.000 | 0.956 | 2.175 |
| 8 | 90.000 | 70.000 | 0.824 | 2.450 |
| 9 | 90.909 | 66.667 | 0.763 | 2.175 |
| 10 | 92.857 | 62.500 | 0.868 | 2.175 |
| Total | 76.923 | 71.429 | 95% CI:
0.724–0.958 |
|
Discussion
EPE is defined as the presence of cancer cells
outside of the prostatic capsule (22). This terminology was introduced in
1996; subsequently, EPE was confirmed to be an adverse prognostic
factor for PCa (22). Several
variables, including the Partin table, MRI, TRUS and digital rectal
examination findings, and prostate cancer antigen 3 score, have
been studied to assess the predictive power of EPE; however,
sensitivities have ranged from only 50–70% (8,9,23–25). Thus,
there is an urgent need to explore novel and more effective
approaches for predicting EPE for determining an appropriate
surgical strategy and also for delineating a favourable
postoperative adjuvant therapy.
The prostate-specific gene TMPRSS2 is fused
with the transcription factor ERG in a large proportion of
cases of PCa; however, its biological relationships with the
clinicopathological parameters of the disease, such as PSA level,
GS, pathological stage and prognosis, are not clear-cut, and the
reported results lack consistency (13–15,26). This
may be due to differences in study design, detection techniques,
sample origin and the intrinsic mechanism of gene rearrangement
(27–29). In our pilot study, no significant
association was identified between ERG status and tumour
stage in a limited cohort of patients (16). However, ERG rearrangement was
found to be positively correlated with advanced tumour stage in a
larger cohort of samples in this study, which is consistent with
previous reports (28–31).
Overall, ERG rearrangement tends to be
positively associated with advanced pathological stage, which has
been further verified in a meta-analysis of 34 series (32). For example, Furusato et al
(33) demonstrated that ERG protein
expression is positively correlated with pathological stage, tumour
grade and metastatic status in Japanese PCa patients. Paulo et
al (34) also reported that a
higher percentage of patients with locally advanced disease (pT3a)
possess ERG rearrangements compared to patients with
organ-confined disease. Minner et al (35) demonstrated that ERG fusion is
positively associated with pathological stage, and
ERG-positive patients tended to have higher GSs. The results
of the present study revealed that ERG rearrangement
detection in biopsy specimens was positively correlated with
advanced stage and was able to predict EPE, with an optimal cut-off
of 2.25% and an AUC of 0.781. In addition, ERG rearrangement
was an independent factor for EPE. In the LOOCV internal validation
model, it was also observed that ERG rearrangement was a
valuable indicator of EPE, with a 95% CI for the AUC ranging from
0.724 to 0.958. The detection of ERG rearrangements may be
useful for guiding decisions related to surgical margins,
regardless of the statuses of other clinicopathological
factors.
In the present study, the clinical stage T2c was
determined to be an independent predictor of EPE. The clinical T
stage has long been considered an important factor for predicting
EPE. For example, the use of a Partin table, including the clinical
T stage, tPSA level and biopsy GS, was investigated for the
prediction of EPE in 1997 (36).
Subsequently, following several updates, other clinical variables
were also incorporated into this model, thereby improving its
predictive capacity (3,37).
In the current study, it was also observed that
biopsy GSs of 7 and ≥8 were positively correlated with EPE, and a
high GS was usually associated with advanced-stage PCa. The
findings also demonstrated that ERG rearrangement had a
similar AUC as biopsy GS (0.781 vs. 0.763); furthermore,
multivariate logistic regression models revealed that biopsy GSs of
7 and ≥8 were independent factors for EPE, which is consistent with
prior publications (12,38–40).
Other preoperative factors investigated in the
present study, such as age, tPSA level, prostate volume and
percentage of PBCs, were not found to have predictive power for
EPE. Although previous studies have reported that these parameters
may individually or collectively be used to predict EPE in PCa
patients from Western countries (41), considering the discrepancies related
to race and study population, it is necessary to further explore
the predictive values of these parameters using a larger sample of
Chinese patients with clinically localised PCa.
In summary, the current findings demonstrated that
approximately 28.1% of patients with clinically localised PCa have
EPE, and that ERG rearrangements in biopsy samples may be
independent factors for predicting ECE, similar to biopsy GSs of
≥7. These results may be useful for determining an appropriate
surgical strategy in PCa patients with organ-confined disease.
Consequently, the TMPRSS2-ERG gene, which is specific to PCa
(18), should be studied further with
regard to its roles in progression and prognosis, and may be a
target for modern personalised therapy for PCa patients.
Acknowledgements
This work was supported by the Program of 5010 of
Sun Yat-Sen University (grant no. 2007028 awarded to Professor Xin
Gao). The authors are grateful to Professor Chun-kui Shao from the
Department of Pathology at Third Affiliated Hospital of Sun Yat-Sen
University.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Heidenreich A, Bellmunt J, Bolla M, Joniau
S, Mason M, Matveev V, Mottet N, Schmid HP, van der Kwast T, Wiegel
T and Zattoni F: European Association of Urology: EAU guidelines on
prostate cancer. Part 1: Screening, diagnosis and treatment of
clinically localised disease. Eur Urol. 59:61–71. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eifler JB, Feng Z, Lin BM, Partin MT,
Humphreys EB, Han M, Epstein JI, Walsh PC, Trock BJ and Partin AW:
An updated prostate cancer staging nomogram (Partin tables) based
on cases from 2006 to 2011. BJU Int. 111:22–29. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hull GW, Rabbani F, Abbas F, Wheeler TM,
Kattan MW and Scardino PT: Cancer control with radical
prostatectomy alone in 1,000 consecutive patients. J Urol.
167:528–534. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Masieri L, Minervini A, Vittori G,
Lanciotti M, Lanzi F, Lapini A, Carini M and Serni S: The role of
free to total PSA ratio in prediction of extracapsular tumor
extension and biochemical recurrence after radical prostatectomy in
patients with PSA between 4 and 10 ng/ml. Int Urol Nephrol.
44:1031–1038. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Magi-Galluzzi C, Evans AJ, Delahunt B,
Epstein JI, Griffiths DF, van der Kwast TH, Montironi R, Wheeler
TM, Srigley JR, Egevad LL and Humphrey PA: ISUP Prostate:
International society of urological pathology (ISUP) consensus
conference on handling and staging of radical prostatectomy
specimens. Working group 3: Extraprostatic extension,
lymphovascular invasion and locally advanced disease. Mod Pathol.
24:26–38. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
van Veggel BA, van Oort IM, Witjes JA,
Kiemeney LA and de Hulsbergen-van Kaa CA: Quantification of
extraprostatic extension in prostate cancer: Different parameters
correlated to biochemical recurrence after radical prostatectomy.
Histopathology. 59:692–702. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roethke MC, Lichy MP, Kniess M, Werner MK,
Claussen CD, Stenzl A, Schlemmer HP and Schilling D: Accuracy of
preoperative endorectal MRI in predicting extracapsular extension
and influence on neurovascular bundle sparing in radical
prostatectomy. World J Urol. 31:1111–1116. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eisenberg ML, Cowan JE, Davies BJ, Carroll
PR and Shinohara K: The importance of tumor palpability and
transrectal ultrasonographic appearance in the contemporary
clinical staging of prostate cancer. Urol Oncol. 29:171–176. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Schwartz DJ, Sengupta S, Hillman DW,
Sargent DJ, Cheville JC, Wilson TM, Mynderse LA, Choo R and Davis
BJ: Prediction of radial distance of extraprostatic extension from
pretherapy factors. Int J Radiat Oncol Biol Phys. 69:411–418. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Brassell SA, Kao TC, Sun L and Moul JW:
Prostate-specific antigen versus prostate-specific antigen density
as predictor of tumor volume, margin status, pathologic stage and
biochemical recurrence of prostate cancer. Urology. 66:1229–1233.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Magheli A, Rais-Bahrami S, Trock BJ,
Humphreys EB, Partin AW, Han M and Gonzalgo ML: Prostate specific
antigen versus prostate specific antigen density as a
prognosticator of pathological characteristics and biochemical
recurrence following radical prostatectomy. J Urol. 179:1780–1784,
1784; discussion. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kumar-Sinha C, Tomlins SA and Chinnaiyan
AM: Recurrent gene fusions in prostate cancer. Nat Rev Cancer.
8:497–511. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tomlins SA, Bjartell A, Chinnaiyan AM,
Jenster G, Nam RK, Rubin MA and Schalken JA: ETS gene fusions in
prostate cancer: From discovery to daily clinical practice. Eur
Urol. 56:275–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Clark JP and Cooper CS: ETS gene fusions
in prostate cancer. Nat Rev Urol. 6:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun QP, Li LY, Chen Z, Pang J, Yang WJ,
Zhou XF, Qiu JG, Su ZL, He D and Gao X: Detection of TMPRSS2-ETS
fusions by a multiprobe fluorescence in situ hybridization assay
for the early diagnosis of prostate cancer: A pilot study. J Mol
Diagn. 12:718–724. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gao X, Li LY, Zhou FJ, Xie KJ, Shao CK, Su
ZL, Sun QP, Chen MK, Pang J, Zhou XF, et al: ERG rearrangement for
predicting subsequent cancer diagnosis in high-grade prostatic
intraepithelial neoplasia and lymph node metastasis. Clin Cancer
Res. 18:4163–4172. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tomlins SA, Rhodes DR, Perner S,
Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J,
Kuefer R, et al: Recurrent fusion of TMPRSS2 and ETS transcription
factor genes in prostate cancer. Science. 310:644–648. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sobin LH, Gospodarowicz MK and Wittekind
C: TNM Classification of Malignant Tumours (7th). Wiley-Blackwell.
Hoboken, NJ: 2010.
|
|
20
|
Epstein JI: An update of the Gleason
grading system. J Urol. 183:433–440. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Epstein JI, Partin AW, Sauvageot J and
Walsh PC: Prediction of progression following radical
prostatectomy. A multivariate analysis of 721 men with long-term
follow-up. Am J Surg Pathol. 20:286–292. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sakr WA, Wheeler TM, Blute M, Bodo M,
Calle-Rodrigue R, Henson DE, Mostofi FK, Seiffert J, Wojno K and
Zincke H: Staging and reporting of prostate cancer-sampling of the
radical prostatectomy specimen. Cancer. 78:366–368. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu JB, Makarov DV, Sharma R, Peschel RE,
Partin AW and Gross CP: Validation of the partin nomogram for
prostate cancer in a national sample. J Urol. 183:105–111. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Karakiewicz PI, Bhojani N, Capitanio U,
Reuther AM, Suardi N, Jeldres C, Pharand D, Péloquin F, Perrotte P,
Shariat SF and Klein EA: External validation of the updated Partin
tables in a cohort of North American men. J Urol. 180:898–902;
discussion 902–903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Pepe P, Fraggetta F, Galia A and Aragona
F: Is PCA3 score useful in preoperative staging of a single
microfocus of prostate cancer diagnosed at saturation biopsy? Urol
Int. 89:143–147. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Xu B, Chevarie-Davis M, Chevalier S,
Scarlata E, Zeizafoun N, Dragomir A, Tanguay S, Kassouf W, Aprikian
A and Brimo F: The prognostic role of ERG immunopositivity in
prostatic acinar adenocarcinoma: A study including 454 cases and
review of the literature. Hum Pathol. 45:488–497. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hermans KG, van Marion R, van Dekken H,
Jenster G, van Weerden WM and Trapman J: TMPRSS2: ERG fusion by
translocation or interstitial deletion is highly relevant in
androgen-dependent prostate cancer, but is bypassed in late-stage
androgen receptor-negative prostate cancer. Cancer Res.
66:10658–10663. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Perner S, Demichelis F, Beroukhim R,
Schmidt FH, Mosquera JM, Setlur S, Tchinda J, Tomlins SA, Hofer MD,
Pienta KG, et al: TMPRSS2: ERG fusion-associated deletions provide
insight into the heterogeneity of prostate cancer. Cancer Res.
66:8337–8341. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Attard G, Clark J, Ambroisine L, Fisher G,
Kovacs G, Flohr P, Berney D, Foster CS, Fletcher A, Gerald WL, et
al: Duplication of the fusion of TMPRSS2 to ERG sequences
identifies fatal human prostate cancer. Oncogene. 27:253–263. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hofer MD, Kuefer R, Maier C, Herkommer K,
Perner S, Demichelis F, Paiss T, Vogel W, Rubin MA and Hoegel J:
Genome-wide linkage analysis of TMPRSS2-ERG fusion in familial
prostate cancer. Cancer Res. 69:640–646. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Rajput AB, Miller MA, De Luca A, Boyd N,
Leung S, Hurtado-Coll A, Fazli L, Jones EC, Palmer JB, Gleave ME,
et al: Frequency of the TMPRSS2: ERG gene fusion is increased in
moderate to poorly differentiated prostate cancers. J Clin Pathol.
60:1238–1243. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Pettersson A, Graff RE, Bauer SR, Pitt MJ,
Lis RT, Stack EC, Martin NE, Kunz L, Penney KL, Ligon AH, et al:
The TMPRSS2: ERG rearrangement, ERG expression and prostate cancer
outcomes: A cohort study and meta-analysis. Cancer Epidemiol
Biomarkers Prev. 21:1497–1509. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Furusato B, van Leenders GJ, Trapman J,
Kimura T, Egawa S, Takahashi H, Furusato M, Visakorpi T and Hano H:
Immunohistochemical ETS-related gene detection in a Japanese
prostate cancer cohort: Diagnostic use in Japanese prostate cancer
patients. Pathol Int. 61:409–414. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Paulo P, Barros-Silva JD, Ribeiro FR,
Ramalho-Carvalho J, Jerónimo C, Henrique R, Lind GE, Skotheim RI,
Lothe RA and Teixeira MR: FLI1 is a novel ETS transcription factor
involved in gene fusions in prostate cancer. Genes Chromosomes
Cancer. 51:240–249. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Minner S, Enodien M, Sirma H, Luebke AM,
Krohn A, Mayer PS, Simon R, Tennstedt P, Müller J, Scholz L, et al:
ERG status is unrelated to PSA recurrence in radically operated
prostate cancer in the absence of antihormonal therapy. Clin Cancer
Res. 17:5878–5888. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Partin AW, Kattan MW, Subong EN, Walsh PC,
Wojno KJ, Oesterling JE, Scardino PT and Pearson JD: Combination of
prostate-specific antigen, clinical stage and Gleason score to
predict pathological stage of localized prostate cancer. A
multi-institutional update. JAMA. 277:1445–1451. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Huang Y, Isharwal S, Haese A, Chun FK,
Makarov DV, Feng Z, Han M, Humphreys E, Epstein JI, Partin AW and
Veltri RW: Prediction of patient-specific risk and percentile
cohort risk of pathological stage outcome using continuous
prostate-specific antigen measurement, clinical stage and biopsy
Gleason score. BJU Int. 107:1562–1569. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Paquette EL, Connelly RR, Sun L, Paquette
LR and Moul JW: Predictors of extracapsular extension and positive
margins in African American and white men. Urol Oncol. 21:33–38.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nishimoto K, Nakashima J, Hashiguchi A,
Kikuchi E, Miyajima A, Nakagawa K, Ohigashi T, Oya M and Murai M:
Prediction of extraprostatic extension by prostate specific antigen
velocity, endorectal MRI and biopsy Gleason score in clinically
localized prostate cancer. Int J Urol. 15:520–523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Giannarini G, Scott CA, Moro U, Pertoldi
B, Beltrami CA and Selli C: Are PSA density and PSA density of the
transition zone more accurate than PSA in predicting the
pathological stage of clinically localized prostate cancer? Urol
Oncol. 26:353–360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Lin S, Zhang Q, Li P, Li Z, Sun Y, Shao Y,
Zhang X and Fu S: Prediction of extraprostatic extension in
patients with clinically organ-confined prostate cancer. Urol Int.
92:282–8. 2014. View Article : Google Scholar : PubMed/NCBI
|