Introduction
Breast cancer is a progressive disease and the most
common neoplasm affecting women of all ages. Similar to all
progressive diseases, early and reliable diagnosis is the key to
successful treatment. Benign epithelial breast disease represents a
growing percentage of the pathological characteristics of this
disease, which include numerous benign entities such cysts,
fibrosis, adenosis and duct ectasia, which require neither surgery
nor follow-up (1,2). Lesions such as as papillomas, sclerosing
adenosis, lobular intraepithelial neoplasia, flat epithelial
atypia, radial scars and atypical hyperplasias, which are all
considered pre-malignant lesions, are signs of an increased risk of
breast cancer (3–7).
It has been demonstrated that human malignant tumors
of the breast have an elevated expression of the Harvey Ras
oncogene when compared to their respective normal tissue samples
(7). In a previous study, the authors
performed a comparative analysis of Harvey Ras oncogene expression
with conventional clinicopathological parameters of breast cancer
(8). In another study, an
immunohistochemical analysis of Ras oncogene expression in human
breast lesions was performed (9). A
high expression of p21 Ras oncogene in breast cancer patients is
considered to have important clinical significance (10).
In breast cancer, the increased expression and/or
activation of Ras are often associated with tumor aggressiveness
(10,11). Available and emerging
immunohistochemical and molecular studies have improved the
classification of breast cancer, as well as the prognostic and
predictive information regarding breast cancer pathology; thus, a
new tumor classification has been proposed (12–17). In
this new classification, the basal-like and the triple-negative
types of breast cancer have a likelihood of distant recurrence and
mortality compared with other types, and have a tend to affect
younger women. For both subtypes, a need for effective biological
markers has been reported in certain studies; thus, Ras expression
may prove to be an effective prognotic marker (13,17).
The complexity of breast cancer pathology remains a
challenge for the scientific community; therefore, protein
biomarkers are needed as indicators of pathological, physiological,
or pharmacological processes. Among these, estrogen receptor (ER)
and Her2/neu are biomarkers that have been approved for the
prognosis, diagnosis and treatment of this disease. Since breast
cancer is a heterogeneous disease, some breast cancer cells lose
their ability to express ERα among other proteins, resulting in a
disease which is therapy-resistant. To identify human breast cancer
biomarkers between ERα-positive and -negative breast cancer,
tissues need to be micro-dissected and protein expression can then
be identified to be compared with either normal ductal epithelium
or ductal epithelium containing ductal carcinoma in situ
lesions. Ras and Her2/neu protein expression have been previously
considered as biomarkers, since they are highly expressed in human
breast cancer (11).
Breast cancer is a heterogeneous complex disease, a
spectrum of many subtypes with distinct biological features, and
this leads to differences in response patterns to various treatment
modalities and clinical outcomes. In this context, the analysis of
breast cancer using immunohistochemical markers remains an
essential component of routine pathological examinations, and plays
an important role in the management of the disease as regards
diagnostic, prognostic and therapeutic strategies (18–22). Ras
family members (H-Ras, K-Ras, N-Ras and M-Ras) are small GTPases
which are activated indirectly by external stimuli. Since the
cloning of HRas, the first human oncogene, the Ras/MPK pathway has
been a preferential subject of cancer research (23,24), and
it is known as an important pathway in the initiation and
progression of cancer. Therefore, the aim of the present study was
to evaluate Ras expression by immunohistochemical analysis in
breast cell lines, as well as in normal, benign and malignant
breast sample biopsies, in order to identify a marker that may be
used as a prognostic tool for breast cancer patients.
Materials and methods
Breast cancer cell lines
An in vitro experimental breast cancer model
(Alpha model), previously developed by our group by exposing the
immortalized human breast epithelial cell line, MCF-10F, to low
doses of high linear energy transfer (LET) α particles radiation
(150 keV/µm) and subsequent growth in the presence or absence of
17β-estradiol (25), was used in this
study. This model consisted of human breast epithelial cells in
different stages of transformation: i) a control cell line,
MCF-10F; ii) a malignant and tumorigenic cell line termed Alpha5
(60 cGy plus estrogen/60 cGy plus estrogen); and iii) Tumor2 cells
derived from cells originating from a tumor following the injection
of the Alpha5 cell line into nude mice. The cells were grown in
DMEM/F-12 (1:1) medium supplemented with antibiotics [100 U/mI
penicillin, 100 µg/ml streptomycin, 2.5 µg/ml amphotericin B (all
from Life Technologies, Grand Island, NY, USA)], 10 µg/m 5% equine
serum (Biofluids, Rockville, MD, USA), 0.5 µg/ml hydrocortisone
(Sigma-Aldrich, St. Louis, MO, USA) and 0.02 µg/ml epidermal growth
factor (Collaborative Research Inc., Bedford, MA, USA).
Protein expression by immunoperoxidase
staining
Exponentially growing cell line cells were plated on
a glass chamber slide (Nunc Inc., Naperville, IL, USA), at a
density of 1×104 cells/ml of medium and allowed to grow
for 2–3 days until 70% confluent. The cells were fixed with
buffered paraformaldehyde at room temperature, incubated with 1%
H2O2 in methanol to block endogenous
peroxidase and again washed twice with buffer solution.
Subsequently, the cell cultures were then covered with normal horse
serum for 30 min at room temperature and incubated with either
anti-mouse or anti-goat monoclonal or polyclonal antibodies: ERα
(mouse, sc-8002), Neu (rabbit, sc-284) and H-Ras (mouse, sc-29)
(all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA) at
1:500 dilution overnight at 4°C and then incubated for 45 min with
diluted biotinylated secondary antibody solution (Vector
Laboratories Inc., Burlingame, CA, USA) and Vectastin Elite ABC
reagent (Vector Laboratories Inc.) was used. The experiments were
repeated twice in cells with identical passages in
vitro.
Breast samples
All samples were obtained from archives entrusted by
Professor P. Maldague and Professor A. Trouet from the School of
Medicine, Saint-Luc Hospital, IMAG Unit (IREC), University of
Louvain, Brussels, Belgium. This study was approved by the Ethics
Committee of the University of Louvain and was conducted in
accordance with institutional guidelines. The study consisted of
the analysis of a total of 40 samples from the archives (all
patients had provided with written informed consent prior to
obtaining the samples). Normal tissues were obtained from 10
patients admitted for reduction mammoplasty with a family history
for cancer; 15 samples had benign breast lesions and 15 specimens
had breast cancer. All samples were diagnosed and classified
according to the World Health Organization classification (26).
Immunohistochemistry
Processing of the breast samples was performed
following a routine pathological examination. All samples
investigated were tested for mouse H-Ras monoclonal antibody (Santa
Cruz Biotechnology, Inc.). Immunolocalization was performed using a
streptavidin-biotin immunoperoxidase method according to the
protocol of the laboratory. Briefly, 5 µm-thick sections obtained
after formalin fixation and paraffin-embedding were deparaffinyzed
in xylene and rehydrated with Tris-buffered saline (TBS). The
sections were then treated with citrate buffer (pH 6.0) and
hydrogen peroxide was used to quench endogenous peroxidase
activity. The slides were then subjected to the primary antibody
solution and placed in a moist chamber overnight at 4°C. After
washing in TBS, a biotinylated link antibody was applied, followed
by washing and the addition of streptavidin peroxidase. The
localization of the antibody was visualized using
3,3′-diaminobenzidine tetrahydrochloride (DAB) and counterstaining
with Mayer's hematoxylin (Sigma-Aldrich).
Results
The established breast cancer model (25) was shown to exhibit several
characteristics of breast carcinogenesis. The normal cell line,
MCF-10F, did not exhibit any of the features that characterize
malignant cells, such as anchorage-independent growth in soft agar,
invasion and tumor growth in nude mice. The Alpha5 cell line
induced the development of mammary gland tumors in animals after
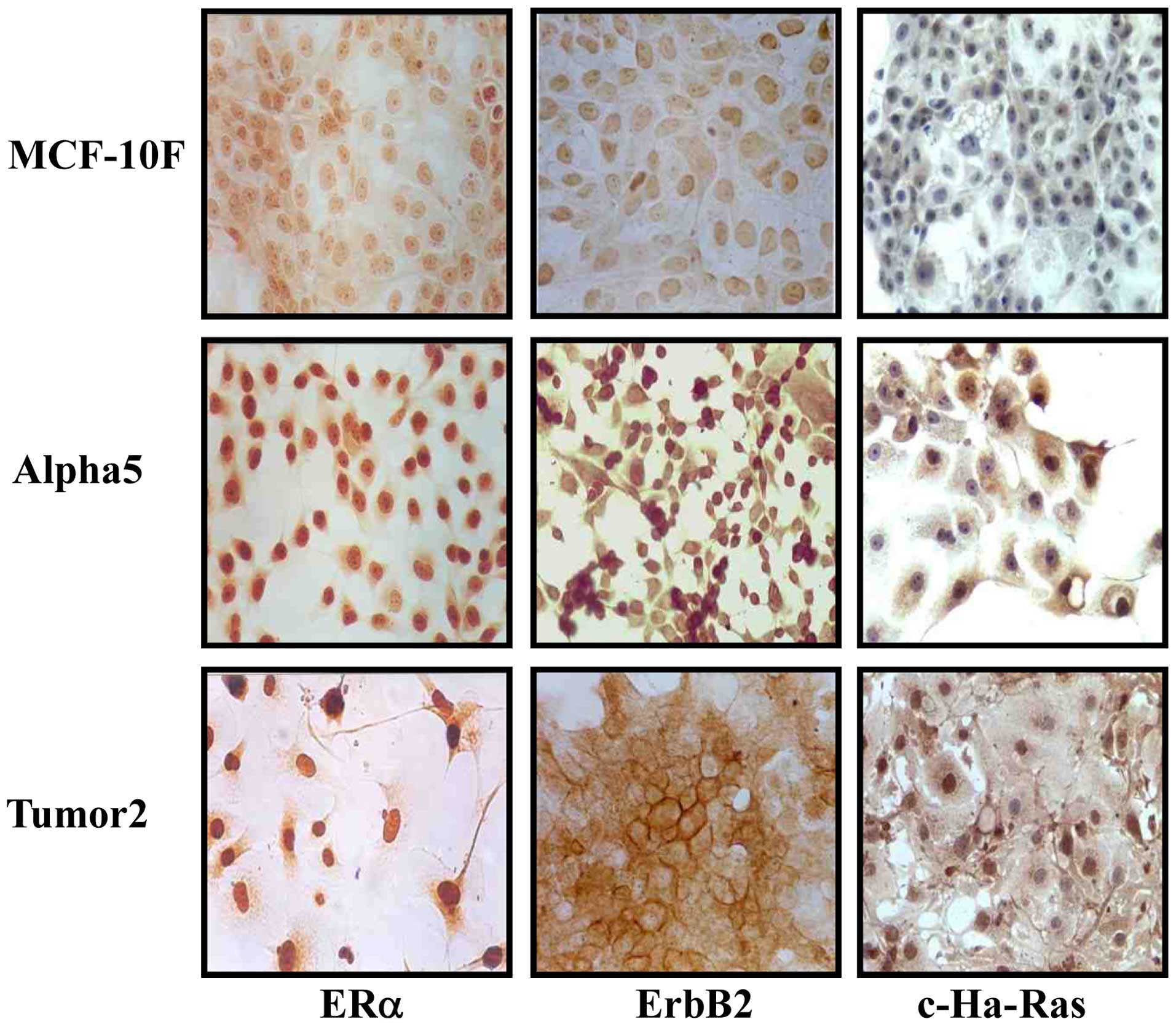
injection and gave rise to the Tumor2 cell line (25). Fig. 1
shows the results of immunoperoxidase staining obtained to detect
ERα, ErbB2 and c-Ha-Ras protein expression in the MCF-10F, Alpha5
and Tumor2 cell lines. The results indicated a positive expression
in the Alpha5 and Tumor2 cell lines for all 3 markers. However, a
negative expression was observed in the MCF-10F cell line. The
Tumor2 malignant cells derived from the mice were positive for ERα,
ErbB2 and c-Ha-Ras protein expression, as with the original cell
line, Alpha5.
An analysis of breast specimens obtained from the
tissue archives of patients with a family history of breast cancer
was also carried out. A routine pathological examination revealed
marked fibrosis, a variety of cysts with or without apocrine
metaplasia, some microcalcifications, some low duct hyperplasias
and sporadic epithelial flat lesions in both groups. Our findings
support the concept of the heterogeneity of expression in
epithelial lesions and the value of immunohistochemical analysis in
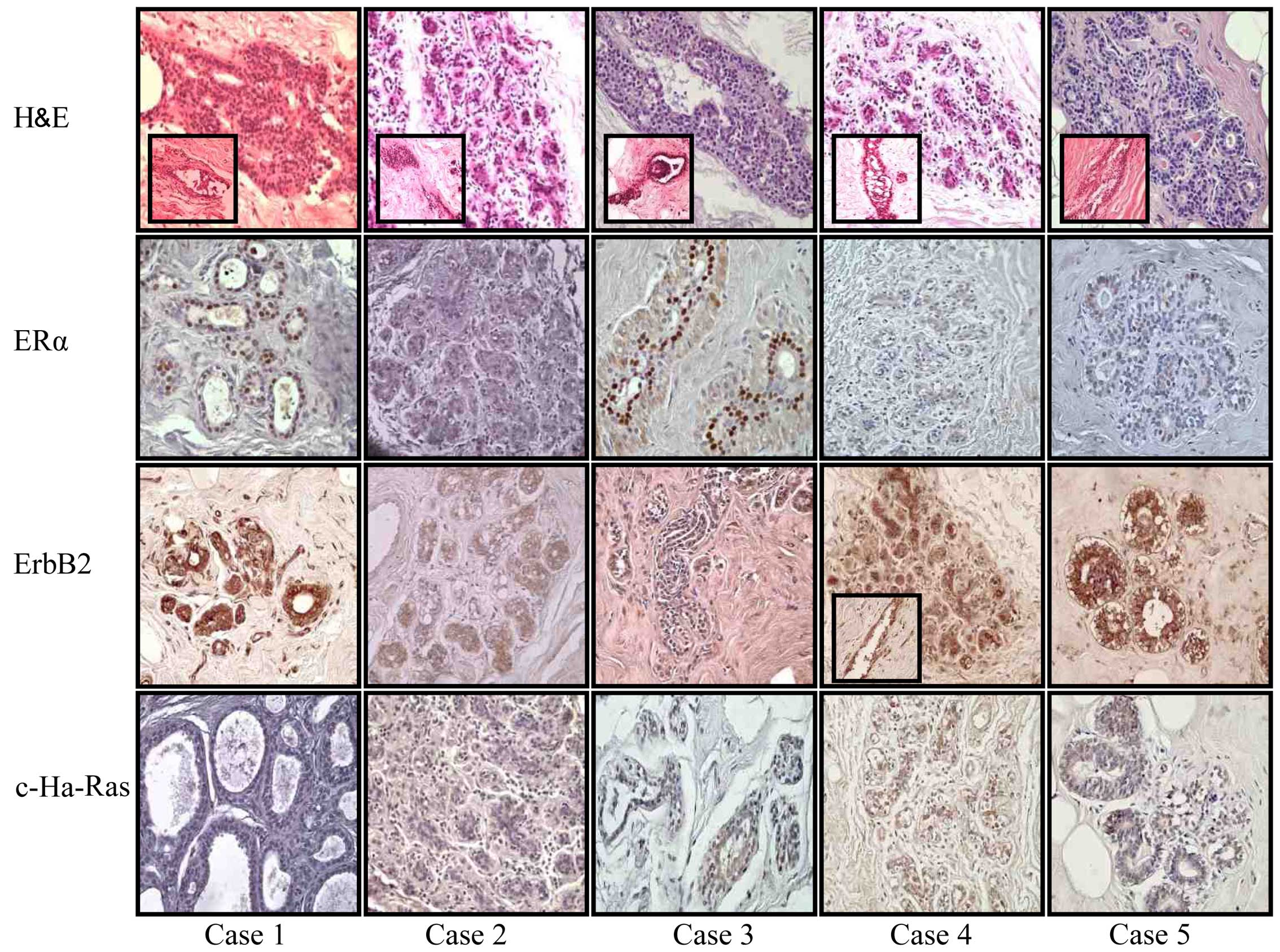
histopathological diagnosis. Representative images are shown in
Figs. 2 and 3. The results indicated that case 1 (without
a family history) was a low ductal hyperplasia. Proliferative ducts
originating from small structures were observed (see inset). Ducts
and ductules were positive for ERα and ErbB2; however, a negative
c-Ha-Ras expression was observed in ducts and enlarged ductules.
Case 2 (without a family history) exhibited lobules that were
negative for ERα, ErbB2 and c-Ha-Ras. Case 3 (with a family
history) exhibited ducts and ductules positive for ERα and ErbB2;
however, the ductules were negative for c-Ha-Ras. Case 4 (with a
family history) exhibited lobules negative for ERα, but positive
for ErbB2 and c-Ha-Ras. Case 5 (with a family history) also
exhibited lobules negative for ERα and c-Ha-Ras, but positive for
ErbB2. Representative images of benign lesions are illustrated in
Fig. 2. Insets (when available for
each image) show other structures also present in that case, as
ducts and ductules.
As shown in Fig. 3,
the results indicated that case 6 (with a family history) was a
lobular hyperplasia where the lobules were negative for ERα, but
positive for ErbB2 and c-Ha-Ras. Case 7 (with a family history)
also had lobules negative for ERα, but positive for ErbB2 and
c-Ha-Ras. Case 8 (with a family history) corresponded to ductal
carcinoma where the cells were positive for ERα, ErbB2 and
c-Ha-Ras. Case 9 (with a family history) exhibited structures
positive for ERα, ErbB2 and c-Ha-Ras. Case 10 (with a family
history) exhibited hyperplastic foci with structures negative for
ERα, ErbB2 and c-Ha-Ras. Representative images of breast lesions
are shown in Fig. 3. The results
revealed normal or benign structures, apart from case 3; however,
the report stated that those patients had malignant lesions. On the
other hand, there was no association with the markers used
clinically, as ER and ErbB2 protein expression were positive in
benign, as well as in malignant breast lesions with or without
hereditary predisposition. The present study revealed that the
percentage of cells positive for ERα was low in normal mammary
glands and non-proliferative benign breast disease or displasias,
but increased in certain malignant lesions.
Representative images of H-Ras protein expression in
normal (Fig. 4A) benign (Fig. 4B) and malignant (Fig. 4C) breast lesions are shown. The
expression of H-Ras was negative in the normal breast samples, but
weakly positive and heterogenous in ductules and ducts. In the
benign breast samples, the cyst epithelium was usually positive,
with some differential patterns. In most cases, it was distributed
in the cytoplasm and occasionally with membrane localization. The
apocrine metaplasia foci exhibited a stronger expression, with
different patterns between the samples analyzed. A significant
increase in immunostaining was observed in the atypical ductal
hyperplastic lesions, associated with epithelial ductal
proliferation. The cancer specimens revealed a more variable
staining pattern. The expression of H-Ras was almost invariably
stronger in the cancer specimens than in the benign epithelium from
the same patients. Some heterogeneity of expression was observed,
with weakly positive or negative cancer cells adjacent to strongly
positive cells. Metastatic cancer cells within the stroma (clusters
and isolated cells) exhibited a variable H-Ras expression. Only the
tumor budding (clumps) directly associated with carcinoma foci were
more homogeneous, with similar patterns of tumor foci. The
expression of H-Ras was similar in lobular and ductal
carcinomas.
Discussion
As with all progressive diseases, the early and
reliable diagnosis is key to successful treatment. In our analyses,
a previously established breast cancer model (25) was used. The normal cell line, MCF-10F,
exhibited a negative protein expression of ER, ErbB2 and c-Ha-Ras,
whereas the malignant Alpha5 and Tumor2 cell line exhibited a
positive expression ER, ErbB2 and c-Ha-Ras. This model of
progression indicated that malignancy can influence the positivity
of these markers, as was also observed in our biopsy specimens.
Immunohistochemistry plays is an important diagnostic adjunct to
morphological examination in the diagnosis of benign and malignant
lesions (2,7,17,20). Markers to help predict the risk of
progression, and to ultimately provide non-surgical treatment
options to those at lower risk would be of great benefit. At
present, there are no applied molecular markers available to
predict the risk of carcinoma in situ progression to
invasive cancer, and therefore, all women diagnosed with such a
malignancy must undergo surgery.
Our results revealed positive c-Ha-Ras protein
expression only in the malignant lesions in comparison to the
benign lesions. The high c-Ha-Ras expression in intraductal
proliferative lesions is very relevant. These epithelial lesions
have a risk of progression to invasive breast cancer and related
issues, particularly atypical ductal hyperplasia (4). However, the use of c-Ha-Ras expression
as a diagnostic and prognostic marker should be selective and
should preferably be used in conjunction with other markers.
According to published data, the major value of c-Ha-Ras expression
is in its clinical correlation with an improved prognosis of
relapse (11,24).
In a previous study, the expression of
transformation phenotypes was examined in the human breast
epithelial cell line, MCF-10F, with the chemical carcinogens,
7,12-dimethylbenz[a]anthracene, N-methyl-N-nitrosourea,
N-methyl-N-nitro-N'-nitrosoguanidine and benzo[a]pyrene (27). This was done to clarify whether
chemically induced neoplastic transformation correlates with
alterations in the Ras gene. MCF-10F cells have two c-Ha-Ras
alleles, identified by 1.0- and 1.2-kb restriction fragments.
Treatment with carcinogens resulted in the loss of one of the
alleles (1.0 kb). Polymerase chain reaction-amplified DNA from all
carcinogen-treated cells was analyzed for point mutations in
c-Ha-Ras at codons 12 and 61. All of the carcinogens induced a
mutation of the remaining allele at the first position of codon 12
(GGC→AGC). Another frequent mutation occurred at the first position
of codon 61 (CAG→GAG). The changes in c-Ha-Ras were associated with
the emergence of colony formation in agar-methocel, but no specific
changes in this gene correlated with the emergence of invasiveness
or tumorigenesis, indicating that other genes may be involved in
the process.
It has been concluded that c-Ha-Ras induces an
additive effect on the expression of tumorigenesis in the human
breast epithelial cell line, MCF-10F, treated with chemical
carcinogens. This has provided a model for analyzing the role of
c-Ha-Ras in human breast cancer (28). Data have also highlighted the
importance of chromosome 11 in the radiation-induced malignant
transformation of human breast epithelial cells and suggest the
usefulness of the model in uncovering specific derangements during
breast cancer progression (29).
Among the loci of chromosome 11, the locus 11p15.5, where the
c-Ha-Ras oncogene is located, had an incidence of allelic imbalance
between 25 and 40%. Furthermore, direct sequencing analysis of
codons 12 and 61 of the c-Ha-Ras oncogene identified various point
mutations (29). On the other hand,
studies have shown that the c-Ha-Ras oncogene is upregulated by the
effect of malathion alone and by the combination of estrogen with
either malathion or parathion (30).
This suggests that pesticides and estrogens affect human breast
cells, inducing molecular changes indicative of transformation
(30).
In this study, Ras protein expression was examined
in samples of normal and hyperplasias, as well as malignant breast
cancer lesions with hereditary predisposition in conjunction with
other markers regularly used in clinical practice. Since Ras is
often mutated in human cancers, much effort has been devoted to
devising means of controling the activity of Ras (23). Breast tumors have been shown to have
an elevated expression of the Harvey Ras oncogene when compared to
their respective normal tissue sections (7). The Harvey Ras oncogene has also been
shown to have a significant correlation with the
clinicopathological characteristics of breast cancer (8). Immunohistochemical analyses of Ras
oncogene expression in human breast lesions have been carried out
(9), as well as of the p21 Ras
oncogene (10,21,22), with
a high expression in breast cancer patients indicating clinical
significance. It can thus be concluded that the oncogene, c-Ha-Ras,
is a good marker to be used in breast cancer in patients and to
predict response to therapy in such patients. The evaluation of Ras
expression by immunohistochemistry in cell lines in an experimental
breast cancer model of transformed cells by environmental
substances and estrogen, as well as in normal, benign and malignant
breast biopsies from patients, provided evidence that it can be
used as good a prognostic tool for breast cancer patients.
Acknowledgements
The technical support of Guiliana Rojas, Georgina
Vargas and secretarial assistance and suggestions of Leodán A.
Crispin and Richard Ponce-Cusi are greatly appreciated. This study
was supported by Grant support FONDECYT no. 1120006 (GMC).
References
|
1
|
Azzopardi JG, Ahmed A and Millis RR:
Problems in breast pathology. Major Probl Pathol. 11:i–xvi, 1–466.
1979.PubMed/NCBI
|
|
2
|
Dupont WD and Page DL: Risk factors for
breast cancer in women with proliferative breast disease. N Engl J
Med. 312:146–151. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Tsuchiya A, Kanno M, Nomizu T, Hatakeyama
Y, Kimijima I and Abe R: Clinical characteristics of breast cancer
patients with family history. Fukushima J Med Sci. 44:35–41.
1998.PubMed/NCBI
|
|
4
|
De Mascarel I and MacGrogan G: Management
of breast epithelial atypia. Ann Pathol. 27:182–194. 2007.(In
French). View Article : Google Scholar : PubMed/NCBI
|
|
5
|
de Mascarel I, Debled M, Brouste V,
Mauriac L, Sierankowski G, Velasco V, Croce S, Chibon F, Boudeau J,
Debant A, et al: Comprehensive prognostic analysis in breast cancer
integrating clinical, tumoral, micro-environmental and
immunohistochemical criteria. Springerplus. 4:5282015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Walker RA, Hanby A, Pinder SE, Thomas J
and Ellis IO: National Coordinating Committee for Breast Pathology
Research Subgroup: Current issues in diagnostic breast pathology. J
Clin Pathol. 65:771–785. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Spandidos DA and Agnantis NJ: Human
malignant tumours of the breast, as compared to their respective
normal tissue, have elevated expression of the Harvey ras oncogene.
Anticancer Res. 4:269–272. 1984.PubMed/NCBI
|
|
8
|
Agnantis NJ, Parissi P, Anagnostakis D and
Spandidos DA: Comparative study of Harvey-ras oncogene expression
with conventional clinicopathologic parameters of breast cancer.
Oncology. 43:36–39. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Agnantis NJ, Petraki C, Markoulatos P and
Spandidos DA: Immunohistochemical study of the ras oncogene
expression in human breast lesions. Anticancer Res. 6:1157–1160.
1986.PubMed/NCBI
|
|
10
|
Efremidis AP, Agnantis NJ, Patra F,
Papadopoulou C and Spandidos DA: Clinical significance of elevated
p21 ras oncogene expression in breast cancer patients. Cancer J.
2:288–291. 1989.
|
|
11
|
Spandidos DA, Yiagnisis M, Papadimitriou K
and Field JK: Ras, c-myc, and c-erbB-2 oncoprotein expression in
human breast carcinomas. Anticancer Res. 9:1385–1394.
1989.PubMed/NCBI
|
|
12
|
Gruver AM, Portier BP and Tubbs RR:
Molecular pathology of breast cancer: The journey from traditional
practice toward embracing the complexity of a molecular
classification. Arch Pathol Lab Med. 135:544–557. 2011.PubMed/NCBI
|
|
13
|
Park S, Koo JS, Kim MS, Park HS, Lee JS,
Lee JS, Kim SI and Park BW: Characteristics and outcomes according
to molecular subtypes of breast cancer as classified by a panel of
four biomarkers using immunohistochemistry. Breast. 21:50–57. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lerwill MF: Current practical applications
of diagnostic immunohistochemistry in breast pathology. Am J Surg
Pathol. 28:1076–1091. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Abdel-Fatah TM, Powe DG, Hodi Z,
Reis-Filho JS, Lee AH and Ellis IO: Morphologic and molecular
evolutionary pathways of low nuclear grade invasive breast cancers
and their putative precursor lesions: Further evidence to support
the concept of low nuclear grade breast neoplasia family. Am J Surg
Pathol. 32:513–523. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moriya T, Kanomata N, Kozuka Y, Fukumoto
M, Iwachido N, Hata S, Takahashi Y, Miura H, Ishida K and Watanabe
M: Usefulness of immunohistochemistry for differential diagnosis
between benign and malignant breast lesions. Breast Cancer.
16:173–178. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dewar R, Fadare O, Gilmore H and Gown AM:
Best practices in diagnostic immunohistochemistry: Myoepithelial
markers in breast pathology. Arch Pathol Lab Med. 135:422–429.
2011.PubMed/NCBI
|
|
19
|
Leong AS and Zhuang Z: The changing role
of pathology in breast cancer diagnosis and treatment.
Pathobiology: Journal of Immunopathology. Mol Cell Biol. 78:99–114.
2011.
|
|
20
|
Omi Y, Yamamoto T, Okamoto T, Obara T and
Kobayashi M: A useful immunohistochemical approach to evaluate
intraductal proliferative lesions of the breast and to predict
their prognosis. Histol Histopathol. 26:79–86. 2011.PubMed/NCBI
|
|
21
|
Candlish W, Kerr IB and Simpson HW:
Immunocytochemical demonstration and significance of p21 ras family
oncogene product in benign and malignant breast disease. J Pathol.
150:163–167. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Going JJ, Anderson TJ and Wyllie AH: Ras
p21 in breast tissue: Associations with pathology and cellular
localisation. Br J Cancer. 65:45–50. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Malumbres M and Barbacid M: RAS oncogenes:
The first 30 years. Nat Rev Cancer. 3:459–465. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Giltnane JM and Balko JM: Rationale for
targeting the Ras/MAPK pathway in triple-negative breast cancer.
Discov Med. 17:275–283. 2014.PubMed/NCBI
|
|
25
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lakhani SR, Ellis IO, Schnitt SJ, Tan PH
and van de Vijver MJ: WHO classification of tumors of the breast.
International Agency for Research on Cancer. Lyon: 2012.
|
|
27
|
Zhang PL, Calaf G and Russo J: Allele loss
and point mutation in codons 12 and 61 of the c-Ha-ras oncogene in
carcinogen-transformed human breast epithelial cells. Mol Carcinog.
9:46–56. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Calaf G, Zhang P, Alvarado M, Estrada S
and Russo J: C-ha-ras enhances the neoplastic transformation of
human breast epithelial-cells treated with chemical carcinogens.
Int J Oncol. 6:5–11. 1995.PubMed/NCBI
|
|
29
|
Roy D, Calaf G and Hei TK: Allelic
imbalance at 11p15.5–15.4 correlated with c-Ha-ras mutation during
radiation-induced neoplastic transformation of human breast
epithelial cells. Int J Cancer. 103:730–737. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Calaf GM and Roy D: Cancer genes induced
by malathion and parathion in the presence of estrogen in breast
cells. Int J Mol Med. 21:261–268. 2008.PubMed/NCBI
|