Article
Network and pathway analysis of microRNAs, transcription factors, target genes and host genes in human glioma
- Authors:
- Ying Zhang
- Shishun Zhao
- Zhiwen Xu
-
View Affiliations / Copyright
Affiliations:
College of Mathematics, Jilin University, Jilin, Changchun 130012, P.R. China, Department of Computer Science and Technology, Jilin University, Jilin, Changchun 130012, P.R. China
-
Pages:
3534-3542
|
Published online on:
March 31, 2016
https://doi.org/10.3892/ol.2016.4398
- Expand metrics +
Metrics:
Total
Views: 0
(Spandidos Publications: | PMC Statistics:
)
Metrics:
Total PDF Downloads: 0
(Spandidos Publications: | PMC Statistics:
)
This article is mentioned in:
Abstract
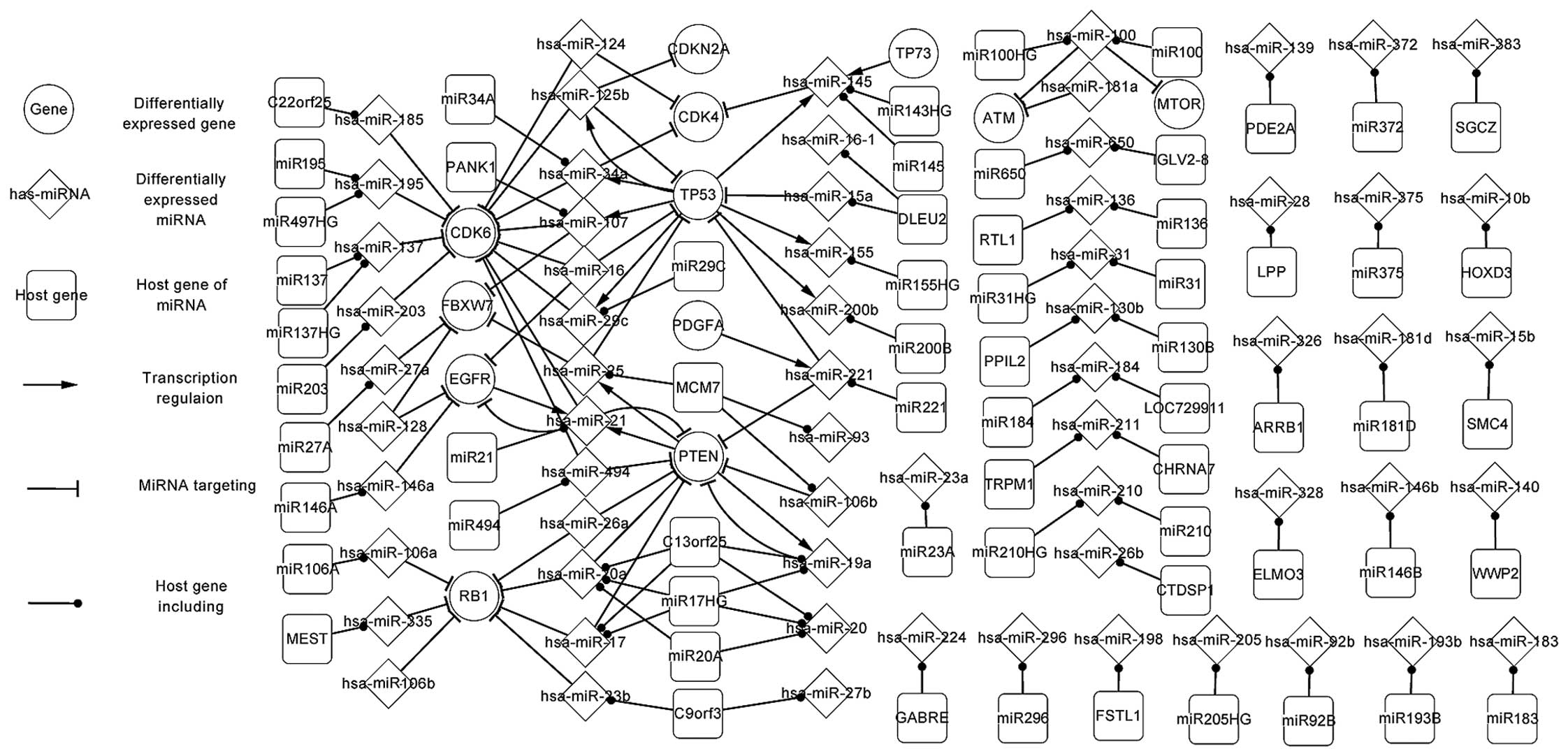
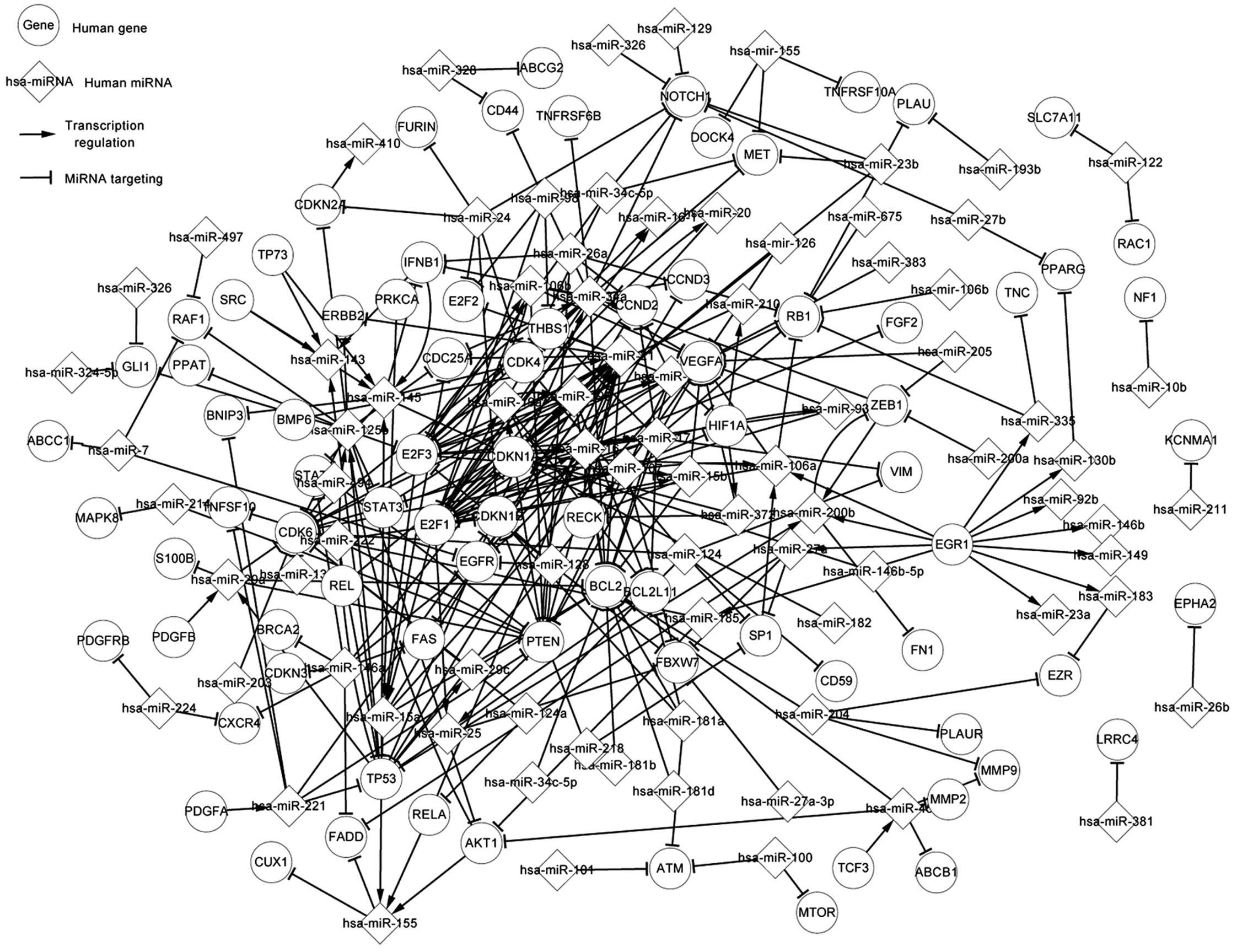
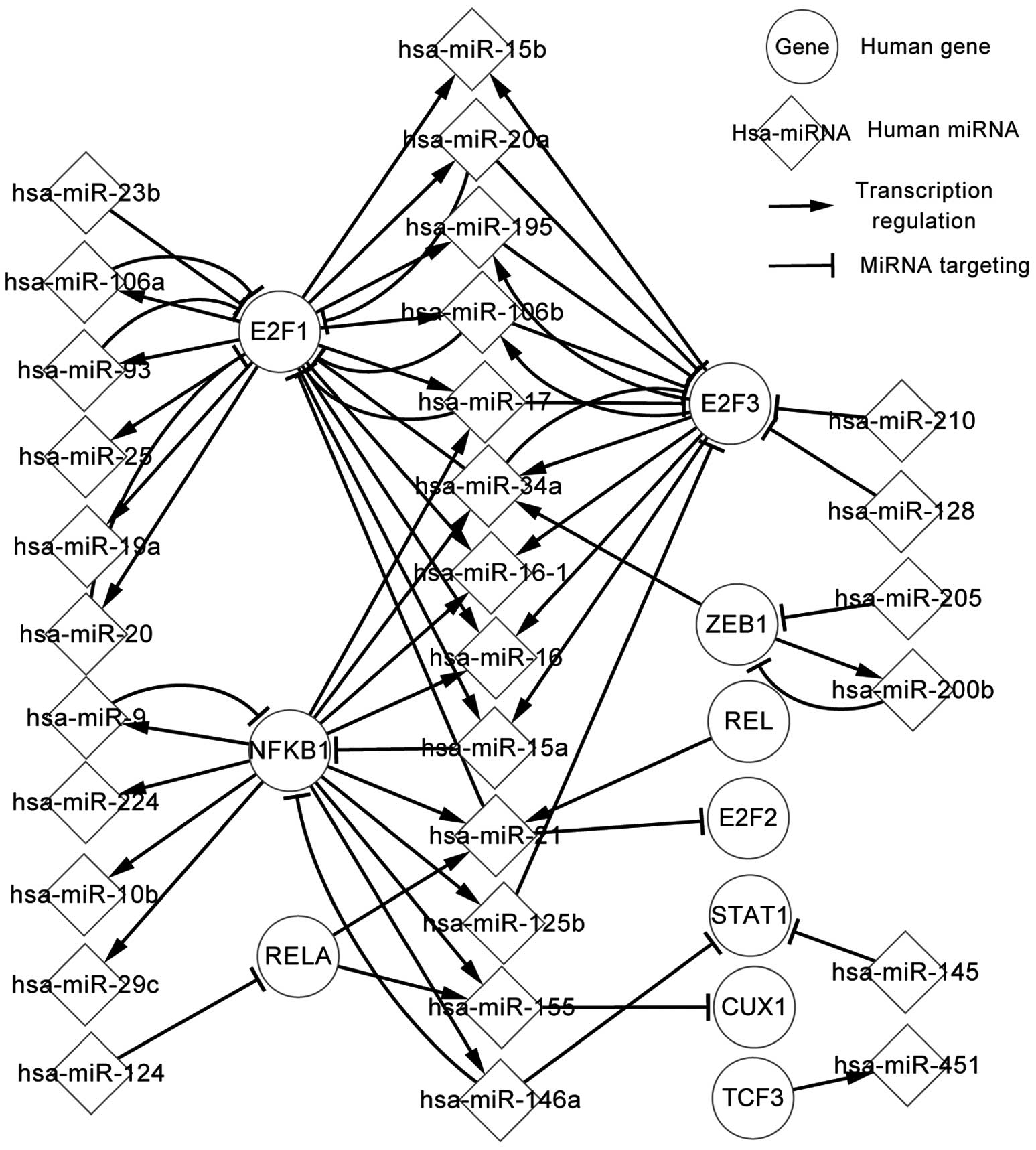
To date, there has been rapid development with regard to gene and microRNA (miR/miRNA) research in gliomas. However, the regulatory mechanisms of the associated genes and miRNAs remain unclear. In the present study, the genes, miRNAs and transcription factors (TFs) were considered as elements in the regulatory network, and focus was placed on the associations between TFs and miRNAs, miRNAs and target genes, and miRNAs and host genes. In order to show the regulatory correlation clearly, all the elements were investigated and three regulatory networks, namely the differentially‑expressed, related and global networks, were constructed. Certain important pathways were highlighted, with analysis of the similarities and differences among the networks. Next, the upstream and downstream elements of differentially‑expressed genes, miRNAs and predicted TFs were listed. The most notable aspect of the present study was the three levels of network, particularly the differentially‑expressed network, since the differentially‑expressed associations that these networks provide appear at the initial stages of cancers such as glioma. If the states of the differentially‑expressed associations can be adjusted to the normal state via alterations in regulatory associations, which were also recorded in the study networks and tables, it is likely that cancer can be regulated or even avoided. In the present study, the differentially‑expressed network illuminated the pathogenesis of glioma; for example, a TF can regulate one or more miRNAs, and a target gene can be targeted by one or more miRNAs. Therefore, the host genes and target genes, the host genes and TFs, and the target genes and TFs indirectly affect each other through miRNAs. The association also exists between TFs and TFs, target genes and target genes, and host genes and host genes. The present study also demonstrated self‑adaption associations and circle‑regulations. The related network further described the regulatory mechanism associated with glioma. These results can be utilized to adjust the states. The present study expounded the regulatory mechanisms of glioma and supplied theoretical data for further studies, in which greater attention should be focused on the highlighted genes and miRNAs.
View References
|
1
|
He J, Zhang W, Zhou Q, Zhao T, Song Y,
Chai L and Li Y: Low-expression of microRNA-107 inhibits cell
apoptosis in glioma by upregulation of SALL4. Int J Biochem Cell
Biol. 45:1962–1973. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lefranc F, Rynknowski M, DeWitte O and
Kiss R: Present and potential future adjuvant issues in high-grade
astrocytic glioma treatment. Adv Tech Stand Neurosurg. 34:3–35.
2009.PubMed/NCBI
|
|
3
|
Yu J, Cai X, He J, Zhao W, Wang Q and Liu
B: Microarray-based analysis of gene regulation by transcription
factors and microRNAs in glioma. Neurol Sci. 34:1283–1289. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dong CG, Wu WK, Feng SY, Wang XJ, Shao JF
and Qiao J: Co-inhibition of microRNA-10b and microRNA-21 exerts
synergistic inhibition on the proliferation and invasion of human
glioma cells. Int J Oncol. 41:1005–1012. 2012.PubMed/NCBI
|
|
6
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38(Database issue): D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kozomara A and Griffiths-Jones S: miRBase
integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39(Database issue): D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Safran M, Dalah I, Alexander J, Rosen N,
Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al:
GeneCards Version 3: The human gene integrator. Database (Oxford).
2010:baq0202010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chekmenev DS, Haid C and Kel AE: P-Match:
Transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33(Web Server issue):
W432–W437. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC Genome Browser database: Update 2011. Nucleic Acids
Res. 39(Database issue): D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z,
Chen Y, Cao X, Jiang C, Yan W and Xu C: MicroRNA-449 and
microRNA-34b/c function redundantly in murine testes by targeting
E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway.
J Biol Chem. 287:21686–21698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ishii N, Maier D, Merlo A, Tada M,
Sawamura Y, Diserens AC and Van Meir EG: Frequent co-alterations of
TP53, p16/CDKN2A, p14 (ARF), PTEN tumor suppressor genes in human
glioma cell lines. Brain Pathol. 9:469–479. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu L, Li G, Feng D, Qin H, Gong L, Zhang J
and Zhang Z: MicroRNA-21 expression is associated with overall
survival in patients with glioma. Diagn Pathol. 8:2002013.
View Article : Google Scholar : PubMed/NCBI
|