Introduction
Hepatocellular carcinoma (HCC) is one of the most
prevalent types of cancer worldwide (1) with an incidence rate of 10.1 cases per
100,000 individuals and a mortality rate of 9.5 cases per 100,000
individuals (2). Previously,
dysfunction of the yes-associated protein (YAP) signaling pathway
has been associated with liver tumorigenesis (3), and liver-specific YAP overexpression in
transgenic mice resulted in tumor development (4). In addition, clinical studies have
demonstrated that YAP is overexpressed and is a predictor of poor
survival in patients with HCC (5).
Due to the key role YAP is considered to play in liver
tumorigenesis, it is extremely important to understand how this
protein is regulated in HCC cells. A previous study established the
role of cAMP response element-binding protein (CREB) in the
upregulation of YAP in HCC cells, and identified a novel CREB
binding site within the YAP promoter region (6). However, whether and how other proteins
stimulate CREB-dependent transcription of YAP remains unclear.
Functioning as a cofactor, YAP increases the
transcriptional activities of various transcription factors,
including TEA domain (TEAD) family (7). Although several YAP target genes that
operate in liver tumorigenesis have been gradually identified
(8,9),
a large number remain unknown. Previously, the membrane protein,
melanoma cell adhesion molecule (MCAM), was established as a novel
HCC-specific YAP target (10). MCAM
serum levels have been revealed to be specifically increased in
HCC, indicating that MCAM serum levels may function as a specific
tumor marker for HCC (10). In
addition, it was demonstrated that MCAM is essential for HCC cell
transformation and survival through the promotion of
pro-tumorigenic c-Jun/c-Fos protein function (10). However, whether and how MCAM
conversely regulates YAP function requires additional
investigation.
The aim of the present study was to demonstrate
whether MCAM regulates YAP in HCC cells and how this mechanism
occurs. The present study demonstrated that MCAM is key in the
stimulation of CREB-dependent transcription of YAP via the MCAM
downstream effector c-Jun/c-Fos, and that YAP is a downstream
effector of MCAM and c-Jun/c-Fos in the maintenance of the
transformative phenotype of HCC cells. The present study
established a complete, positive, auto-regulatory feedback loop, in
which YAP is able to stimulate MCAM gene transcription and
MCAM conversely enhances YAP gene transcription.
Materials and methods
Cell culture and vectors
Human HCC Bel-7402, SMMC-7721 and Huh7 cells lines
were purchased from Cell Bank of Type Culture Collection of Chinese
Academy of Sciences (Shanghai, China) and cultured in Dulbecco's
modified Eagle's medium (DMEM) supplemented with 10% fetal bovine
serum (FBS) (Gibco®; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) at 37°C in an incubator with 5% CO2.
Small hairpin RNAs (shRNAs) against MCAM (sh1 and sh2), c-Fos and
c-Jun were purchased from GeneChem Co., Ltd. (Shanghai, China). The
cDNA fragments encoding human MCAM were purchased from OriGene
Technologies, Inc. (Beijing, China), and subcloned into a
pcDNA3.1(+) vector (Invitrogen™; Thermo Fisher Scientific, Inc.) as
previously described (10). The human
c-Fos- and c-Jun-expressing vectors were purchased from OriGene
Technologies, Inc.
Luciferase reporters containing either a CREB
binding site (CREB-reporter) or a core YAP promoter region
(YAP-promoter) were constructed as described previously
(6,11). The TEAD-Gal4/pUAS-Luc system was
provided by Dr Junhao Mao (University of Massachusetts, Worcester,
MA, USA) from the authors' previous study (12), and was used to indirectly test YAP
activity, since YAP binds to the TEAD transcription factor. The
YAP-expressing plasmids were constructed as described in a previous
study (6). Briefly, the open reading
frame of YAP was polymerase chain reaction (PCR)-amplified using
the following primers: YAP, forward
5′-ATGCGCTAGCGATCCCGGGCAGCAGCCGCCGCCTC-3′ and reverse
5′-ATGCGGCCGCCTATAACCATGTAAGAAAGCTTTCTTTATC-3′. The cDNA of
Bel-7402 cells was used as the template, and a
pGIZ2a-lentiviral-based vector (provided by Dr Junhao Mao) was used
as the backbone.
Immunofluorescence
Glass slides were seeded with Bel-7402 cells and
were fixed by 4% paraformaldehyde (Scigen Ltd, Shanghai, China) for
15 min, followed by washing with phosphate-buffered saline (PBS)
for 5 min three times. Subsequently, the cells were incubated with
blocking buffer [PBS solution containing 3% FBS, 1% goat serum
(Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA)
and 0.1% Triton X-100 (Sangon Biotech Co., Ltd., Shanghai, China)]
for 2 h at room temperature prior to incubation with primary
antibodies overnight at 4°C. The following primary antibodies were
used at a dilution of 1:200, and were purchased from Cell Signaling
Technology, Inc. (Danvers, MA, USA): Rabbit polyclonal anti-YAP
(catalog no., 4912) and mouse monoclonal anti-FLAG (catalog no.,
8146). The slides were then washed in PBS three times prior to
incubation with Alexa Fluor®-488/555 fluorescent
conjugated secondary antibodies (mouse/rabbit monoclonal; catalog
nos., 4408 and 4413, respectively; Cell Signaling Technology, Inc.)
for 1 h in the dark. Subsequently, the slides were washed three
times with PBS prior to being mounted with ProLong® Gold
Antifade Reagent with DAPI (Molecular Probes, Eugene, OR, USA). The
sides were observed using a LSM 800 Confocal Microscope (Carl Zeiss
AG, Oberkochen, Germany).
Western blot analysis
Cells were harvested and lysed using Western/IP
buffer (Beyotime Institute of Biotechnology, Haimen, China) with
protease inhibitor (Roche Diagnostics, Indianapolis, IN, USA). The
supernatants of cell lysates were collected by centrifugation at
15,000 × g for 10 min. The proteins was quantified by a BSA kit
(Beyotime Institute of Biotechnology). Protein (30 µg each sample)
were resolved using 12% sodium dodecyl sulfate (SDS) polyacrylamide
gel electrophoresis. The proteins were electrophoretically
transferred onto Immobilon P Membrane (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA). The blots were blocked with 5% non-fat milk in
Tris-buffered saline (TBS; pH 7.4; Sangon Biotech Co., Ltd.) for 1
h at room temperature and subsequently incubated with primary
antibodies at 4°C overnight. The primary antibodies were used at a
dilution of 1:1,000 and were as follows: Rabbit monoclonal
anti-MCAM (catalog no., 2505-1) rabbit monoclonal anti-YAP (catalog
no., 2060-1), and rabbit monoclonal anti-c-Jun (catalog no.,
1254-1) (Epitomics, Burlingame, CA, USA); mouse monoclonal
anti-FLAG (catalog no., F3165; Sigma-Aldrich, St. Louis, MO, USA);
rabbit polyclonal anti-FLAG (catalog no., 2368), rabbit monoclonal
anti-CREB (catalog no., 9197), rabbit monoclonal anti-phospho-CREB
(catalog no., 9198), rabbit monoclonal anti-glyceraldehyde
3-phosphate dehydrogenase (GAPDH; catalog no., 5174) and rabbit
monoclonal anti-c-Fos (catalog no., 2250) (Cell Signaling
Technology, Inc.). The membranes were washed with TBS and incubated
with secondary antibodies conjugated with horseradish peroxidase
(catalog no., 7074 and 7076; Cell Signalling Technology, Inc.) for
1 h at room temperature. After washing with TBS, the membranes were
visualized using Pierce ECL Western Blotting Substrate (Pierce™;
Thermo Fisher Scientific, Inc.) and recorded on X-ray films. YAP
protein levels were normalized to those of GAPDH and p-CREB levels
were normalized to those of total CREB. Densitometry of the blots
was performed using ImageJ version 1.47 software (National
Institutes of Health, Bethesda, MD, USA).
Luciferase reporter analysis
Luciferase reporter plasmids were transiently
transfected into Bel-7402, SMMC-7721 or Huh7 cells using
Lipofectamine® 2000 (Invitrogen™; Thermo Fisher
Scientific, Inc.). pRL-TK (Promega Corporation, Madison, WI, USA)
was co-transfected as an internal control to calibrate transfection
efficiency. Following additional cultivation for 24 h, the
transfected cells were harvested and lysed used 1X passive lysis
buffer, centrifuged at 15,000 × g for 10 min and the pellet
subjected to luciferase assay. Luciferase activity was measured as
chemiluminescence using a plate reader (Σ960; Metertech, Inc.,
Taipei, Taiwan) and the Dual-Luciferase® Reporter Assay
System (Promega Corporation), according to the manufacturer's
protocol. Firefly luciferase activities were normalized to those
from Renilla.
Reverse transcription-quantitative PCR
(qPCR)
Total RNA from Bel-7402, SMMC-7721 and Huh7 cells
was extracted using TRIzol Reagent (Invitrogen™; Thermo Fisher
Scientific, Inc.). In total, 20 ml reaction mixture was used to
reverse transcribe 1 mg total RNA to cDNA using PrimeScript RT
Reagent kit (Takara, Bio, Inc., Otsu, Japan). qPCR was performed on
the cDNA using primers specific for YAP. The primers were purchased
from BGI Shenzhen (Shenzhen, China) and were as follows: YAP,
forward 5′-CCTCGTTTTGCCATGAACCAG-3′ and reverse
5′-GTTCTTGCTGTTTCAGCCGCAG-3′; GAPDH, forward
5′-ATCATCCCTGCCTCTACTGG-3′ and reverse 5′-GTCAGGTCCACCACTGACAC-3′.
SYBR Green Mix kit (Takara Bio, Inc.) was used for the qPCR. The
conditions for qPCR were as follows: Activation at 94°C for 5 min,
and then 40 cycles of denaturation at 94°C for 30 sec and combined
annealing and extension at 60°C for 30 sec. The RT-qPCR was
performed on an ABI 7500 Real-time PCR System (Applied
Biosystems®; Thermo Fisher Scientific, Inc.). The
fluorescence of each sample was determined following every cycle.
Relative expression levels were calculated as the ratios normalized
against those of GAPDH, using the ∆Cq method. The ∆Cq value was
determined by subtracting the GAPDH ∆Cq value from the YAP gene ∆Cq
value. The ∆Cq of the stimulated cells (∆Cqt) was subtracted from
the ∆Cq of the untreated cells (∆Cqu) as follows: ∆∆Cq = ∆Cqt -
∆Cqu. The expression level for the YAP gene in the stimulated cells
compared with the level in the untreated cells was calculated as
follows: x-fold of unstimulated control = 2-∆∆Cq
(13).
Chromatin immunoprecipitation
(ChIP)
ChIP assays were performed with the
ChIP-IT® Express kit (catalog no., 53008; Active Motif,
Carlsbad, CA, USA), according to the manufacturer's protocol.
Briefly, 1×107 cells were fixed with 1% formaldehyde,
washed with cold PBS and lysed in 1X lysis buffer. Nuclei were
sonicated (JY92-IIN; Scientz Biotech Co., Ltd., Ningbo, China) to
shear DNA, and the lysates were pelleted and precleared using A/G
beads (Novex®; Thermo Fisher Scientific, Inc.). The
beads were separated by centrifugation at 15,000 × g for 20 min and
the protein-DNA complexes were incubated with 3 µg rabbit
monoclonal anti-CREB antibody and 10 µl protein A/G beads in a
total reaction mixture of 200 µl overnight at 4°C. The protein-DNA
complexes were eluted in 1% SDS/0.1 M NaHCO3 and
cross-links were reversed at 65°C. Finally, DNA was subjected to
qPCR analysis following recovery using a SYBR Green Mix kit
(Takara, Bio, Inc.) and an ABI 7500 Real-time PCR System (Applied
Biosystems®; Thermo Fisher Scientific, Inc.) with the
following cycling conditions: 94°C for 5 min, then 40 cycles of
94°C for 30 sec, and 60°C for 30 sec. The primers were purchased
from BGI Shenzhen and were used for detection of CREB binding on
the YAP promoter (R1 and R2), as follows: R1, forward
5′-CCAACAACTATGAGGTAGGTG-3′ and reverse
5′-GAGTTTTGAGTTCAGGTCCCAGTTC-3′; R2, forward
5′-GCAGACTAACAGATAAGTGAAAC-3′ and reverse
5′-TCCGCTCCGCTCGGCCCCTTTTC-3′. The fluorescence of each sample was
determined following every cycle. Relative enrichments of CREB
binding within the YAP promoter were calculated as the ratios
normalized against those of the input sample, using the ΔCq method.
The ΔCq value was determined by subtracting the input sample ΔCq
value from the CREB ChIP sample ΔCq value. The percentage of input
(%) was calculated as follows: 2-∆∆Cq × 100% (11).
Soft-agar assays
Anchorage-independent soft-agar growth assays were
performed as previously described (6). Briefly, control (Bel-7402, SMMC-7721 or
Huh7 without treatment), Bel-7402, SMMC-7721 or Huh7 cells with
MCAM, c-Jun or c-Fos knocked-down, using shRNAs, in the presence or
absence of overexpression of YAP were seeded onto 6-well agar
plates at a density of 5×103 cells/well and maintained
in DMEM supplemented with 1 µg/ml puromycin (Mediatech, Inc.,
Manassas, VA, USA) for 4 weeks until foci were evident. Colonies
were subsequently counted.
Statistical analysis
Student's t-test was used to examine the differences
between groups using STATA version 11.0 software (StataCorp LP,
College Station, TX, USA). P<0.05 was considered to indicate a
statistically significant difference. Results are expressed as the
mean ± standard deviation of three independent experiments.
Results
MCAM conversely regulates YAP
To investigate whether MCAM conversely regulates
YAP, YAP protein expression was analyzed prior to and following
knockdown or overexpression of MCAM. The results demonstrated that
YAP protein levels were downregulated by MCAM-knockdown through two
independent MCAM shRNAs (sh1 and sh2, respectively), whilst
overexpression of MCAM resulted in upregulated YAP protein levels
(Fig. 1A and B). Since TEAD
transcription factors mediate YAP-dependent transcriptional
activity (7), the present study
utilized a previously reported pUAS-Luc-TEAD-Gal4 system (12) to indirectly test YAP activity prior to
and following knockdown or overexpression of MCAM. It was
demonstrated that TEAD-controlled downstream luciferase activities
could be reduced significantly by MCAM-sh1 and -sh2, and could be
induced by ectopically-expressed MCAM (Fig. 1C), thus suggesting that MCAM also
induces YAP activity. In addition, it was observed that MCAM had a
similar effect on YAP mRNA as that on YAP protein (Fig. 1D), indicating that MCAM regulates YAP
expression primarily through its effect on YAP transcription.
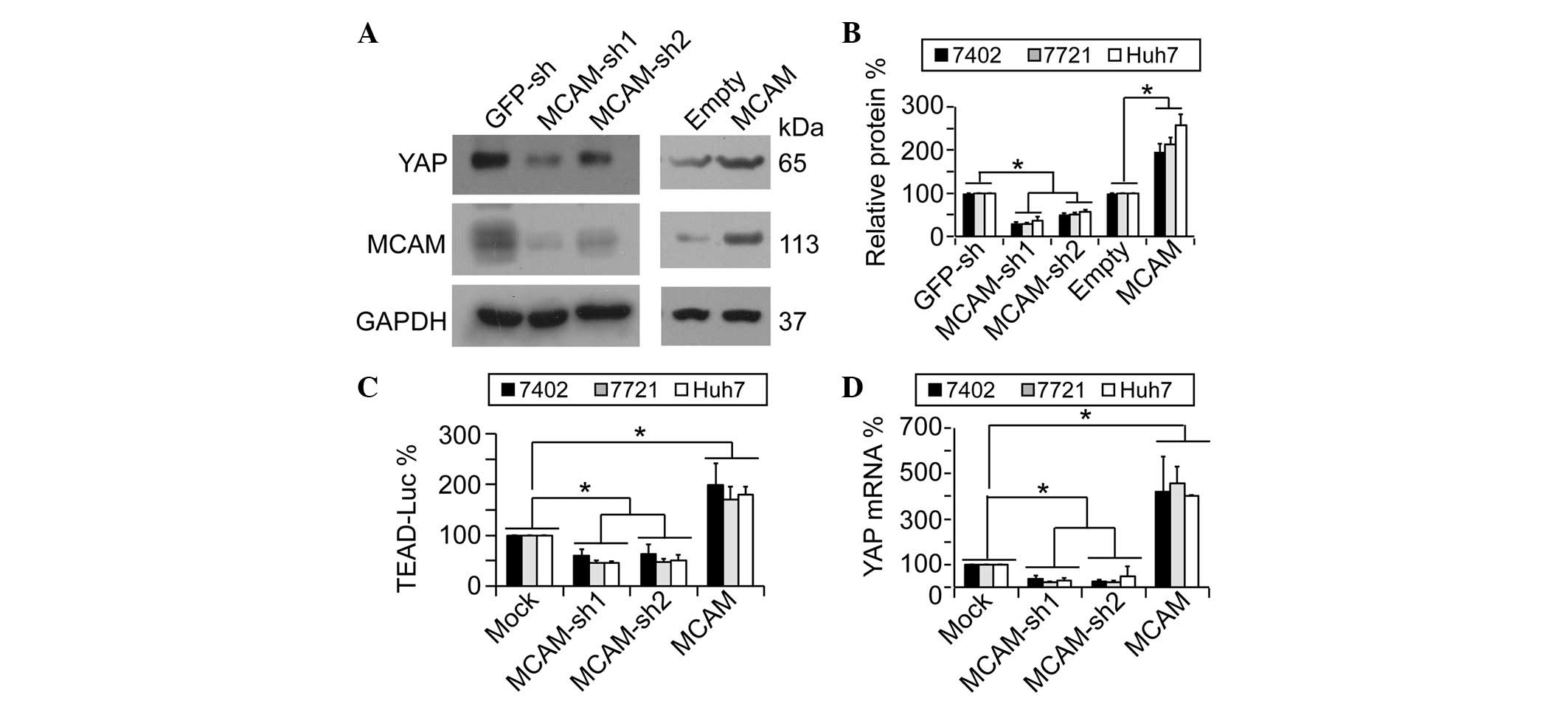 | Figure 1.YAP is positively regulated by MCAM.
(A and B) Representative western blotting images and quantification
of western blots of YAP and MCAM in human HCC Bel-7402 cells under
various treatments. The results demonstrated that MCAM regulated
YAP protein expression. (C) MCAM affected YAP activity. The
pUAS-Luc-TEAD-Gal4 luciferase reporter systems were transfected
into human HCC Bel-7402, SMMC-7721 or Huh7 cells with or without
MCAM-knockdown or overexpression. (D) MCAM regulated YAP mRNA
levels. mRNA levels of YAP were measured by quantitative polymerase
chain reaction in Bel-7402, SMMC-7721 or Huh7 cells with or without
MCAM-knockdown or overexpression. Data are presented as the mean ±
standard deviation of three independent experiments. Mock and
GFP-sh groups were arbitrarily set to 100%. *P<0.05. YAP,
yes-associated protein; MCAM, melanoma cell adhesion molecule;
GAPDH, glyceraldehyde 3-phosphate dehydrogenase; sh, small hairpin;
GFP, green fluorescent protein; TEAD, TEA domain; Luc, luciferase;
HCC, hepatocellular carcinoma; MOCK, control cells without
treatment. |
YAP is regulated by c-Jun/c-Fos
It was previously reported that c-Jun/c-Fos proteins
function as key downstream effectors of MCAM in the maintenance of
a transformative phenotype in HCC cells (10). Therefore, the present study
investigated whether MCAM regulates YAP through c-Jun/c-Fos. The
results demonstrated that the overexpression of c-Jun and c-Fos
resulted in a significant upregulation of YAP protein expression,
and these effects were synergized by a co-overexpression of c-Jun
and c-Fos (Fig. 2A and B; P<0.05).
Subsequently, whether c-Jun/c-Fos regulates YAP mRNA levels was
analyzes, and it was observed that YAP mRNA levels were also
upregulated by c-Jun/c-Fos (Fig. 2C).
By contrast, knocking down c-Jun and c-Fos led to a significant
downregulation of YAP protein (Fig. 2D
and E; P<0.05). Data from the pUAS-Luc-TEAD-Gal4 system
indicated that TEAD-controlled downstream luciferase activities may
be upregulated in a dose-dependent manner by transfection of
increasing concentrations of c-Jun/c-Fos, in addition to
demonstrating that the simultaneous effects of c-Jun and c-Fos were
synergic (Fig. 2F). Conversely,
TEAD-controlled downstream luciferase activities could be
downregulated by c-Jun/c-Fos-knockdown (Fig. 2F). Furthermore, immunofluorescence
assays demonstrated that YAP expression levels were increased in
Bel-7402 cells with a successful transfection of
c-Jun/c-Fos-expressing plasmids compared with untransfected control
cells (Fig. 2G). These results
suggest that the MCAM downstream effectors, c-Jun/c-Fos, may be
critical in the MCAM-induced upregulation of YAP.
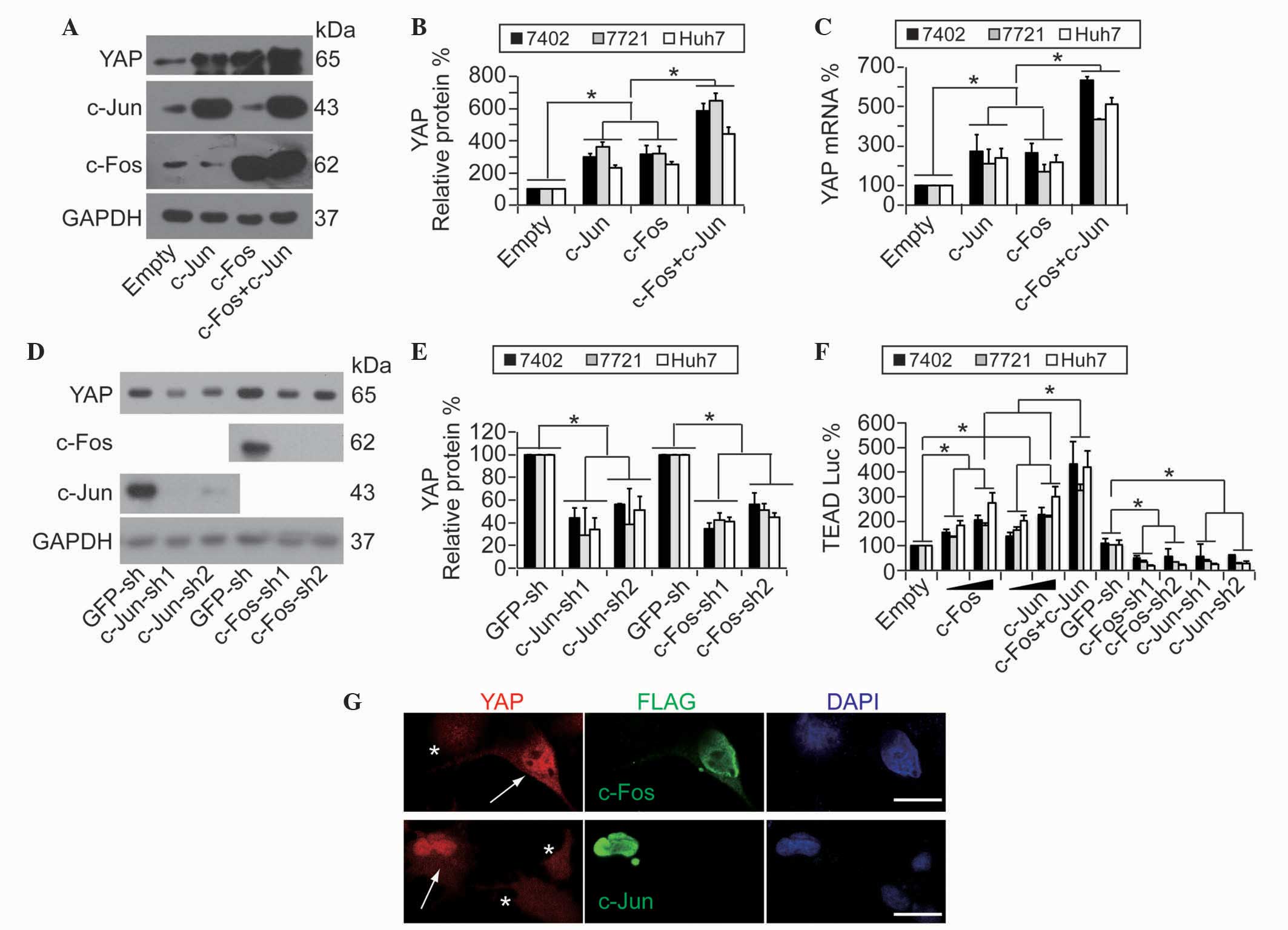 | Figure 2.Activity and expression of YAP is
regulated by c-Jun/c-Fos. (A and B) Overexpression of c-Jun/c-Fos
upregulated YAP protein expression as shown by (A) representative
western blotting images and (B) quantification of YAP, c-Jun and
c-Fos in human HCC Bel-7402 cells with or without c-Jun/c-Fos
overexpression. (C) c-Jun/c-Fos upregulated YAP mRNA levels. (D and
E) Knockdown of c-Jun/c-Fos reduced YAP protein expression as shown
by (D) representative western blotting images and (E)
quantification of YAP, c-Fos and c-Jun in Bel-7402 cells with or
without c-Jun/c-Fos-knockdown. (F) YAP activity was regulated by
c-Jun/c-Fos. The pUAS-Luc-TEAD-Gal4 luciferase reporter systems
were transfected into human HCC Bel-7402, SMMC-7721 or Huh7 cells
with or without c-Jun/c-Fos-knockdown or overexpression
(transfected with 0.5–1.5 µg plasmids). (G) Endogenous cellular YAP
expression was increased by overexpression of c-Jun/c-Fos, as
measured by immunofluorescence in Bel-7402 cells transfected with
exogenous c-Fos-FLAG or c-Jun-FLAG-expressing plasmids. Arrows
indicate cells with c-Jun/c-Fos transfection, while asterisks
indicate cells without successful transfection. Scale bar, 20 µm.
Data are presented as the mean ± standard deviation of three
independent experiments. Empty and GFP-sh groups were arbitrarily
set to 100%. *P<0.05. YAP, yes-associated protein; GAPDH,
glyceraldehyde 3-phosphate dehydrogenase; sh, small hairpin; GFP,
green fluorescent protein; TEAD, TEA domain; Luc, luciferase; DAPI,
4′,6-diamidino-2-phenylindole; HCC, hepatocellular carcinoma. |
YAP transcription is upregulated by
c-Jun/c-Fos via activation of CREB
The present study demonstrated that c-Jun/c-Fos
upregulated YAP by enhancing its transcription, and it was also
reported previously that CREB is critical in the transcription of
YAP (6). Therefore, the present study
hypothesized that c-Jun/c-Fos may upregulate the transcription of
YAP via CREB. To investigate this, CREB-dependent transcriptional
activities were analyzed using a luciferase reporter containing a
CREB binding site (CREB-reporter). It was demonstrated that
overexpression of either c-Jun or c-Fos resulted in upregulation of
CREB-dependent luciferase activities, and such effects could be
synergized by simultaneous overexpression of c-Jun and c-Fos
(Fig. 3A). By contrast,
c-Jun/c-Fos-knockdown resulted in significantly reduced
CREB-dependent luciferase activities (Fig. 3A; P<0.05). In a similar manner to
the CREB-reporter, c-Jun/c-Fos upregulated luciferase activities
from the reporter containing the core YAP promoter regions
(YAP-promoter) (Fig. 3A).
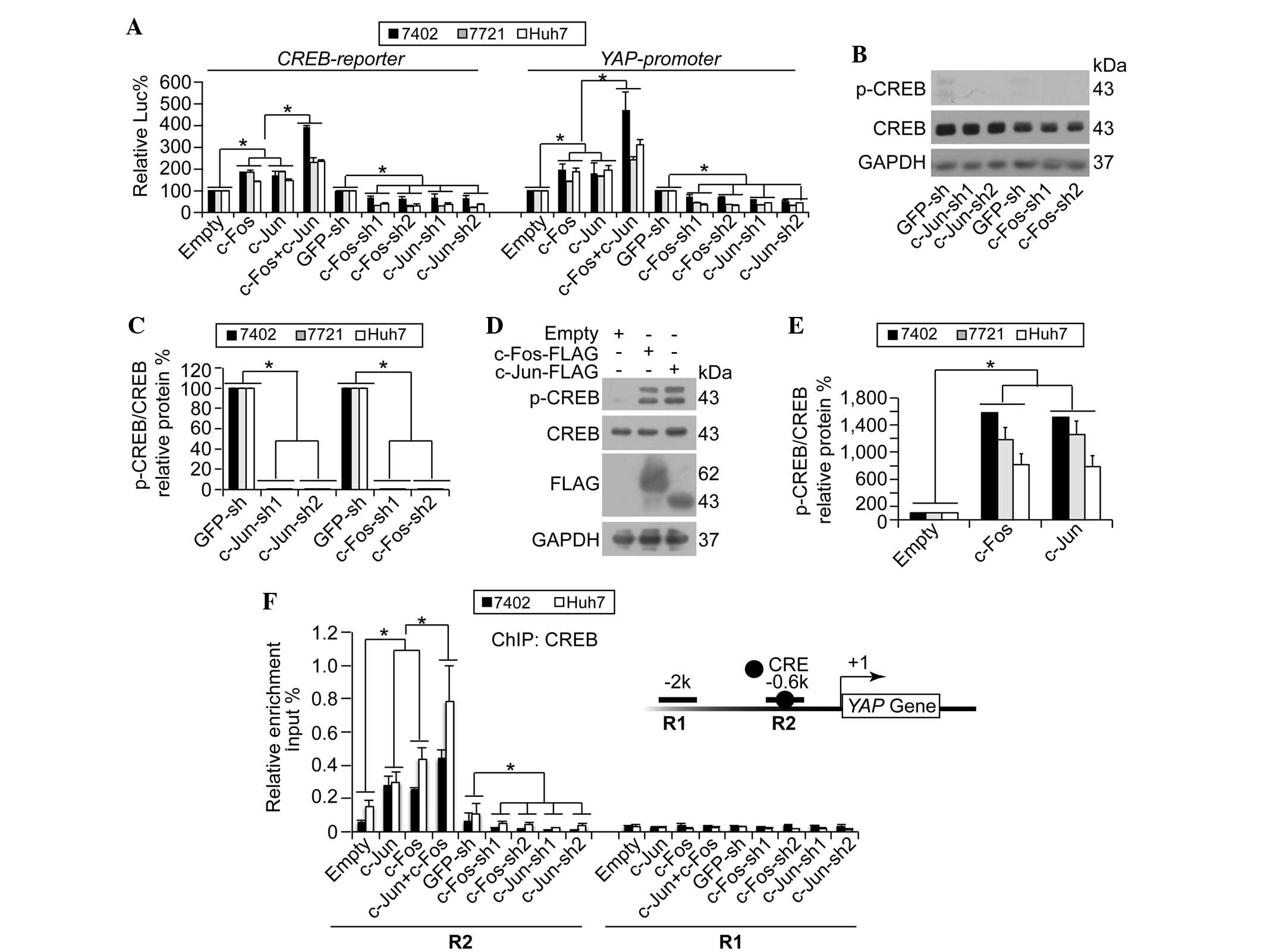 | Figure 3.YAP transcription is regulated by
c-Jun/c-Fos via CREB. (A) c-Jun/c-Fos upregulated CREB-dependent
transcription and YAP promoter activities. Luciferase
reporter plasmids, containing a CREB binding site (CREB-reporter)
or YAP promoter regions (YAP-promoter), were
co-transfected with pRL-TK-Renilla reporter plasmids into human
hepatocellular carcinoma Bel-7402, SMMC-7721 or Huh7 cells with
various treatments. (B and C) Knockdown of c-Jun/c-Fos reduced CREB
phosphorylation as shown by (B) representative western blotting
images and (C) quantification of p-CREB and CREB in Bel-7402 cells
with or without c-Jun/c-Fos-knockdown. Even with a long exposure,
p-CREB was hardly detected. (D and E) Overexpression of c-Jun/c-Fos
facilitated phosphorylation of CREB as shown by (D) representative
western blotting images and (E) quantification of p-CREB and CREB
in Bel-7402 cells with or without c-Jun/c-Fos overexpression. (F)
c-Jun/c-Fos stimulated CREB binding to the YAP gene promoter
region. ChIP was performed using anti-CREB antibodies in Bel-7402
and Huh7 cells under various treatments, using primer sets
encompassing R2 and R1 regions within the YAP promoter.
Insert shows YAP promoter region, with the black circle
indicating a CRE site. The relative enrichments of CREB were
normalized to the percentage of the input chromosomal DNA. Data are
presented as the mean ± standard deviation of three independent
experiments. Empty and GFP-sh groups were arbitrarily set to 100%.
*P<0.05. CREB, cAMP response element-binding protein; p-CREB,
phospho-CREB; YAP, yes-associated protein; Luc, luciferase; GAPDH,
glyceraldehyde 3-phosphate dehydrogenase; sh, small hairpin; GFP,
green fluorescent protein; ChIP, chromatin immunoprecipitation;
CRE, cAMP response element. |
As the phosphorylation of CREB at Ser-133 (p-CREB)
is a characteristic of CREB activation (14), the present study investigated whether
c-Jun/c-Fos affects the phosphorylation of CREB. The results
demonstrated that c-Jun/c-Fos-knockdown significantly downregulated
p-CREB (Fig. 3B and C; P<0.05),
whilst overexpression of c-Jun/c-Fos resulted in significantly
upregulated levels of p-CREB (Fig. 3D and
E; P<0.05). These results suggest that c-Jun/c-Fos
stimulates activation of CREB.
It was subsequently investigated whether c-Jun/c-Fos
could directly stimulate recruitment of CREB onto the YAP
promoter. It was observed that the co-overexpression of c-Jun and
c-Fos induced a significant enrichment of CREB onto the R2 region,
which contains a known CREB binding site, of the YAP
promoter compared with single overexpression of c-Jun and c-Fos
(Fig. 3F). However, no CREB
enrichment was detected at the R1 region (Fig. 3F), which is a CREB-unrelated region.
By contrast, c-Jun- and c-Fos-knockdown reduced recruitment of CREB
onto the YAP promoter at the R2 region, whereas this had no
effect on the R1 region (Fig. 3F).
These results indicate that c-Jun/c-Fos stimulates YAP
transcription through the induction of CREB recruitment onto the
YAP promoter.
Associations between MCAM, c-Jun/c-Fos
and YAP are critical in liver tumorigenesis
The present study analyzed whether the associations
between MCAM, c-Jun/c-Fos and YAP are important for the maintenance
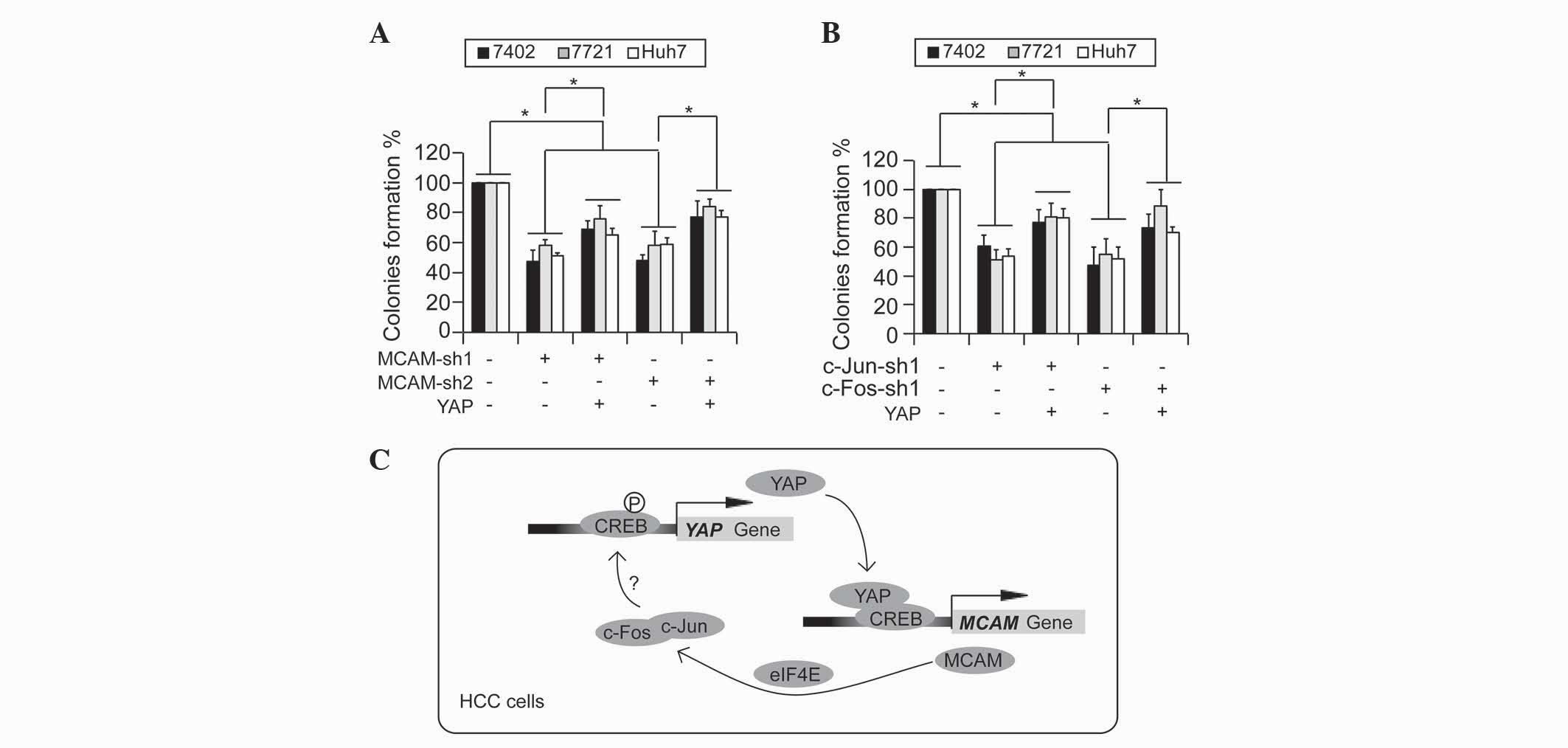
of a transformative phenotype in HCC cells. It was observed that
knockdown of MCAM significantly impaired the ability of the
Bel-7402, SMMC-7721 and Huh7 cells to form colonies in soft-agar,
which could be partially reversed by simultaneous overexpression of
YAP (Fig. 4A). Furthermore,
c-Jun/c-Fos-knockdown induced downregulation of colony formation,
which could also be partially rescued by overexpression of YAP
(Fig. 4B). These results indicate
that YAP is a potential downstream target of the MCAM-c-Jun/c-Fos
signaling cascade, and the associations between MCAM, c-Jun/c-Fos
and YAP may be critical during liver tumorigenesis.
Discussion
Recently, MCAM was identified as a direct target of
YAP in HCC cells (10). Additionally,
in the present study, it was demonstrated that YAP expression could
be conversely regulated by MCAM through its downstream effector,
c-Jun/c-Fos. c-Jun and c-Fos proteins are able to form
heterodimers, which are capable of promoting proliferation and
neoplastic transformation (15).
Notably, c-Jun/c-Fos dimers bind with high affinity to cAMP
response elements (CREs), which are CREB binding sites (15). Furthermore, CREB is able to
efficiently form a heterodimeric complex with c-Jun, and the
interaction between the two proteins increases the repertoire of
possible regulatory complexes that are important in the regulation
of transcription (16). Although no
previous study has yet established a direct interaction between
CREB and c-Fos, the present study observed that the synergic
effects of c-Fos and c-Jun resulted in the recruitment of CREB onto
the YAP promoter and the upregulation of YAP expression.
Therefore, the present study hypothesizes that c-Jun, c-Fos and
CREB may scaffold into a complex, and the close interactions
between these three proteins may be fundamental for the
transcriptional regulation of certain genes, including YAP, in the
promotion of tumorigenesis.
The present study hypothesizes that the critical
step during the regulation of YAP by MCAM is the phosphorylation of
CREB at Ser-133, which is facilitated by c-Jun/c-Fos. Ser-133 may
be phosphorylated by various kinases, including protein kinase B,
ribosomal protein S6 kinase, mitogen- and stress-activated protein
kinase and protein kinase C (17,18), with
its phosphorylation promoting the activation of CREB (19,20).
Despite no direct evidence to suggest that c-Jun/c-Fos proteins
possess a kinase function, c-Jun/c-Fos are able to be
phosphorylated by other kinases. It has been demonstrated that
constitutive activation of the extracellular-signal-regulated
kinase (ERK) signaling pathway activates c-Jun by phosphorylation,
and thus contributes to tumorigenesis by increasing cell
proliferation (21). Furthermore, ERK
signaling catalyzes the phosphorylation of c-Fos at Ser-374
(22,23). Notably, it was reported that ERK
phosphorylation is associated with increased phosphorylation of
CREB (24). As ERK functions as a
signal transmitter between the cytoplasm and nucleus, the
overexpression of membrane-bound MCAM in HCC may attract ERK to
translocate from the cytoplasm to CRE sites in the nucleus. With
the aid of c-Jun/c-Fos, ERK may add a phosphate group onto CREB and
consequently stimulate CREB-dependent YAP transcription. However,
further investigation is required to fully elucidate the
associations between MCAM, c-Jun/c-Fos and CREB.
In conclusion, the current study established a
complete, positive, auto-regulatory loop between MCAM and YAP.
Inhibition of this loop may serve as a possible target for the
treatment of HCC.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant no.
81301689), Yangfan Project of the Shanghai Committee of Science and
Technology (Shanghai, China; grant no. 14YF1412300), Young College
Teachers' Training Scheme of Shanghai (Shanghai, China; grant no.
ZZjdyx13007), Outstanding Youth Training Program of Tongji
University (Shanghai, China; grant no. 1501219080) and Shanghai
Tenth People's Hospital Climbing Training Program (Shanghai, China;
grant no. 04.01.13024).
References
|
1
|
Farazi PA and DePinho RA: Hepatocellular
carcinoma pathogenesis: From genes to environment. Nat Rev Cancer.
6:674–687. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 36:E359–E386. 2015.
View Article : Google Scholar
|
|
3
|
Zeng Q and Hong W: The emerging role of
the hippo pathway in cell contact inhibition, organ size control
and cancer development in mammals. Cancer Cell. 13:188–192. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dong J, Feldmann G, Huang J, Wu S, Zhang
N, Comerford SA, Gayyed MF, Anders RA, Maitra A and Pan D:
Elucidation of a universal size-control mechanism in
Drosophila and mammals. Cell. 130:1120–1133. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu MZ, Yao TJ, Lee NP, Ng IO, Chan YT,
Zender L, Lowe SW, Poon RT and Luk JM: Yes-associated protein is an
independent prognostic marker in hepatocellular carcinoma. Cancer.
115:4576–4585. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Ma L, Weng W, Qiao Y, Zhang Y, He
J, Wang H, Xiao W, Li L, Chu Q, et al: Mutual interaction between
YAP and CREB promotes tumorigenesis in liver cancer. Hepatology.
58:1011–1020. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu
J, Lin JD, Wang CY, Chinnaiyan AM, et al: TEAD mediates
YAP-dependent gene induction and growth control. Genes Dev.
22:1962–1971. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Urtasun R, Latasa MU, Demartis MI, Balzani
S, Goñi S, Garcia-Irigoyen O, Elizalde M, Azcona M, Pascale RM, Feo
F, et al: Connective tissue growth factor autocriny in human
hepatocellular carcinoma: Oncogenic role and regulation by
epidermal growth factor receptor/yes-associated protein-mediated
activation. Hepatology. 54:2149–2158. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Wolfe A, Septer S, Edwards G, Zhong
X, Abdulkarim AB, Ranganathan S and Apte U: Deregulation of Hippo
kinase signalling in human hepatic malignancies. Liver Int.
32:38–47. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang J, Tang X, Weng W, Qiao Y, Lin J, Liu
W, Liu R, Ma L, Yu W, Yu Y, et al: The membrane protein melanoma
cell adhesion molecule (MCAM) is a novel tumor marker that
stimulates tumorigenesis in hepatocellular carcinoma. Oncogene.
34:5781–5795. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wang J, Liu X, Wu H, Ni P, Gu Z, Qiao Y,
Chen N, Sun F and Fan Q: CREB up-regulates long non-coding RNA,
HULC expression through interaction with microRNA-372 in liver
cancer. Nucleic Acids Res. 38:5366–5383. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Park JS, Wei Y, Rajurkar M, Cotton
JL, Fan Q, Lewis BC, Ji H and Mao J: TRIB2 acts downstream of
Wnt/TCF in liver cancer cells to regulate YAP and C/EBPα function.
Mol Cell. 51:211–225. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Johannessen M, Delghandi MP and Moens U:
What turns CREB on? Cell Signal. 16:1211–1227. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
van Dam H and Castellazzi M: Distinct
roles of Jun: Fos and Jun: ATF dimers in oncogenesis. Oncogene.
20:2453–2464. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Benbrook DM and Jones NC: Heterodimer
formation between CREB and JUN proteins. Oncogene. 5:295–302.
1990.PubMed/NCBI
|
|
17
|
Mayr B and Montminy M: Transcriptional
regulation by the phosphorylation-dependent factor CREB. Nat Rev
Mol Cell Biol. 2:599–609. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Böhm M, Moellmann G, Cheng E,
Alvarez-Franco M, Wagner S, Sassone-Corsi P and Halaban R:
Identification of p90RSK as the probable CREB-Ser133 kinase in
human melanocytes. Cell Growth Differ. 6:291–302. 1995.PubMed/NCBI
|
|
19
|
Chrivia JC, Kwok RP, Lamb N, Hagiwara M,
Montminy MR and Goodman RH: Phosphorylated CREB binds specifically
to the nuclear protein CBP. Nature. 365:855–859. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kundu TK, Palhan VB, Wang Z, An W, Cole PA
and Roeder RG: Activator-dependent transcription from chromatin in
vitro involving targeted histone acetylation by p300. Mol Cell.
6:551–561. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Smalley KS: A pivotal role for ERK in the
oncogenic behaviour of malignant melanoma? Int J Cancer.
104:527–532. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okazaki K and Sagata N: The Mos/MAP kinase
pathway stabilizes c-Fos by phosphorylation and augments its
transforming activity in NIH 3T3 cells. EMBO J. 14:5048–5059.
1995.PubMed/NCBI
|
|
23
|
Murphy LO, Smith S, Chen RH, Fingar DC and
Blenis J: Molecular interpretation of ERK signal duration by
immediate early gene products. Nat Cell Biol. 4:556–564.
2002.PubMed/NCBI
|
|
24
|
Mao L, Yang L, Tang Q, Samdani S, Zhang G
and Wang JQ: The scaffold protein Homer1b/c links metabotropic
glutamate receptor 5 to extracellular signal-regulated protein
kinase cascades in neurons. J Neurosci. 25:2741–2752. 2005.
View Article : Google Scholar : PubMed/NCBI
|