Introduction
Gliomas are the most common type of tumors of the
primary central nervous system in adults (1). Their highly invasive nature precludes
complete resection, resulting in significant neurological morbidity
and mortality (2). Despite progress
in surgical, radio- and chemotherapeutic approaches for the
treatment of glioma, its prognosis remains poor (3). For instance, patients with glioblastoma
multiforme (GBM), which is the most malignant and frequently
reported histological type of glioma, present a median life
expectancy of only 14.6 months following diagnosis (4). Thus, a greater understanding of the
molecular mechanisms of gliomagenesis may lead to more effective,
individualized treatments.
Casitas B-lineage lymphoma (c-Cbl) is a really
interesting new gene finger-type E3 ubiquitin ligase in the
ubiquitin-proteasome pathway (5). Cbl
proteins play important roles in the inhibition of growth factor
receptors (6). For example,
Cbl-mediated ubiquitination of active receptors is essential for
their degradation and the termination of receptor-induced signal
transduction (7–9). As such, the ubiquitin-proteasome pathway
is important for maintaining cellular homeostasis, and mutations in
the components of this pathway may result in tumorigenesis
(10). Indeed, the expression of
c-Cbl has been reported to be upregulated in various solid tumors,
including gastric carcinoma (11),
primary colorectal cancer (12),
prostate cancer (13) and non-small
cell lung cancer (14).
The association between c-Cbl expression and the
clinicopathological features of glioma has not been investigated to
date, nor has been the prognostic significance of c-Cbl
overexpression in this type of tumor. These questions were
addressed in the present study by examining the expression levels
of c-Cbl in samples derived from patients with glioma via reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
immunohistochemistry and western blotting, and the correlation
between c-Cbl expression, glioma stage and patient survival was
assessed.
Materials and methods
Patients and specimens
The present study protocol was approved by the
Ethics Committee of The First Hospital of China Medical University
(Shenyang, China). Paraffin-embedded specimens of 136 glioma cases
were obtained from The First Hospital of China Medical University
from January 2007 to December 2009. The cases comprised 73 men and
63 women, with a mean age of 53.3 years (range, 35–72 years). The
clinicopathological features of the study population are summarized
in Table I. Written informed consent
was obtained from each patient enrolled in the study.
 | Table I.Correlation between the protein
expression levels of c-Cbl and clinicopathological features of
patients with glioma. |
Table I.
Correlation between the protein
expression levels of c-Cbl and clinicopathological features of
patients with glioma.
|
|
| c-Cbl protein
expression levels |
|
|---|
|
|
|
|
|
|---|
| Variable | n | Low (n=40) | High (n=96) | P-value |
|---|
| Age (years) |
|
|
| 0.509 |
| ≥45 | 84 | 23 | 61 |
|
|
<45 | 52 | 17 | 35 |
|
| Gender |
|
|
| 0.340 |
| Male | 73 | 24 | 49 |
|
|
Female | 63 | 16 | 47 |
|
| Extent of
resection |
|
|
| 0.634 |
|
Partial | 37 | 10 | 27 |
|
|
Total | 99 | 30 | 69 |
|
| WHO grade |
|
|
| <0.001 |
| I/II | 72 | 38 | 34 |
|
|
III/IV | 64 | 2 | 62 |
|
| KPS score |
|
|
| <0.001 |
| ≥80 | 76 | 36 | 40 |
|
|
<80 | 60 | 4 | 56 |
|
Immunohistochemistry
Samples were fixed in 10% formaldehyde solution
(Santa Cruz Biotechnology, Inc., Dallas, TX, USA) and embedded in
paraffin (Sigma-Aldrich, St. Louis, MO, USA) blocks, which were
subsequently cut at a thickness of 4 µm. Sections were mounted on
glass slides, deparaffinized in xylene and rehydrated in a graded
series of alcohol (Sigma-Aldrich), followed by boiling in 10 mmol/l
citrate buffer (Santa Cruz Biotechnology, Inc.) at pH 6.0 for 10
mins for antigen retrieval. Following inhibition of endogenous
peroxidase activity by incubation in methanol containing 0.3%
H2O2 (Santa Cruz Biotechnology, Inc.) for 30
min, sections were blocked with 2% bovine serum albumin
(Sigma-Aldrich) for 30 min and incubated overnight at 4°C with
rabbit anti-human c-Cbl monoclonal antibody (cat. no. ab32446;
dilution, 1:200; Abcam, Cambridge, UK). Upon washing three times
with phosphate-buffered saline, sections were incubated with
horseradish peroxidase-conjugated mouse anti-rabbit immunoglobulin
(Ig)G (cat. no. 2357; dilution; 1:200; Santa Cruz Biotechnology,
Inc.) for 30 min, followed by reaction with 3,3′-diaminobenzidine
(Sigma-Aldrich) and counterstaining with hematoxylin (Abcam). For
the negative controls, the primary antibody was substituted with a
nonspecific rabbit IgG antibody (cat. no. sc-2027; Santa Cruz
Biotechnology, Inc.). A Eclipse 90i microscope was used to view the
sections (Nikon Corporation, Tokyo, Japan).
Immunoreactivity was evaluated and scored
semi-quantitatively by two pathologists who were blinded to the
patients' clinical data. Using the double scoring system (staining
intensity multiplied by staining area), the staining intensity was
evaluated as follows: 0, no staining; 1, definite but weak
staining; 2, moderate staining; and 3, strong staining. The
staining area was scored as follows: 1, <35% of cells were
stained; 2, 35–75% of cells were stained; and 3, >75% of cells
were stained. High c-Cbl expression was defined as a score ≥4,
whereas low c-Cbl expression was defined as a score <4.
Western blot analysis
Whole cell lysates were prepared from glioma tissue,
and western blotting was performed as previously described
(15). Protein concentration was
determined with the Protein Quantitation kit (Bradford assay;
Abcam), using bovine serum albumin as a standard. Lysates (20 mg)
were solubilized in Laemmli sample buffer (Abcam) by boiling, and
then resolved by 10% sodium dodecyl sulfate polyacrylamide gel
electrophoresis, followed by electrotransfer (45V for 15 h) onto a
nitrocellulose membrane (Amersham; GE Healthcare Life Sciences,
Chalfont, UK), which was then incubated with the rabbit anti-c-Cbl
antibody at 4°C overnight, followed by incubation with the
peroxidase-conjugated anti-rabbit IgG at room temperature for 1 h.
Rabbit anti-mouse monoclonal α-tubulin (cat. no. ab52866; dilution,
1:2,000; Abcam) antibody served as a loading control. The immune
complexes were visualized with an enhanced chemiluminescence
western blotting detection system (Amersham; GE Healthcare Life
Sciences). Quantity One 4.6 software (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used for desitometry analysis.
RT-qPCR
Total RNA was isolated from tissues using TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), according to the manufacturer's protocol. RNA was reverse
transcribed using SuperScript First-Strand Synthesis System
(Invitrogen; Thermo Fisher Scientific, Inc.), and PCR amplification
was performed using the following sets of sense and antisense
primers: c-Cbl, sense 5′-CGCTAAAGAATAGCCCACCTTAT-3′ and antisense
5′-ATGGCCTCCAGCCCAGAACTGAT-3′; and glyceraldehyde-3-phosphate
dehydrogenase (GAPDH), sense 5′-TGCACCACCAACTGCTTAGC-3′ and
antisense 5′-GGCATGGACTGTGGTCATGAG-3′. Primers were designed using
the Primer premier software 5.0 (Premier Biosoft International,
Palo Alto, CA, USA) and synthesized by Invitrogen (Thermo Fisher
Scientific, Inc.). The amplification reaction consisted of 40
cycles of 94°C for 30 sec, 60°C for 30 sec and 72°C for 30 sec, and
was conducted on an Applied Biosystems 7900HT Fast Real-Time PCR
System (Thermo Fisher Scientific, Inc.) with 1.0 µl of
complementary DNA and SYBR Green Master Mix (Takara Bio, Inc.,
Otsu, Japan). RNA from 3 non-cancerous brain tissue samples,
obtained from patients that had undergone surgery for
drug-resistant temporal epilepsy at The First Hospital of China
Medical University, were used as the control. Data were collected
and analyzed using SDS version 2.3 software (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The expression levels of the
target gene were normalized to those of GAPDH, and determined using
the 2−ΔΔCq method (16). The
experiment was performed in triplicate, and repeated three
times.
Statistical analysis
Statistical analysis was performed with SPSS version
19.0 (IBM SPSS, Armonk, NY, USA). The χ2 test was used
to assess the association between c-Cbl expression and
clinicopathological parameters. Patient survival curves were
generated using the Kaplan-Meier method, and Cox regression
analysis and log-rank test were used to identify independent
prognostic factors. Data were expressed as the mean ± standard
deviation, and statistical significance was defined as a two-tailed
P<0.05.
Results
High c-Cbl expression is associated
with clinicopathological features of glioma
RT-qPCR analysis of the messenger (m)RNA expression
levels of c-Cbl in high-grade [anaplastic astrocytoma (AA)
and GBM] and low-grade [low-grade astrocytoma (LGA)] glioma
revealed that the c-Cbl transcript was upregulated in
high-grade glioma samples, compared with low-grade glioma samples
(P<0.05; Fig. 1A). Similarly, the
protein levels of c-Cbl were increased in AA and GBM, compared with
LGA (Fig. 1B).
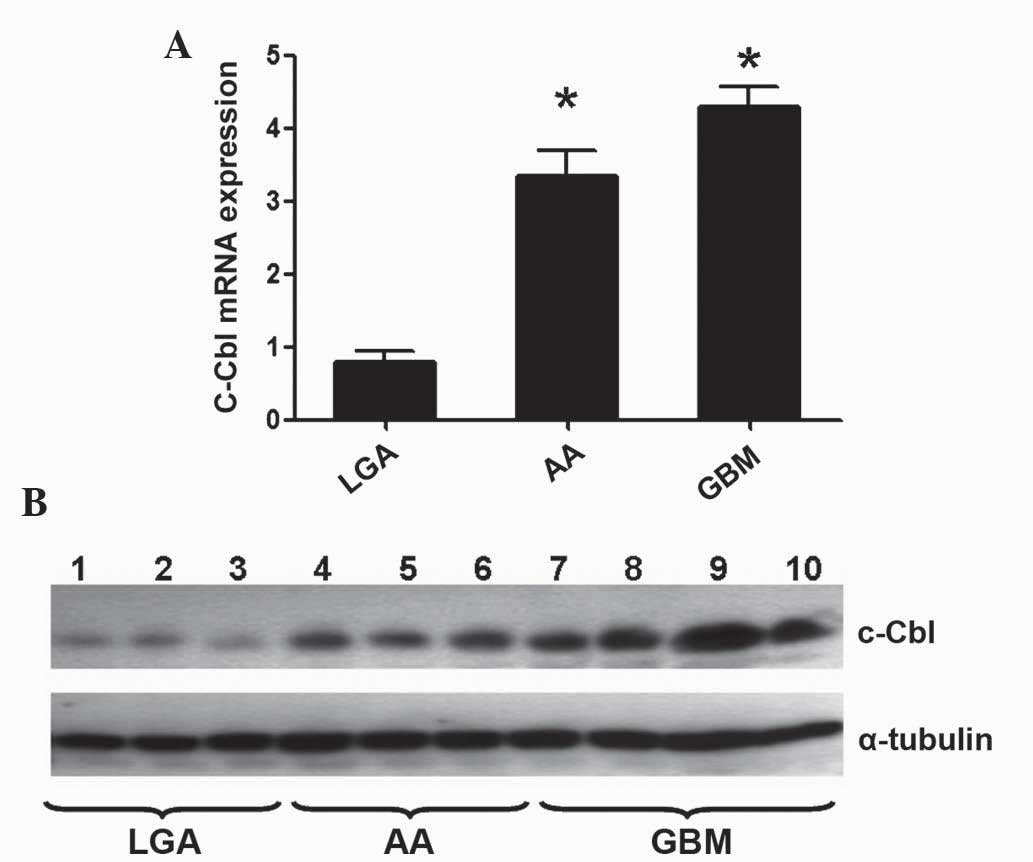 | Figure 1.c-Cbl expression in glioma. (A) Higher
messenger RNA expression levels of c-Cbl were detected in
high-grade (AA and gGBM) glioma samples, compared with low-grade
(LGA) glioma samples, as determined by reverse
transcription-quantitative polymerase chain reaction. The graph
represents the expression levels of c-Cbl, relative to the
expression levels of glyceraldehyde-3-phosphate dehydrogenase.
*P<0.01. (B) c-Cbl protein expression in LGA (n=3), AA (n=3) and
GBM (n=4) frozen samples, as determined by western blotting.
α-tubulin was used as a loading control. c-Cbl, casitas B-lineage
lymphoma; AA, anaplastic astrocytoma; GBM, glioblastoma multiforme;
LGA, low-grade astrocytoma; mRNA, messenger RNA. |
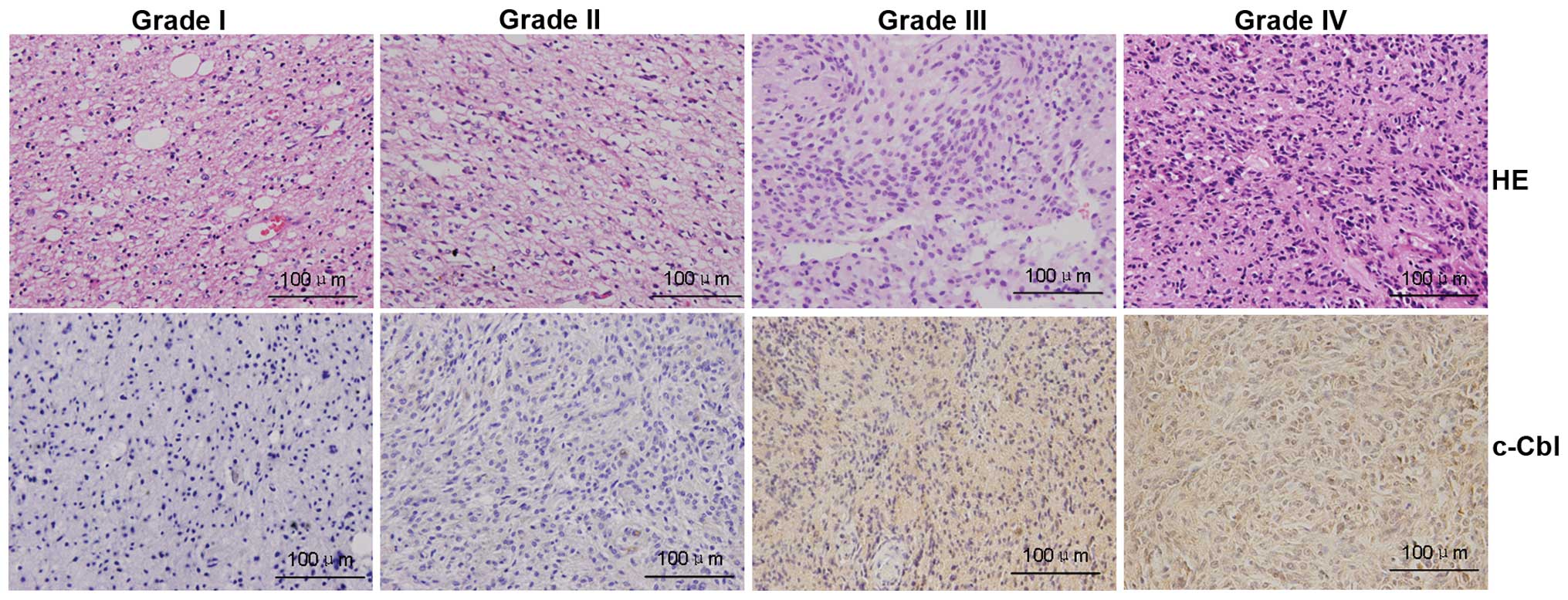
Immunohistochemical analysis demonstrated that c-Cbl
was mainly localized in the cytoplasm of malignant cells, and c-Cbl
immunoreactivity was higher in high- vs. low-grade glioma (Fig. 2). The protein levels of c-Cbl were
upregulated in 96 of 136 (70.6%) patients with glioma, and high
c-Cbl expression was correlated with World Health Organization
(WHO) grade (P<0.001) and Karnofsky performance status (KPS)
score (P<0.001). There were no significant differences in c-Cbl
expression with respect to gender, age or extent of resection
(P>0.05, Table I).
High c-Cbl protein expression levels
are correlated with poor clinical outcome
Six patients were not available for follow-up, and
therefore were excluded from the survival analyses. The remaining
130 patients were followed-up for 6–68 months. Multivariate
analysis revealed that c-Cbl expression was independently
associated with overall survival [hazard ratio (HR)=4.923, 95%
confidence interval (CI)=3.163–7.662; P<0.001], and that c-Cbl
protein expression and WHO grade were independent prognostic
factors of progression-free survival (HR=6.181, 95% CI=3.854–9.915;
P<0.001, and HR=10.247, 95% CI=9.009–11.655; P<0.001,
respectively) (Table II). The
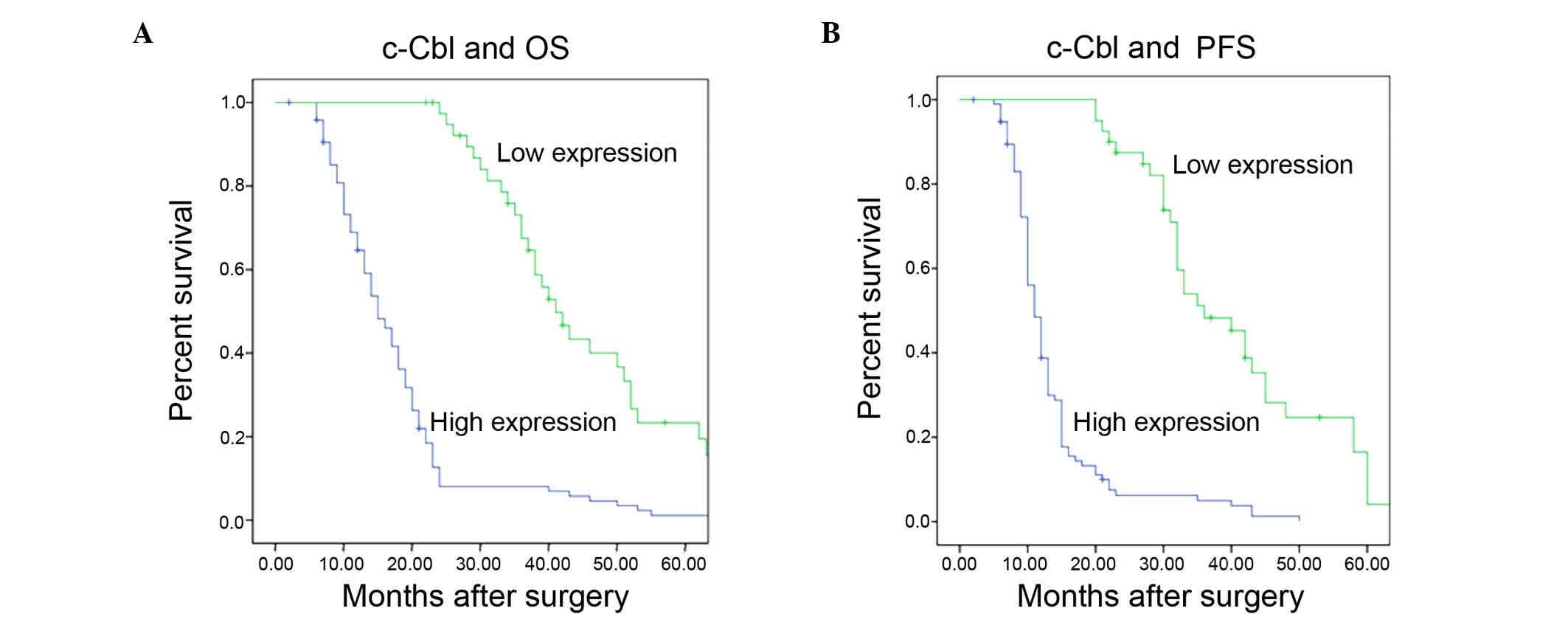
Kaplan-Meier analysis with log-rank test indicated that high c-Cbl
protein expression was associated with poor overall (P<0.001;
Fig. 3A) and progression-free
survival (P<0.001; Fig. 3B).
 | Table II.Cox regression model for multivariate
analysis of glioma prognostic factors. |
Table II.
Cox regression model for multivariate
analysis of glioma prognostic factors.
|
| Overall survival | Progression-free
survival |
|---|
|
|
|
|
|---|
| Variable | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Age (≥45 vs. <45
years) | 1.763 | 1.222–2.543 | 0.002 | 1.747 | 1.210–2.532 | 0.003 |
| Gender (male vs.
female) | 0.516 | 0.471–0.546 | <0.001 | 0.531 | 0.485–0.581 | <0.001 |
| Extent of resection
(partial vs. total) | 3.668 | 3.242–4.149 | <0.001 | 3.475 | 3.076–3.927 | <0.001 |
| WHO grade (III/IV vs.
I/II) | 8.842 | 7.827–9.989 | <0.001 | 10.247 | 9.009–11.655 | <0.001 |
| KPS score (<80 vs.
≥80) | 0.984 | 0.905–1.069 | 0.701 | 0.926 | 0.852–1.008 | 0.067 |
| c-Cbl expression
(high vs. low) | 4.923 | 3.163–7.662 | <0.001 | 6.181 | 3.854–9.915 | <0.001 |
Discussion
The results of the current study indicate that
glioma progression is associated with the upregulation of c-Cbl
expression. Western blotting and immunohistochemical analysis of
tissue samples derived from patients with glioma revealed a
significant correlation between c-Cbl expression and WHO glioma
grade. Furthermore, survival analysis demonstrated that
overexpression of c-Cbl is a predictor of poor prognosis in these
patients.
c-Cbl is a E3 ubiquitin ligase and multifunctional
adaptor protein that regulates cell growth, invasion, apoptosis and
angiogenesis in various human tumors, and mutations in the c-Cbl
gene may lead to tumorigenesis and metastasis in non-small cell
lung cancer (14). In hematological
malignancies, mutations in the genes of the Cbl family result in
the failure of tyrosine kinase signaling of protein degradation,
which is linked to poor prognosis (17). In previous studies, c-Cbl was
expressed in 67% of gastric carcinoma cells, and was associated
with the degree of tumor invasion and lymph node metastasis
(11). c-Cbl was also demonstrated to
promote tumor invasion (18).
Furthermore, c-Cbl gene deficiency decreased osteoclast (19) and macrophage (20) migration, and modulated glioma cell
invasion via regulation of matrix metalloproteinase 2 (21).
In the present study, the protein levels of c-Cbl
were significantly associated with WHO grade and KPS score,
suggesting that c-Cbl expression is associated with glioma
development and progression. The present findings revealed that
patients with higher c-Cbl expression in tumor tissue exhibited
worse overall and progression-free survival than those expressing
lower levels of the protein, indicating that c-Cbl upregulation in
glioma may increase tumor malignancy, and thereby lead to worse
prognosis. Notably, the present results demonstrate that high
levels of c-Cbl in human glioma tissues are associated with lower
KPS scores and higher pathological grade. Survival analysis
demonstrated that high c-Cbl expression in glioma tissues is
correlated with, and is a prognostic factor for, lower
progression-free and overall survival. A subgroup analysis
suggested that c-Cbl may be an independent prognostic factor for
high (III and IV), but not low (I and II) histopathological grade.
These findings demonstrate that the expression levels of c-Cbl may
be used to predict prognosis in patients with glioma following
surgery.
The mechanisms underlying the oncogenic functions of
c-Cbl in glioma remain to be investigated. A previous study
reported a link between c-Cbl-mediated epidermal growth factor
receptor signaling, tumor progression and metastasis, and poor
prognosis in human gastric carcinoma (6). Identifying c-Cbl target mRNAs and
binding partners will provide additional insight into the role of
c-Cbl in gliomagenesis.
Considering the poor prognosis of patients with
glioma with the currently available therapies, the development of
novel treatment approaches is necessary, which requires a better
understanding of the pathophysiological and molecular properties of
gliomas. The findings of the present study suggest that high c-Cbl
expression is a prognostic biomarker for glioma malignancy, and
provide a basis for developing novel treatments.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant nos.
81101917 and 81270036), and the Natural Science Foundation of
Liaoning Province (Shenyang, China; grant no. 2013021045).
References
|
1
|
Ma R, de Pennington N, Hofer M, Blesing C
and Stacey R: Diagnostic and prognostic markers in gliomas - an
update. Br J Neurosurg. 27:311–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hentschel SJ and Lang FF: Current surgical
management of glioblastoma. Cancer J. 9:113–125. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Diamond EL, Corner GW, De Rosa A,
Breitbart W and Applebaum AJ: Prognostic awareness and
communication of prognostic information in malignant glioma: A
systematic review. J Neurooncol. 119:227–234. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clarke J, Butowski N and Chang S: Recent
advances in therapy for glioblastoma. Arch Neurol. 67:279–283.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andoniou CE, Thien CB and Langdon WY:
Tumour induction by activated abl involves tyrosine phosphorylation
of the product of the cbl oncogene. EMBO J. 13:4515–4523.
1994.PubMed/NCBI
|
|
6
|
Thien CB and Langdon WY: Cbl: Many
adaptations to regulate protein tyrosine kinases. Nat Rev Mol Cell
Biol. 2:294–307. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Di Fiore PP and De Camilli P: Endocytosis
and signaling. An inseparable partnership. Cell. 106:1–4. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Joazeiro CA and Hunter T: Biochemistry.
Ubiquitination - more than two to tango. Science. 289:2061–2062.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Waterman H and Yarden Y: Molecular
mechanisms underlying endocytosis and sorting of ErbB receptor
tyrosine kinases. FEBS Lett. 490:142–152. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Mohapatra B, Ahmad G, Nadeau S, Zutshi N,
An W, Scheffe S, Dong L, Feng D, Goetz B, Arya P, et al: Protein
tyrosine kinase regulation by ubiquitination: Critical roles of
Cbl-family ubiquitin ligases. Biochim Biophys Acta. 1833:122–139.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ito R, Nakayama H, Yoshida K, Matsumura S,
Oda N and Yasui W: Expression of Cbl linking with the epidermal
growth factor receptor system is associated with tumor progression
and poor prognosis of human gastric carcinoma. Virchows Arch.
444:324–331. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cristóbal I, Manso R, Rincón R, Caramés C,
Madoz-Gúrpide J, Rojo F and García-Foncillas J: Up-regulation of
c-Cbl suggests its potential role as oncogene in primary colorectal
cancer. Int J Colorectal Dis. 29:6412014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Knight JF, Shepherd CJ, Rizzo S, Brewer D,
Jhavar S, Dodson AR, Cooper CS, Eeles R, Falconer A, Kovacs G, et
al: TEAD1 and c-Cbl are novel prostate basal cell markers that
correlate with poor clinical outcome in prostate cancer. Br J
Cancer. 99:1849–1858. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tan YH, Krishnaswamy S, Nandi S, Kanteti
R, Vora S, Onel K, Hasina R, Lo FY, El-Hashani E, Cervantes G, et
al: CBL is frequently altered in lung cancers: Its relationship to
mutations in MET and EGFR tyrosine kinases. PLoS One.
5:e89722010.Hsu HH [added]. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shibuya N, Inoue K, Tanaka G, Akimoto K
and Kubota K: Augmented pentose phosphate pathway plays critical
roles in colorectal carcinomas. Oncology. 88:309–319. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Makishima H, Cazzolli H, Szpurka H, Dunbar
A, Tiu R, Huh J, Muramatsu H, O'Keefe C, Hsi E, Paquette RL, et al:
Mutations of e3 ubiquitin ligase cbl family members constitute a
novel common pathogenic lesion in myeloid malignancies. J Clin
Oncol. 27:6109–6116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nam JM, Onodera Y, Mazaki Y, Miyoshi H,
Hashimoto S and Sabe H: CIN85, a Cbl-interacting protein, is a
component of AMAP1-mediated breast cancer invasion machinery. EMBO
J. 26:647–656. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chiusaroli R, Sanjay A, Henriksen K,
Engsig MT, Horne WC, Gu H and Baron R: Deletion of the gene
encoding c-Cbl alters the ability of osteoclasts to migrate,
delaying resorption and ossification of cartilage during the
development of long bones. Dev Biol. 261:537–547. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Caveggion E, Continolo S, Pixley FJ,
Stanley ER, Bowtell DD, Lowell CA and Berton G: Expression and
tyrosine phosphorylation of Cbl regulates macrophage chemokinetic
and chemotactic movement. J Cell Physiol. 195:276–289. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lee H and Tsygankov AY: c-Cbl regulates
glioma invasion through matrix metalloproteinase 2. J Cell Biochem.
111:1169–1178. 2010. View Article : Google Scholar : PubMed/NCBI
|