Introduction
Xeroderma pigmentosum (XP) is a sun-toxicity
disease. A total of 8 subtypes (from XP-A to XP-G and XP-V) of this
disease have been identified by their different pathogenic genes
(1,2).
The pathogenic mechanisms of almost all these subtypes result from
a defect in nucleotide excision repair (3), except XP-V subtype, which results from a
translesion synthesis (TLS) defect (4). XP-V is a common subtype (21%) in XP
disease, and has a similar phenotype to other subtypes (2), including sun sensitivity, photophobia,
early onset of freckling, and subsequent neoplastic changes in
sun-exposed skin (5,6). The majority of studies have demonstrated
that XP-V disease is a result of mutations in the POLH gene
(encoding DNA polymerase η). Polymerase η is the main DNA
polymerase responsible for TLS, and its defect could apparently
reduce TLS efficiency and increase mismatch in DNA replication.
These phenomena result in genomic instability, leading to a high
incidence of tumors in patients (7–15). It has
been previously demonstrated that polymerase η has defective
expression in XP-V cells and that certain other polymerases
involving TLS are unusually expressed, such as polymerase κ and ζ
(encoded by POLK and REV3 l, respectively) (16). An additional polymerase, polymerase θ
(encoded by POLQ), also has low expression in XP-V cells and
tumor tissue and has the same function as polymerase η, which is to
generate A/T mutations during the somatic hypermutation of
immunoglobulin (Ig) genes (16,17). Given
that a number of polymerases change their expression in XP-V cells
and tumor tissue, certain factors may co-regulate the expression of
these polymerases.
MicroRNAs (miRNAs) are endogenous, small non-coding
RNAs that regulate translation and degradation of mRNAs at the
post-transcriptional level (18).
Protein expression from hundreds of genes are directly suppressed,
albeit relatively mildly, by a single miRNA (19). Dysregulated miRNAs are correlated with
various cancers and may function as tumor suppressors or oncogenes,
depending on the function of their targets and cellular context
(20). Therefore, certain miRNAs with
unusual expression may explain the changes in expression of these
polymerases in XP-V tumor that accelerate DNA mismatch.
Previous studies have mainly verified POLH
mutation as an etiological factor of developing XP-V tumors
(7–9,14). In the
present study, polymerase-suppressive miRNAs associated with XP-V
tumor were identified by analyzing miRNAs that may directly
regulate DNA polymerases with unusual expression in XP-V tumor
cells. miR-20b-5p was identified to be a polymerase suppressor by
directly targeting POLK and POLQ.
Materials and methods
Prediction of miRNA as co-suppressor
of POLK, REV3 l, and POLQ
POLK, REV3 l and POLQ all
demonstrate low expression in XP-V tumor cells (16). Accordingly, Targetscan (http://www.targetscan.org), miRDB (http://mirdb.org/miRDB), and miRanda (http://www.microrna.org) were used to predict miRNA
co-targeting these three genes.
Cell culture
All cells including XP-V tumor fibroblast cell
lines, human skin fibroblasts (HSFs), and HeLa cells were cultured
in DMEM supplemented with 20% FBS (HyClone, Logan, UT, USA). HeLa
cells and HSFs were purchased from the cell bank of the Chinese
Academy Of Sciences (Beijing, China). XP-V tumor fibroblast cell
lines (XP30RO, XP1CH, and XP1SF) were purchased from The Coriell
Institute (Camden, NJ, USA). Cells were incubated at 37°C in 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) for candidate miRNAs in XP-V
cells
QIAgen miScript miRNA PCR Arrays kit (QIAgen Inc.,
Hilden, Germany) was used to extract, reverse transcribe and
amplify total miRNAs in XP-V cell lines and HSFs according to the
manufacturer's protocol. U6 was used as an endogenous control to
normalize the amount of total miRNA in each sample. ABI 7500
Real-time PCR System (Applied Biosystems, Carlsbad, CA, USA) was
used to analyze the data. Primers were synthesized by GenePharma
(Shanghai, China) and the sequences are presented in Table I. To identify differences in miRNA
expression, samples of HSF cells were defined as reference samples,
and the quantity of all tested miRNAs in the reference sample was
defined as ‘1.0.’ Student's t-test was used to compare relative
expression levels between XP-V cell lines and HSF control
cells.
 | Table I.Primers sequences. |
Table I.
Primers sequences.
| Primer name | Primer sequence,
5′-3′ |
|---|
| Primers for
quantifying miRNA |
|
|
miR-520b |
AAGTGCTTCCTTTTAGAGGGA |
|
miR-520e |
GGTGCTTCCTTTTTGAGGG |
|
miR-302a-3p |
TGCTTCCATGTTTTGGTGA |
|
miR-302b-3p |
GCGTGCTTCCATGTTTTAGTA |
|
miR-302c-3p |
TGCTTCCATGTTTCAGTGG |
|
miR-302d-3p |
AGTGCTTCCATGTTTGAGTGT |
|
miR-93-5p |
GTGCTGTTCGTGCAGGTAG |
|
miR-373-3p |
GCTTCGATTTTGGGGTGT |
|
miR-548k |
AAAGTACTTGCGGATTTTGCT |
|
miR-20a-5p |
CGTCAGGCCTAAAGTGCTTAT |
|
miR-20b-5p |
CAAAGTGCTCATAGTGCAGGTAG |
|
miR-106a-5p |
AGTCAGGCCAAAGTGCTTAC |
|
miR-106b-5p |
GTAAAGTGCTGACAGTGCAGA |
| Primers for
mutagenesis |
|
| Forward
Primer for mutagenesis in POLK UTR |
TTAAGCTAACTACTATTAAGCTGTCTTCTTTCACAAATATTAATATTTCACCTGATAGAAATGTAACTAAGATACATAATGTGTTTTAATACACAT |
| Reverse
Primer for mutagenesis in POLK UTR |
ATGTGTATTAAAACACATTATGTATCTTAGTTACATTTCTATCAGGTGAAATATTAATATTTGTGAAAGAAGACAGCTTAATAGTAGTTAGCTTAA |
| Forward
Primer for mutagenesis in POLQ UTR |
CATGGTTTACCCAGACAGATGTGGAACCTTTCACCTAAGTGCATATTTCAAGCATCTGTTCT |
| Reverse
Primer for mutagenesis in POLQ UTR |
AGAACAGATGCTTGAAATATGCACTTAGGTGAAAGGTTCCACATCTGTCTGGGTAAACCATG |
Transfection
HeLa cells were transfected with 200 nM candidate
miRNA, miR-NC mimics, or miRNA inhibitor (GenePharma, Shanghai,
China) using Turbofect transfection reagent (Thermo Fisher
Scientific, Waltham, MA, USA) when cells reached 70–80%
confluence.
Western blot analysis
All cells were harvested using RIPA lysis buffer
(Beyotime, Shanghai, China). Then, 1% PMSF (Bioprimacy Co., Ltd.,
Wuhan, China) was added directly prior to use. Protein
concentration was measured using BCA protein assay (Thermo Fisher
Scientific, Inc.). Protein was loaded onto 10% SDS-PAGE gel
(Beyotime, Shanghai, China) and then transferred to PVDF membrane
(Bio-Rad Laboratories, Inc., Hercules, CA, USA). The blot was
blocked with 5% skim milk for 2 h and then probed with primary
mouse monoclonal polymerase κ (dilution, 1:6,000; catalog no.,
ab57070), rabbit polyclonal polymerase θ (dilution, 1:3,000;
catalog no., ab80906), and mouse monoclonal β-actin (dilution,
1:8,000; catalog no., ab8226) antibodies (Abcam, Cambridge, UK).
After incubation at 4°C overnight, the blot was washed with TBST
and incubated in secondary goat anti-mouse IgG-horseradish
peroxidase antibody (dilution, 1:10,000; catalog no., sc-2005;
Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) and goat
anti-rabbit IgG-horseradish peroxidase antibody (dilution,
1:10,000; catalog no., sc-2004; Santa Cruz Biotechnology, Inc.) for
1 h at 25°C. The signal was developed with ECL reagent (Advansta,
Inc., Menlo Park, CA, USA).
Luciferase reporter assay
The 3′-untranslated regions (UTRs) of POLK and POLQ
were amplified using PCR from human genomic DNA and then ligated
into pMIR-report (Ambion, Thermo Fisher Scientific, Inc.). Then,
QuikChange Lightning site-directed mutagenesis kit (Stratagene
Agilent Technologies, Santa Clara, CA, USA) was used to induce
miR-20b-5p target sequences (complementary to the seed region for
miR-20b-5p) to mutate TACTTT to GTGAAA in POLK and CACTTT to GTGAAA
in POLQ. All constructs were confirmed by sequencing. Primers used
are summarized in Table I. HeLa cells
were co-transfected with wild-type or mutant 3′UTR luciferase
reporter construct, the Renilla luciferase construct pRL-TK, and
either miR-20b-5p or miR-NC mimics. Then, 48 h after transfection,
luciferase activities were measured using the Dual Luciferase
Reporter Assay System (Promega Corporation, Madison, WI, USA) and
normalized by dividing the firefly luciferase activity with Renilla
luciferase activity.
Statistical analyses
Values are expressed as mean ± standard deviation
(SD) from triplicate experiments. Student's t-test was used to
compare relative expression levels. Statistical analyses were
performed by SPSS software, version 16.0 (SPSS Inc., Chicago, IL,
USA). P<0.05 was considered to indicate a statistically
significant difference.
Results
Using three web software predictions, it was found
that no miRNA was predicted to target all three genes. Certain
miRNAs were predicted to co-target POLQ and POLK but
not REV3 L (Fig. 1). To find
miRNAs co-regulating polymerases in XP-V tumor cells, only miRNAs
that matched both POLK and POLQ from more than two
software prediction results were selected. All other miRNAs were
predicted to match only one of three genes, which were removed from
the subsequent analysis. miR-520b, miR-520e, miR-302a, miR-302b,
miR-302c, miR-302d, miR-93, miR-373, miR-548k, miR-20a, miR-20b,
miR-106a, and miR-106b were chosen as candidate miRNAs.
The RT-qPCR results demonstrated that only miR-20a,
miR-20b, miR-106a, miR-106b, and miR-548k were expressed at
significantly different levels between XP-V cell lines and HSFs
(Fig. 2).
The western blot analysis results verified that
polymerase κ and θ were expressed at lower levels in XP-V tumor
cell lines compared to the normal control cell line. Furthermore,
when the above five miRNAs were transfected into HeLa cells, only
miR-20b transfection resulted in reduced polymerase κ and θ levels
(Fig. 3).
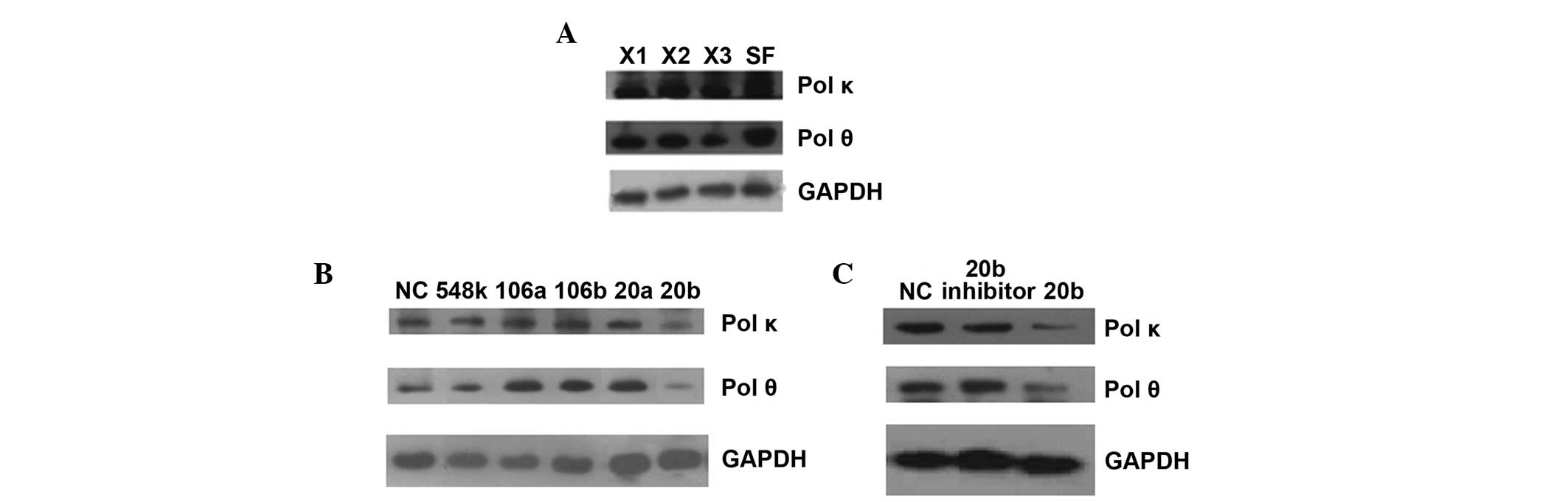 | Figure 3.Western blot results of endogenous Pol
κ and θ protein in different cell lines. (A) Compared with HSF
cells, three XP-V cell lines had lower expression of Pol κ and θ.
X1, XP30RO; X2, XP1CH; X3, XP1SF. (B) When HeLa cells were
transfected with candidate miRNA mimics, only miR-20b-5p in
candidate miRNAs could decrease Pol κ and θ expression. NC,
miR-negative control; 548k, miR-548k; 106a, miR-106a-5p; 106b,
miR-106b-5p; 20a, miR-20a-5p; 20b, miR-20b-5p. (C) Verification of
miR-20b inhibition for Pol κ and θ expression in HeLa cells
transfected by miR-20b-5p mimics. 20b inhibitor, the inhibitor of
miR-20b-5p. GAPDH was used as endogenous control to normalize each
sample. pol κ and θ, polymerase κ and θ; HSF, human skin
fibroblasts. |
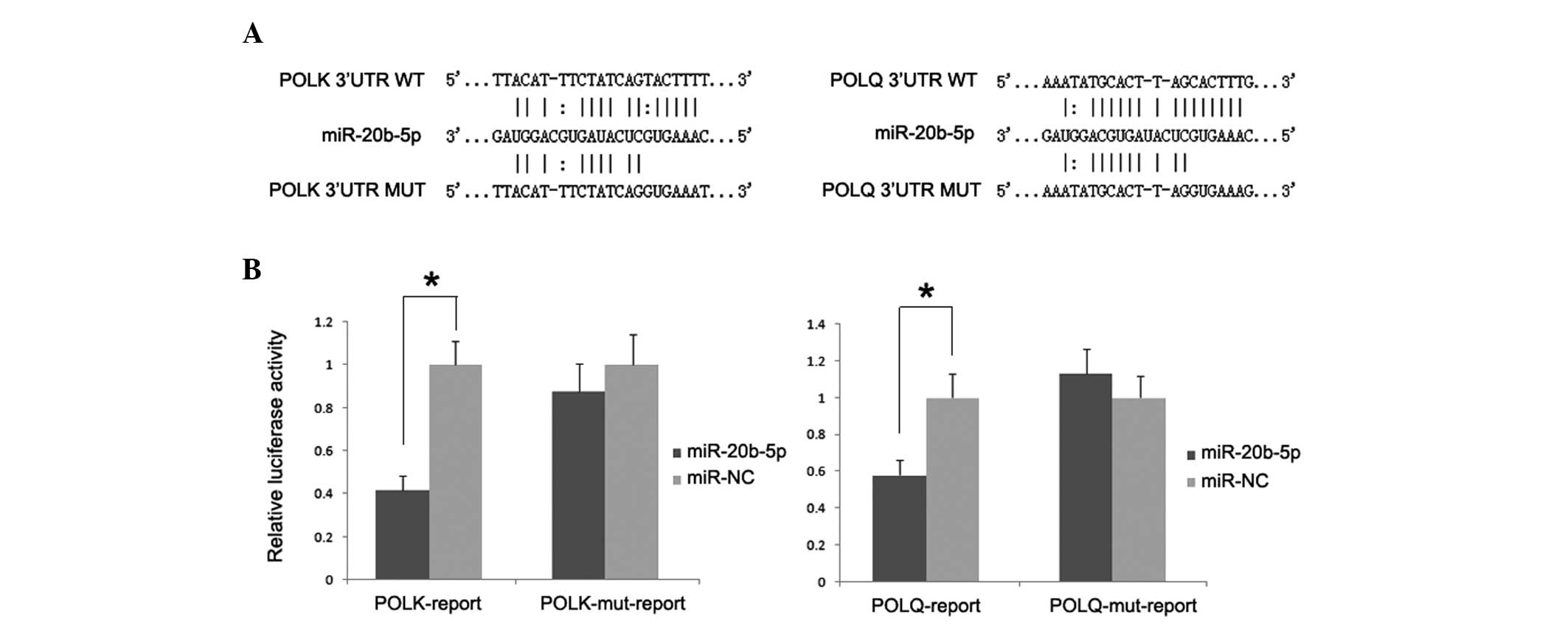
To determine whether such an inhibitory effect on
translation was mediated by specific and direct interaction of
miR-20b-5p with POLK and POLQ target site, luciferase
reporter plasmids containing 3′UTR of both genes were constructed.
The dual-luciferase assay demonstrated that the introduction of
miR-20b-5p significantly reduced luciferase activity with respect
to miR-NC, whereas such inhibitory effect was absent in cells
transfected with reporter plasmids containing the mutant 3′UTR of
both genes (Fig. 4).
Discussion
Low expression levels of polymerases in XP-V cells
such as polymerase η, κ, and ζ may lead to a significant reduction
in the accuracy of TLS in XP-V cells (21). Polymerase θ has also been indicated to
serve a role in base excision repair, and lower expression of
polymerase θ may also seem unfavorable for DNA replication repair
(22). In XP-V tumor cells,
polymerases ζ, κ, and θ are indeed expressed at low levels, in
addition to the dysfunction of polymerase η that disrupts DNA
lesion replication and promotes genetic instability (16,21). In
the present study, no miRNA was predicted to co-regulate
POLK and REV3 L expression, although these two genes
both belonged to the Y-family of DNA polymerases (23). However, miR-20-5p was verified to
function as co-suppressor of POLK and POLQ depending
on its targets. The high expression of miR-20-5p in XP-V tumor
cells could obviously decrease the expression of POLK and
POLQ. Moreover, in XP-V tumor cells, these two polymerases
with low expression may explain abnormal DNA replication repair
apart from polymerase η dysfunction (21). Therefore, miR-20-5p may also serve an
important role in XP-V tumors, accelerating DNA instability by
down-regulating POLK and POLQ.
In summary, the current study demonstrated
miR-20b-5p may co-regulate POLK and POLQ.
Furthermore, miRNA may also be a novel factor that affect
error-prone DNA replication in XP-V tumor cells.
Acknowledgements
The study was supported by grants from the National
Natural Science Foundation of China (grant nos. 81400492 and
31400839).
References
|
1
|
Kraemer KH, Lee MM and Scotto J: Xeroderma
pigmentosum. Cutaneous, ocular and neurologic abnormalities in 830
published cases. Arch Dermatol. 123:241–250. 1987. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kraemer KH and DiGiovanna JJ; Pagon RA,
Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJH, Bird TD,
Fong CT, Mefford HC, Smith RJH and Stephens K: Xeroderma
Pigmentosum. University of Washington. 1993–2015. 2003.
|
|
3
|
Berneburg M and Lehmann AR: Xeroderma
pigmentosum and related disorders: Defects in DNA repair and
transcription. Adv Genet. 43:71–102. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cleaver JE: Xeroderma pigmentosum:
Variants with normal DNA repair and normal sensitivity to
ultraviolet light. J Invest Dermatol. 58:124–128. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Maher VM, Ouellette LM, Curren RD and
McCormick JJ: Frequency of ultraviolet light-induced mutations is
higher in xeroderma pigmentosum variant cells than in normal human
cells. Nature. 261:593–595. 1976. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Myhr BC, Turnbull D and DiPaolo JA:
Ultraviolet mutagenesis of normal and xeroderma pigmentosum variant
human fibroblasts. Mutat Res. 62:341–353. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Masutani C, Kusumoto R, Yamada A, Dohmae
N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K and Hanaoka F: The
XPV (xeroderma pigmentosum variant) gene encodes human DNA
polymerase eta. Nature. 399:700–704. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Johnson RE, Kondratick CM, Prakash S and
Prakash L: hRAD30 mutations in the variant form of xeroderma
pigmentosum. Science. 285:263–265. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Inui H, Oh KS, Nadem C, Ueda T, Khan SG,
Metin A, Gozukara E, Emmert S, Slor H, Busch DB, et al: Xeroderma
pigmentosum-variant patients from America, Europe and Asia. J
Invest Dermatol. 128:2055–2068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gratchev A, Strein P, Utikal J and Sergij
G: Molecular genetics of Xeroderma pigmentosum variant. Exp
Dermatol. 12:529–536. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Broughton BC, Cordonnier A, Kleijer WJ,
Jaspers NG, Fawcett H, Raams A, Garritsen VH, Stary A, Avril MF,
Boudsocq F, et al: Molecular analysis of mutations in DNA
polymerase eta in xeroderma pigmentosum-variant patients. Proc Natl
Acad Sci USA. 99:815–820. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kannouche P, Broughton BC, Volker M,
Hanaoka F, Mullenders LH and Lehmann AR: Domain structure,
localization and function of DNA polymerase eta, defective in
xeroderma pigmentosum variant cells. Genes Dev. 15:158–172. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yuasa M, Masutani C, Eki T and Hanaoka F:
Genomic structure, chromosomal localization and identification of
mutations in the xeroderma pigmentosum variant (XPV) gene.
Oncogene. 19:4721–4728. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tanioka M, Masaki T, Ono R, Nagano T,
Otoshi-Honda E, Matsumura Y, Takigawa M, Inui H, Miyachi Y,
Moriwaki S and Nishigori C: Molecular analysis of DNA polymerase
eta gene in Japanese patients diagnosed as xeroderma pigmentosum
variant type. J Invest Dermatol. 127:1745–1751. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Johnson RE, Prakash S and Prakash L:
Efficient bypass of a thymine-thymine dimer by yeast DNA
polymerase, Poleta. Science. 283:1001–1004. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Guo J, Zhou G, Zhang W, Song Y and Bian Z:
A novel mutation causes XP-V disease and XP-V tumor proneness may
involve imbalance of numerous DNA polymerases. Oncol Lett.
6:1583–1590. 2013.PubMed/NCBI
|
|
17
|
Masuda K, Ouchida R, Hikida M, Kurosaki T,
Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F and O-Wang J: DNA
polymerases eta and theta function in the same genetic pathway to
generate mutations at A/T during somatic hypermutation of Ig genes.
J Biol Chem. 282:17387–17394. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Baek D, Villén J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Esquela-Kerscher A and Slack FJ:
Oncomirs-microRNAs with a role in cancer. Nat Rev Cancer.
6:259–269. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ziv O, Geacintov N, Nakajima S, Yasui A
and Livneh Z: DNA polymerase zeta cooperates with polymerases kappa
and iota in translesion DNA synthesis across pyrimidine photodimers
in cells from XPV patients. Proc Natl Acad Sci USA.
106:11552–11557. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshimura M, Kohzaki M, Nakamura J,
Asagoshi K, Sonoda E, Hou E, Prasad R, Wilson SH, Tano K, Yasui A,
et al: Vertebrate POLQ and POLbeta cooperate in base excision
repair of oxidative DNA damage. Mol Cell. 24:115–125. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ohmori H, Friedberg EC, Fuchs RP, Goodman
MF, Hanaoka F, Hinkle D, Kunkel TA, Lawrence CW, Livneh Z, Nohmi T,
et al: The Y-family of DNA polymerases. Mol Cell. 8:7–8. 2001.
View Article : Google Scholar : PubMed/NCBI
|