Introduction
Human hepatocellular carcinoma (HCC) is one of the
most common types of cancer, causing almost 600,000 mortalities
annually, worldwide (1). Chronic
inflammatory liver disease including viral infection and exposure
to chemical carcinogens are the common prologues of HCC. Surgical
removal of the tumor and liver transplant are currently the most
effective treatment options for HCC, although metastasis and
recurrence are extremely common in patients who have undergone
resection. Therefore, understanding the molecular mechanisms of HCC
tumorigenesis is an essential step in identifying more effective
treatments.
The Hippo signaling pathway is an evolutionary
conserved cascade, and it was previously reported to play a
critical role in liver regeneration and, more significantly, HCC
tumorigenesis (2). The machinery of
the Hippo signaling pathway consists of Mst1/2, WW45, Lats1/2 and
Mob1, and the downstream effectors are Yes-associated protein 1
(YAP1), TAZ and transcriptional enhancer activation domain family
members (TEADs) (1). When the
upstream effectors are activated they recruit YAP1-TAZ complex,
which is transferred into the nucleus to interact with TEADs, and
drives target gene expression to promote cell proliferation and
suppress apoptosis (3). The
disruption of YAP1-TEAD interaction by YAP-like peptides leads to a
significant decrease in the tumor growth rate (4,5). The
overexpression of YAP1 in transgenic mice was observed to lead to
liver outgrowth and HCC (6). YAP1 was
reported to be upregulated in over 60% of HCC patients, and is an
independent predictor for patient survival (7). These studies clearly suggest that YAP1
is an essential downstream effector of the Hippo signaling pathway
and furthermore a promising drug target for the inhibition of HCC
tumorigenesis.
microRNAs (miRNAs or miRs) are a class of 21–25
nucleotide long, small non-coding RNA molecules, which recognize
specific complementary sequences predominantly identified in the
3′-untranslated region (UTR) of target mRNAs. miRNAs either repress
translation or degrade target mRNAs (8,9). miRNAs
have been implicated in various forms of cancer, including liver
cancer, by altering the expression of oncogenes or tumor suppressor
genes (10,11). One of the most extensively studied
microRNAs targeting YAP1 is miR-375. It was initially identified as
a negative regulator of YAP1 by targeting its 3′-UTR in HCC patient
tissues and human HCC cell lines (12). It was later confirmed that miR-375
also exhibited the same functions in HCC mouse and rat animal
models (13,14). Besides miR-375, there have been no
other studies on miRNA regulation of YAP1.
In our study, we aimed to identify novel microRNAs
regulating YAP1. By in silico analysis we first identified
miR-186 as a potential regulator of YAP1 mRNA, and observed
that miR-186 was downregulated in several human HCC cell lines.
Further analysis confirmed that YAP1 mRNA was a bona
fide target of miR-186, and that mRNA and protein levels of
YAP1 in HCC cells were downregulated by overexpression of miR-186.
Finally, we demonstrated that the induction of miR-186 inhibited
migration, invasion and proliferation in HCC cells, suggesting
miR-186 as a potential therapeutic target in treating liver
cancer.
Materials and methods
Cell lines
The human normal liver cell lines THLE-2 and THLE-3,
and human liver cancer cell lines HepG2, Hep3B and SNU398 were
obtained from the American Type Culture Collection (ATCC, Manassas,
VA, USA). Cells were cultured according to ATCC instructions and
passaged for less than 6 months after receipt for completion of the
studies.
miRNA transfection
Has-miR-186 mirVana miRNA mimic was purchased from
Applied Biosystems Life Technologies (assay ID MC11753; Pleasanton,
CA, USA) along with mirVana miRNA mimic negative control #1, and
transfected into cell lines according to the manufacturer's
instructions.
Luciferase reporter assay
The putative miR-186 binding site at the 3′-UTR of
YAP1 was cloned downstream of a cytomegalovirus promoter-driven
firefly luciferase cassette in a pMIR-REPORT vector (Applied
Biosystems). Mutant forms of the luciferase constructs were also
generated using standard PCR-based overlap-extension protocols. For
luciferase reporter assay, HepG2 cells (3×104) were
plated in a 24-well plate and then co-transfected with 400 ng of
either has-miR-186 or miR-control (Applied Biosystems), 200 ng of
either wild-type or mutant luciferase constructs, and 40 ng of
pRL-TK (Promega Corporation, Madison, WI, USA), using Lipofectamine
2000 (Invitrogen Life Technologies, Carlsbad, CA, USA) according to
the manufacturer's instruction. Cells were collected 48 h after
transfection and analyzed using the dual-luciferase reporter assay
system (Promega Corporation). The pRL-TK vector provided the
constitutive expression of Renilla luciferase, and was used
as an internal control to correct the differences in transfection
and harvest efficiencies. Data are shown as the means ± standard
error of the mean (SEM) of three independent experiments.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total mRNA was extracted with the mirVANA miRNA
isolation kit (Ambion Life Technologies, Carlsbad, CA, USA)
according to the manufacturer's instructions. The expression level
of miR-186 was quantified using miRNA-specific TaqMan Pri-miRNA
assays (assay ID Hs03303293_pri; Applied Biosystems) and normalized
by RNU48 control miRNA assay (P/N 001006; Applied Biosystems).
Total RNA was purified using the RNeasy MiniPrep kit (Qiagen,
Valencia, CA, USA). Total RNA (1 µg) was reverse transcribed with
the Superscript II First-Strand synthesis kit (Life Technologies)
as recommended by the manufacturer. GAPDH mRNA levels were measured
for normalization.
Protein isolation and western blot
analysis
Protein lysates were extracted using RIPA buffer [50
mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 1%
sodium deoxycholate, 0.1% sodium dodecyl sulphate (SDS) and
protease inhibitors]. Lysates were centrifuged at 16,000 × g for 10
min at 4°C. Proteins were analyzed by SDS-polyacrylamide gel
electrophoresis and transferred to nitrocellulose membrane using a
semi-dry transfer unit (Bio-Rad Laboratories, Inc., Hercules, CA,
USA). The membrane was then immersed in blocking buffer
(phosphate-buffered saline and 0.1% Tween-20) with 5% nonfat milk
for 20 min and then incubated with rabbit anti-YAP1 (ab52771;
Abcam, Cambridge, UK) overnight at 4°C. After washing with blocking
buffer, blots were incubated with horseradish peroxidase-conjugated
secondary antibody (Life Technologies, Grand Island, NY, USA) for
20 min, washed again with blocking buffer, and visualized using
Luminata Forte Western HRP substrate (EMD Millipore, Billerica, MA,
USA).
Wound-healing assay
Cells (1×105) transfected with mirVana
miRNA mimic negative control #1 and has-miR-186 were seeded into
six-well plates following transfection. A linear wound was
carefully made using a 100 µl sterile pipette tip across the
confluent cell monolayer, and the cell debris was removed by
washing with phosphate. The migrated distance of the growing edge
on the monolayers was measured 24 h after being wounded.
Cell invasion assay
Cell invasion assay was performed in a 24-well plate
with 8 µm pore size chamber inserts (BD Biosciences, Franklin
Lakes, NJ, USA). Cells (1×105) transfected with mirVana
miRNA mimic negative control #1 and has-miR-186 were placed into
the upper chamber of each well with Matrigel-coated membrane, which
was diluted with serum-free culture medium. The lower compartment
was filled with 500 µl medium containing 10% fetal bovine serum as
a chemo-attractant. The cells were incubated at 37°C in a 5%
CO2 humidified incubator for 24 h. Then cells were
exposed to 20 µM 5-ethynyl-2′-deoxyuridine for an additional 4 h at
37°C. Membrane inserts were removed from the plate and stained
using the ENU kit (Invitrogen Life Technologies). The cells were
counted in six random microscopic fields for each well, using NIH
ImageJ software (http://imagej.nih.gov/ij/).
Cell proliferation and growth
assay
Growth and inhibition of growth were assessed by
standard MTT assay at time intervals of 24 h. The number of cells
was scored as a percentage relative to the mock treatment group.
Data are shown as the means ± SEM of three independent
experiments.
Statistical analysis
All data are presented as the means ± SEM. A
two-tailed Student's t-test was used to establish significant
differences between groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-186 expression is downregulated in
HCC cell lines
Firstly, using the microRNA.org
resource, we identified miR-186 as a potential regulator of YAP1
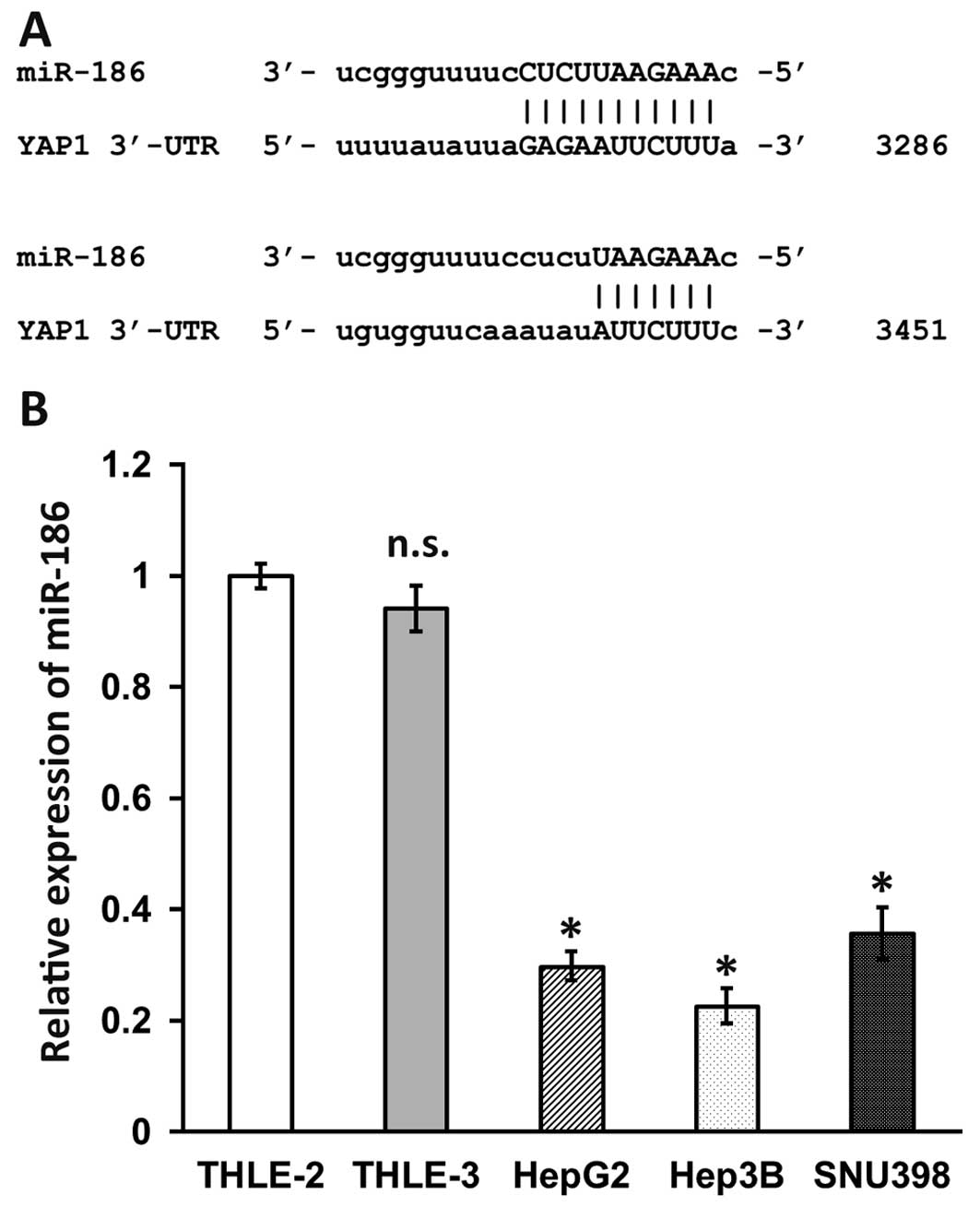
mRNA. As shown in Fig. 1A, miR-186
recognizes two loci in the 3′-UTR of YAP1 (NM_006106) at 3286 and
3451. In order to investigate the role of miR-186 in human HCC, we
next examined miR-186 expression in several human HCC cell lines
(HepG2, Hep3B and SNU398) and normal human liver cells (THLE-2 and
THLE-3) using RT-qPCR (Fig. 1B).
miR-186 levels in THLE-2 and THLE-3 were similar. However, miR-186
expression in Hep3B was 30% lower than that in THLE-2. HepG2 and
SNU398 were slightly higher, but still only 30% and 36% of the
level in THLE-2. These results clearly indicated that compared with
normal human liver cells, miR-186 levels were significantly
downregulated in human HCC cell lines.
miR-186 directly targets and
downregulates YAP1 levels in HCC cell lines
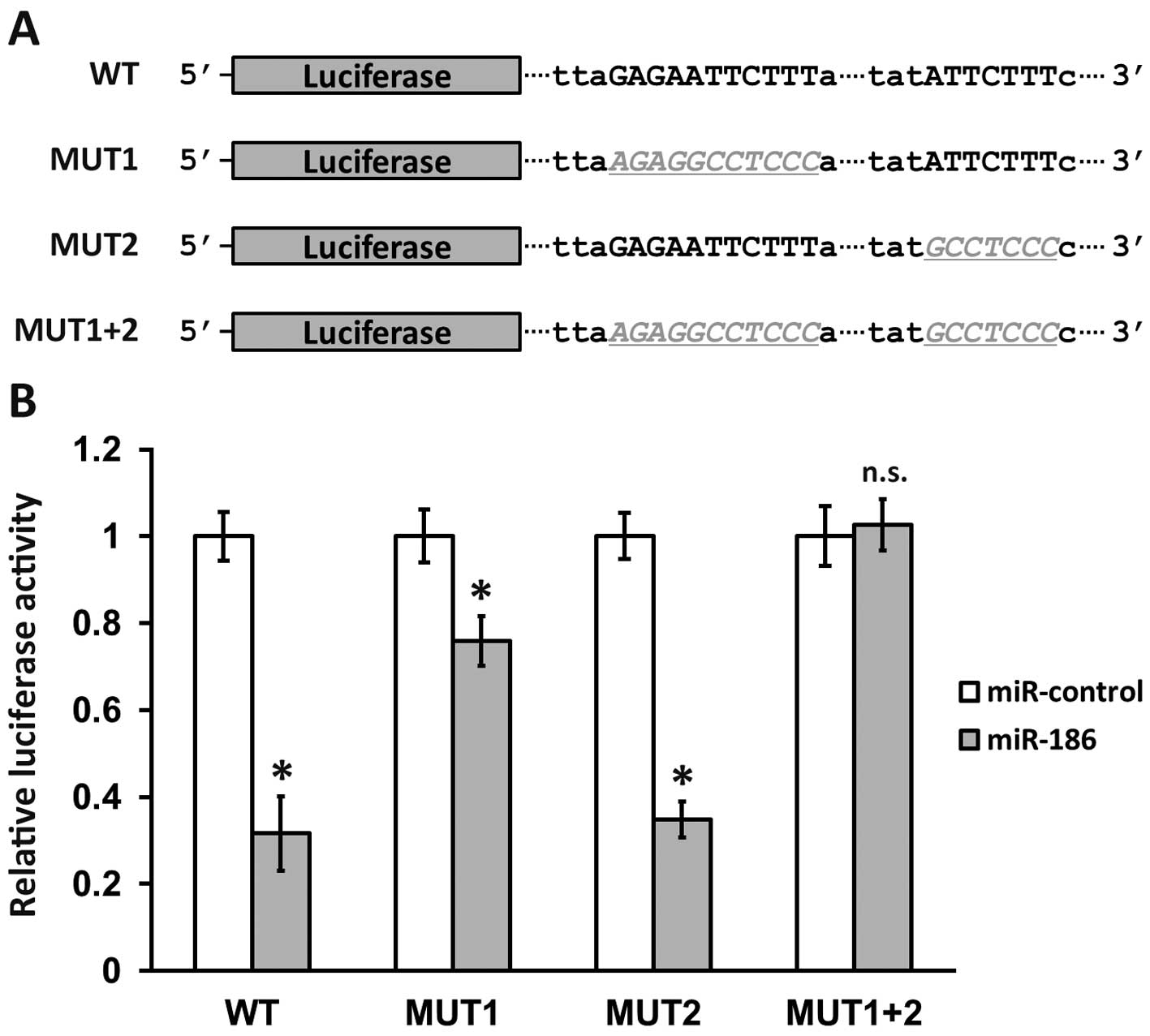
To confirm that YAP1 is bona fide target of miR-186,
we performed a luciferase reporter assay, using sequences from the
original 3′-UTR on YAP1 mRNA as well as three mutated versions
(Fig. 2A). MUT1 contained mutations
that disrupted only the first potential binding site on YAP1
3′-UTR, while MUT2 mutations affected only the second site, and the
MUT1+2 construct contained a combination of both mutated sites. As
a result, wild-type luciferase activity was significantly reduced
by co-transfection of miR-186, to less than 40% of miR-control
transfected levels, indicating that YAP1 3′-UTR was indeed a direct
target of miR-186. Notably, the activity of MUT1 construct was
almost 80% of that of the control, while the activity of MUT2 was
reduced to less than 40% of the control. This suggested that
miR-186 targeting to YAP1 mRNA was almost entirely through binding
to the first site on the 3′-UTR (Fig.
1A, locus 3286), rather than the second site (Fig. 1A, locus 3451). As expected, the
combination of mutations disrupting both binding sites completely
blocked the downregulation of luciferase activity of the MUT1+2
construct (Fig. 2B, MUT1+2 n.s.).
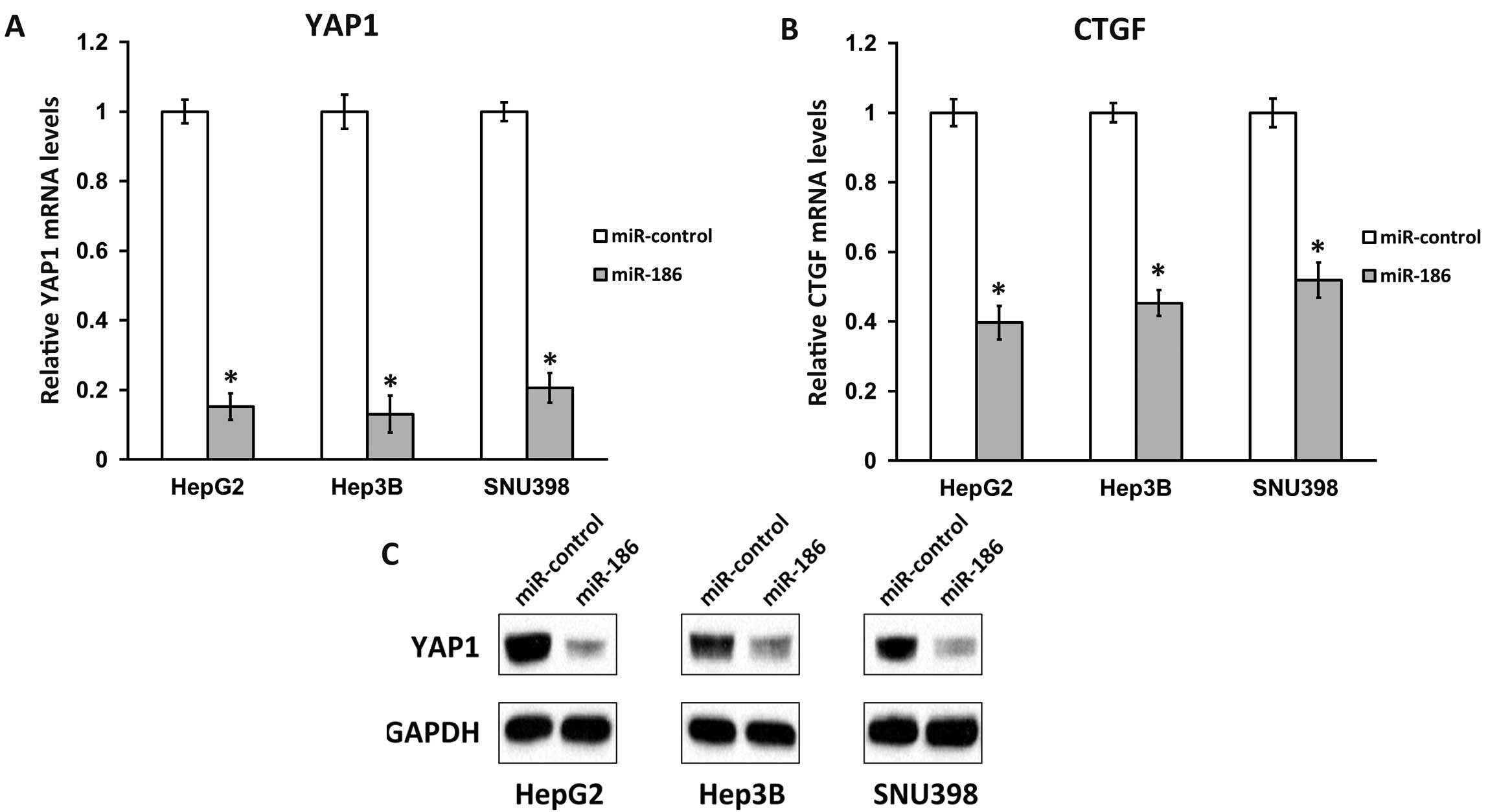
We next transfected miR-186 into HCC cell lines and
analyzed the mRNA and proteins levels of YAP1. Compared with
miR-control transfection, introducing miR-186 significantly reduced
the mRNA levels of YAP1 in all three HCC cell lines examined
(Fig. 3A), suggesting that regulation
of YAP1 by miR-186 occurred mainly through mRNA degradation
rather than translation repression. Using an antibody against YAP1,
we were then able to detect that its protein levels were
consistently downregulated in all three miR-186-transfected HCC
cell lines, but not in miR-control-transfected cells (Fig. 3B). The expression of connective tissue
growth factor (CTGF), a YAP1 downstream effector (3), was also significantly reduced by miR-186
transfection in all three HCC cell lines (Fig. 3C). Taken together, the above results
clearly demonstrated that miR-186 downregulated Hippo signaling by
directly targeting YAP1 mRNA in vivo in HCC cell
lines.
miR-186 inhibits proliferation,
migration and invasion of HCC cells
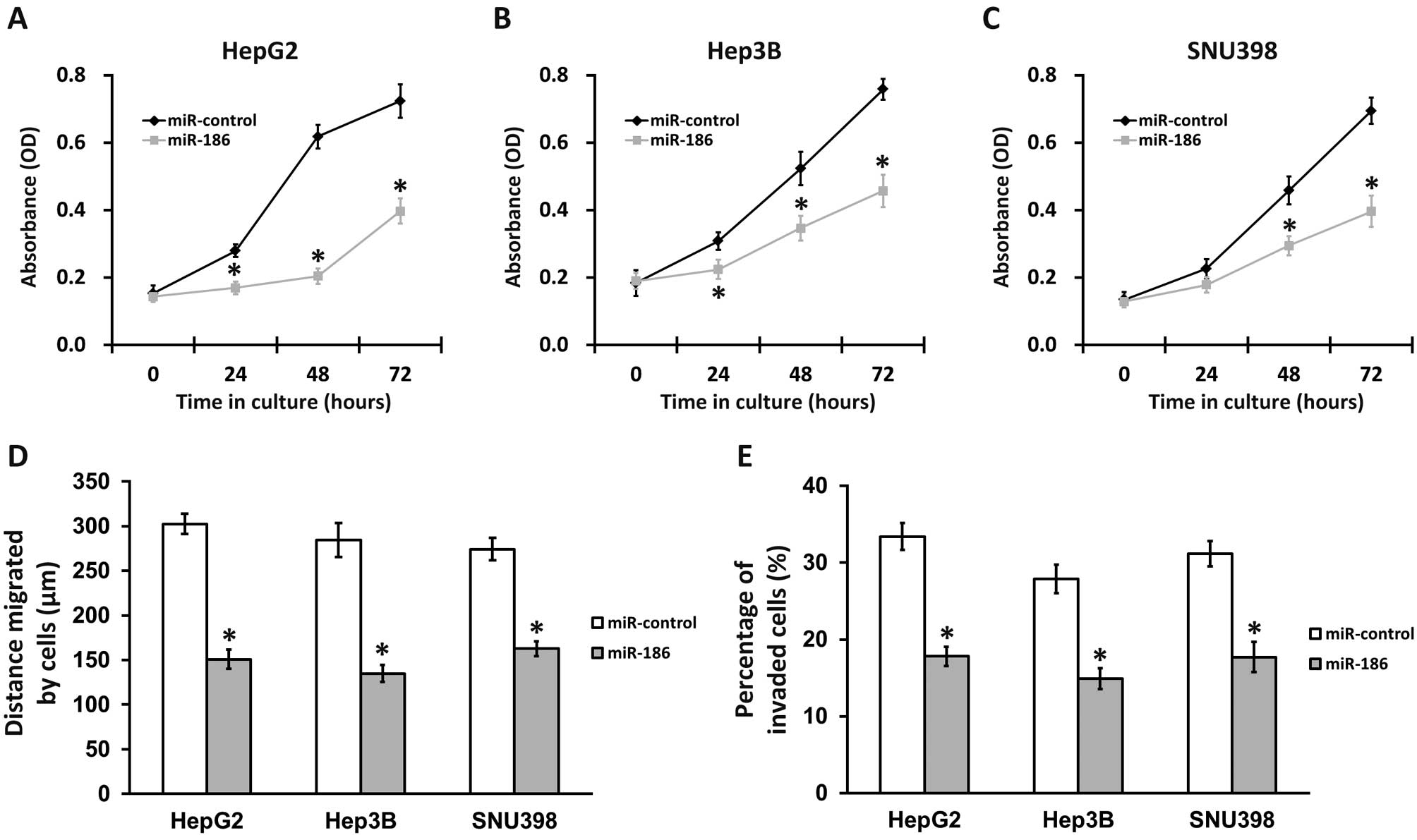
As high levels of YAP1 promote metastasis and
invasive growth in liver cancer (1,2,13), decreasing YAP1 expression by
overexpressing miR-186 in HCC cells should lead to reduced cancer
cell proliferation, migration and invasion. To test this
hypothesis, we performed an MTT proliferation and growth assay,
wound-healing assay and cell invasion assay in miR-186 and
miR-control-transfected HCC cell lines (Fig. 4). All three HCC cell lines exhibited a
significantly slower growth rate following transfection with
miR-186, compared with miR-control experiments (Fig. 4A to C). Moreover, in wound-healing
assays, HepG2, Hep3B and SNU398 cells transfected with miR-control
migrated on average 302, 285 and 274 µm, respectively, after 24 h.
In contrast, when these cells were transfected with miR-186, the
distance migrated was notably decreased to 152, 135 and 163 µm for
HepG2, Hep3B and SNU398 cells, demonstrating a reduction of at
least 40% compared with the control (Fig.
4B). As expected, HCC cells transfected with miR-186 also
exhibited severe defects in the cell invasion assay compared with
those in the control experiments (Fig.
4C). The above results strongly support the role of miR-186 in
inhibiting HCC tumorigenesis.
Discussion
In the present study, we have identified the Hippo
signaling pathway effector YAP1 as being a direct target of
miR-186, where two sites on the 3′-UTR of YAP1 mRNA may be
recognized by miR-186 with different affinity. miR-186 was usually
downregulated in several HCC cell lines we examined, and
overexpression of miR-186 in these HCC cells significantly and
consistently reduces YAP1 mRNA and protein levels, and in
turn downregulates Hippo signaling, which is evidenced by the
decreased expression of downstream effector CTGF. Furthermore,
introducing miR-186 into HCC cells significantly inhibited their
migration, invasion and proliferation.
Various other studies have indicated that miR-186
exhibits multiple functions in a number of aspects of development.
It has been noted to downregulate the expression of the
pro-apoptotic purinergic P2x7 receptor (15). miR-186 also inhibits muscle cell
differentiation through myogenin regulation (16). Plasma miR-186 may be a biomarker for
focal segmental glomerulosclerosis with nephrotic proteinuria
(17). Moreover, miR-186 surfaced in
a previous study as a regulator of acetylcholine packaging and
degradation in neuroinflammation-related disorders (18). In a previous study, miR-186 was also
implicated as a cancer biomarker. miR-186, together with other
miRNAs, was reported to serve as a potential biomarker for oral
squamous cell carcinoma (19). Of
particular interest, Li et al observed that in a mouse model
of human non-small cell lung cancer (NSCLC), miR-186 is able to
downregulate pituitary tumor transforming gene PTTG1 and inhibit
invasion of NSCLC cells (20).
However, there have been no reports of miR-186 involvement in liver
cancer.
To conclude, our study has provided the first
demonstration that miR-186 functions as a tumor suppressor in HCC
cell lines. miR-186 is able to inhibit HCC tumorigenesis by
directly targeting YAP1 and downregulating Hippo signaling, which
supports its role as a potential application in liver cancer
therapy.
Acknowledgments
This study was supported by the Talented Doctor
Funding program (grant no. 20120101).
References
|
1
|
Jie L, Fan W, Weiqi D, Yingqun Z, Ling X,
Miao S, Ping C and Chuanyong G: The hippo-yes association protein
pathway in liver cancer. Gastroenterol Res Pract. 2013:1870702013.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hong L, Cai Y, Jiang M, Zhou D and Chen L:
The Hippo signaling pathway in liver regeneration and
tumorigenesis. Acta Biochim Biophys Sin (Shanghai). 47:46–52. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu
J, Lin JD, Wang CY, Chinnaiyan AM, et al: TEAD mediates
YAP-dependent gene induction and growth control. Genes Dev.
22:1962–1971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sudol M, Shields DC and Farooq A:
Structures of YAP protein domains reveal promising targets for
development of new cancer drugs. Semin Cell Dev Biol. 23:827–833.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhou Z, Hu T, Xu Z, et al: Targeting Hippo
pathway by specific interruption of YAP-TEAD interaction using
cyclic YAP-like peptides. FASEB J. 29:724–732. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in Drosophila and
mammals. Cell. 130:1120–1133. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT,
Zender L, Lowe SW, Poon RT and Luk JM: Yes-associated protein is an
independent prognostic marker in hepatocellular carcinoma. Cancer.
115:4576–4585. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu AM, Poon RT and Luk JM: MicroRNA-375
targets Hippo-signaling effector YAP in liver cancer and inhibits
tumor properties. Biochem Biophys Res Commun. 394:623–627. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kowalik MA, Saliba C, Pibiri M, Perra A,
Ledda-Columbano GM, Sarotto I, Ghiso E, Giordano S and Columbano A:
Yes-associated protein regulation of adaptive liver enlargement and
hepatocellular carcinoma development in mice. Hepatology.
53:2086–2096. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Perra A, Kowalik MA, Ghiso E,
Ledda-Columbano GM, Di Tommaso L, Angioni MM, Raschioni C, Testore
E, Roncalli M, Giordano S and Columbano A: YAP activation is an
early event and a potential therapeutic target in liver cancer
development. J Hepatol. 61:1088–1096. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou L, Qi X, Potashkin JA, Abdul-Karim FW
and Gorodeski GI: MicroRNAs miR-186 and miR-150 down-regulate
expression of the pro-apoptotic purinergic P2×7 receptor by
activation of instability sites at the 3′-untranslated region of
the gene that decrease steady-state levels of the transcript. J
Biol Chem. 283:28274–28286. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Antoniou A, Mastroyiannopoulos NP, Uney JB
and Phylactou LA: miR-186 inhibits muscle cell differentiation
through myogenin regulation. J Biol Chem. 289:3923–3935. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang C, Zhang W, Chen HM, Liu C, Wu J,
Shi S and Liu ZH: Plasma MicroRNA-186 and proteinuria in focal
segmental glomerulosclerosis. Am J Kidney Dis. 65:223–232. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nadorp B and Soreq H: Predicted
overlapping microRNA regulators of acetylcholine packaging and
degradation in neuroinflammation-related disorders. Front Mol
Neurosci. 7:92014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ries J, Vairaktaris E, Agaimy A, Kintopp
R, Baran C, Neukam FW and Nkenke E: miR-186, miR-3651 and miR-494:
Potential biomarkers for oral squamous cell carcinoma extracted
from whole blood. Oncol Rep. 31:1429–1436. 2014.PubMed/NCBI
|
|
20
|
Li H, Yin C, Zhang B, Sun Y, Shi L, Liu N,
Liang S, Lu S, Liu Y, Zhang J, et al: PTTG1 promotes migration and
invasion of human non-small cell lung cancer cells and is modulated
by miR-186. Carcinogenesis. 34:2145–2155. 2013. View Article : Google Scholar : PubMed/NCBI
|