Introduction
Breast cancer is one of the most common malignant
tumors worldwide (1). At present,
chemotherapy remains an important method for the treatment of
breast cancer. However, chemoresistance has become a non-negligible
factor for hindering the prognosis of patients (2). Treatments that increase the
chemosensitivity of breast cancer, and therefore increase the
patient survival rate, are urgently required.
Flap endonuclease GEN homolog 1 (GEN1) was first
proposed by Furukawa et al (3)
and belongs to a new class of the Rad2/xeroderma pigmentosum
complementation group G (XPG) nuclease family, class IV (4). Previous studies have demonstrated that
GEN1 is able to resolve a particular structure termed Holliday
junctions (HJs), which are formed during DNA strand exchange, as a
central intermediate in the process of homologous recombination
(5–7).
Numerous studies have hypothesized that resolving HJs properly is
the key to correct DNA repair (8,9). DNA
damaging drugs, such as 5-fluorouracil (5-FU), injure tumor cells
by damaging the DNA of the cells. During this process, numerous HJs
are produced. Identifying HJ resolvases in tumor cells is essential
for an improved understanding to tumor self-repair (10). At present, certain enzymes have been
identified, including Bloom syndrome, RecQ helicase-like/slow
growth suppressor 1, MUS81 structure-specific endonuclease subunit
(Mus81)-Mms4/essential meiotic structure-specific endonuclease 1,
Rad1-Rad10 and structure-specific endonuclease subunit
SLX1-structure-specific endonuclease subunit SLX4 (Slx4) (5,11). Certain
evidence has indicated that the ability of tumor cells to process
HJ determined the sensitivity of the cells to DNA-damaging drugs
(12–14). It has been confirmed that subsequent
to suppressing the HJ resolvase Slx4, the sensitivity of tumor
cells to DNA damaging agents increased significantly (12). Previous studies also confirmed that
targeting Mus81 increases sensitivity to DNA damaging drugs, such
as 5-FU, in breast cancer cells (13). However, it remains unknown whether
GEN1 affects the chemosensitivity of tumor cells, such as Slx4 and
Mus81. It has been shown that GEN1 interference increases the
pharmaceutical sensitivity of yeast and Drosophila alone or
in combination with other genes (11,15).
Therefore, the present study aimed to explore the effect of GEN1
interference on the chemosensitivity of breast cancer MCF-7 and
SKBR3 cell lines.
Materials and methods
Reagents
The breast MCF-7 and SKBR3 cell lines were purchased
from the Shanghai Cell Bank of Chinese Academy of Sciences
(Shanghai, China). HyClone Minimum essential medium (MEM) and
Roswell Park Memorial Institute-1640 medium (RPMI-1640) were
purchased from GE Healthcare Life Sciences (Logan, UT, USA). Gibco
fetal bovine serum (FBS), penicillin and streptomycin were
purchased from Thermo Fisher Scientific (Waltham, MA, USA). 5-FU
was purchased from Hangzhou Bioer Technology Co., Ltd. (Hangzhou,
China). Lipofectamine 2000 and TRIzol reagent were purchased from
Thermo Fisher Scientific. The short hairpin (sh)RNA interference
plasmid and the plasmid containing shRNA without RNA interference
were purchased from Shanghai Genechem Co., Ltd. (Shanghai, China).
The Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide
(PI) kit and the Cell Counting kit-8 (CCK-8) were purchased from
Nanjing KeyGen Biotech Co., Ltd. (Nanjing, China) for cell
apoptosis detection and cell viability assays. For reverse
transcription-quantitative polymerase chain reaction (RT-qPCR),
First-Strand cDNA Synthesis kit and 2X Taq PCR Mix were purchased
from Biomiga (San Diego, CA, USA). The LightCycler 480 PCR
apparatus was purchased from Hoffmann-La Roche (Basel,
Switzerland). In addition, for western blot analysis, primary
antibodies against GEN1 (dilution, 1:75), Mus81 (dilution, 1:1,000)
and β-actin (dilution, 1:5,000) were purchased from Biorbyt
(Cambridge, UK), GeneTex (Irvine, CA, USA) and Abcam (Cambridge,
UK), respectively. The secondary antibody against rabbit
immunoglobulin (Ig)G conjugated to horseradish peroxidase
(dilution, 1:3,000) was purchased from Hangzhou HuaAn Biotechnology
Co., Ltd. (Hangzhou, China). The ECL-Plus chemiluminescence
detection kit was purchased from Beyotime Institute of
Biotechnology (Haimen, China), and the PowerPac HC TRANS-BLOT
equipment was purchased from Bio-Rad Laboratories (Hercules, CA,
USA). The CKX41 inverted fluorescence microscope was purchased from
Olympus Corporation (Tokyo, Japan).
Cell culture
The MCF-7 cells were cultured in MEM and SKBR3 cells
were cultured in RPMI-1640. MEM and RPMI-1640 were each
supplemented with 10% FBS, 100 U/ml penicillin and 100 mg/ml
streptomycin. The cells were cultured at 37°C in a 5%
CO2 atmosphere.
shRNA transfection
The MCF-7 and SKBR3 cells were grown in 6-well
plates at a density of 1×106 cells per well in MEM and
RPMI-1640 with 10% FBS, but without antibiotics, as antibiotics
would cause cell damage in the transfection process. When the cells
had reached 70–80% confluency in the antibiotic-free medium, the
medium was changed to MEM and RPMI-1640 without FBS or antibiotics,
to prepare for transfection. The expression of GEN1 and Mus81 was
knocked-down by transfection with an shRNA interference plasmid
directed against GEN1 and Mus81, with the addition of 4 µg plasmid
per well. The plasmid containing shRNA without RNA interference was
used as a negative control. Cells were transfected in serum-free
and antibiotic-free MEM and RPMI-1640 medium for 6 h. The medium
was then changed to complete medium (MEM and RPMI-1640 with 10% FBS
and 1% antibiotic) to cultivate continuously. Transfection was
performed using Lipofectamine 2000, according to the manufacturer's
protocol. Interference was determined by RT-qPCR and western
blotting. The shRNA sequences were as follows: GEN1 shRNA
(sh-GEN1),
5′-CCGGGCAAATTAAAGCTGTCAGTAACTCGAGTTACTGACAGCTTTAATTTGCTTTTTG-3′;
Mus81 shRNA (sh-Mus81),
5′-CCGGGAGTTGGTACTGGATCACATTCTCGAGAATGTGATCCAGTACCAACTCTTTTTG-3′;
and control shRNA (sh-Ctrl),
5′-TTTCTCCGAACGTGTCACGTTTCAAGAGAACGTGACACGTTCGGAGAATTTTTTC-3′.
RT-qPCR
Cells were grown in 6-well- plates at a density of
1×106 cells per well in medium containing 10% FBS at
37°C in a 5% CO2 atmosphere. Subsequent to the
transfection of cells with interference plasmids containing sh-GEN1
or sh-Mus81, and control plasmid containing sh-Ctrl for 24 h, total
RNA was extracted. Total cellular RNA was isolated with TRIzol
reagent, followed by RT with the First-Strand cDNA Synthesis kit,
according to the manufacturer's protocol. RT-qPCR was performed
using the LightCycler 480 PCR apparatus. The abundance of the GEN1
and Mus81 transcripts was expressed relative to the control of
β-actin. The experiments were performed three times independently.
The Mus81 and β-actin primer sequences were previously described
(13), while the GEN1 primer sequence
was identified from a previous study (16). The primer sequences are listed in
Table I.
 | Table I.Sequences of the primers used for
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequences of the primers used for
reverse transcription-quantitative polymerase chain reaction.
| Gene | Primer sequence,
5′-3′ | Product size, bp | Tm, °C |
|---|
| GEN1 |
| 117 | 60 |
|
Forward |
CCACATGACTATGAATACTGCTGTCCTT |
|
|
|
Reverse |
TGGGAATCCCTCACAACAGCAAGC |
|
|
| Mus81 |
| 319 | 60 |
|
Forward |
TGTGGACATTGGCGAGAC |
|
|
|
Reverse |
GCTGAGGTTGTGGACGGA |
|
|
| β-actin |
| 108 | 60 |
|
Forward |
ACCCACACTGTGCCCATCTAC |
|
|
|
Reverse |
TCGGTGAGGATCTTCATGAGGTA |
|
|
Western blotting
The cells were harvested and rinsed with
phosphate-buffered saline (Solarbio Biotechnology, Beijing, China).
Total proteins were extracted using radioimmunoprecipitation assay
lysis buffer (Biomiga). The protein concentration was determined
using the bicinchoninic acid assay (Beyotime Institute of
Biotechnology). Equal amounts of the proteins were separated using
10% gel electrophoresis and electrophoretically transferred to
polyvinylidene difluoride (PVDF) membranes (Mylab China, Beijing,
China). The membranes were blocked with 5% skim milk for 2 h at
room temperature. The PVDF membranes were incubated with primary
antibodies against GEN1 (dilution, 1:75), Mus81 (dilution, 1:1,000)
and β-actin (dilution, 1:5,000) overnight at 4°C, followed by
incubation in secondary antibodies against rabbit IgG conjugated to
horseradish peroxidase (dilution, 1:3,000) for 1 h at room
temperature. The blots were then developed using the ECL-Plus
chemiluminescence detection kit, according to the manufacturer's
protocol. Subsequently, the blots were exposed to a radiographic
film (Kodak, Rochester, NY, USA). β-actin expression was used as a
control. The gray scale was scanned by ChemiDocTM XRS gel imaging
system (Bio-Rad Laboratories) and ‘gray value’ semi-quantitative
analysis was performed using Quantity One-4.6.2 software (Bio-Rad
Laboratories). The abundance of the GEN1 and Mus81 proteins was
expressed relative to the expression of the β-actin control.
Relative gene expression was calculated by normalization to that of
β-actin. The calculation formula was as follows: Relative
expression of target gene = Gray value of the target gene / Gray
value of β-actin.
Determination of the half maximal
inhibitory concentration (IC50) values for 5-FU in SKBR3
and MCF-7 cells
Cells in a good condition (firmly adherent, clear
pseudopodia and clear cytoplasm without impurities) were seeded
onto 96-well plates at a density of 1×104 cells per
well. Subsequent to 24 h, the cells were incubated with 5-FU at
concentrations ranging between 0.625 and 10 µg/ml for 48 h. Cells
not treated with 5-FU were used as a negative control. Finally, 10
µl water-soluble tetrazolium salt-8 (Nanjing KeyGen Biotech Co.,
Ltd.) was added into each well. Optical density (OD) values were
measured at a wavelength of 450 nm (OD450) using a
microplate reader (Anthos 2000; Anthos Labtec Instruments GmbH,
Salzburg, Austria). Cell survival was calculated as follows:
Survival of cells (%) = Drug-treated group OD450 /
control group OD450 × 100. The IC50 value was
calculated as follows: logIC50 = Xm - I [P - (3 - Pm -
Pn) / 4], where Xm was the log maximum dose, I was the log (maximum
dose/adjacent dose), P was the sum of the positive response rate,
Pm was the maximum positive response rate and Pn was the minimum
positive response rate.
Cell survival assays
Subsequent to transfection with shRNA for 24 h, the
cells were planted into 96-well plates at a density of
1×104 cells per well. Subsequent to 24 h, the cells were
incubated with 5-FU at concentrations ranging between 0.625 µg/ml
and 10 µg/ml for 48 h. Cells without 5-FU treatment were used as
the negative control. Finally, 10 µl CCK-8 was added to each well.
The OD values were measured at a wavelength of 450 nm (OD450). Cell
survival was calculated as aforementioned.
Apoptosis assay
The cells were transfected with shRNA for 24 h, then
planted into 6-well plates at a density of 1×106 cells
per well. Subsequent to adhering to the wall, the cells were
incubated with 4 µg/ml 5-FU for 24 h. For the slide method, cells
from each group were diluted twice with phosphate-buffered saline.
Then, 5 µl FITC and 5 µl PI were added to 500 µl binding buffer
(Nanjing KeyGen Biotech Co., Ltd.), and the resulting mixture was
added to the cell surface directly. Subsequent to incubation of the
cells in the dark for 5–15 min at room temperature, the
fluorescence was observed under the inverted fluorescence
microscope. For flow cytometry, the cells were harvested and
diluted twice with phosphate-buffered saline. Then, 5 µl FITC and 5
µl PI were added to 500 µl of the cell suspension (final cell
density, ~6×105 cells/ml). Subsequent to incubation in
the dark for 5–15 min at room temperature, flow cytometry was
performed using a flow cytometer (FC500; Beckman Coulter, Inc.,
Brea, CA, USA). The experiments were performed independently three
times.
Statistical analysis
All data were expressed as the mean ± standard
deviation, and SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA)
was used for statistical analyses. One-way analysis of variance and
Student's t-test were used to analyze the significance of
differences between groups. Multiple comparisons between the groups
was performed using the Student-Newman-Keuls method subsequent to
one-way analysis of variance. P<0.05 was considered to indicate
a statistically significant difference.
Results
IC50 values of 5-FU against
SKBR3 and MCF-7 breast cancer cell lines
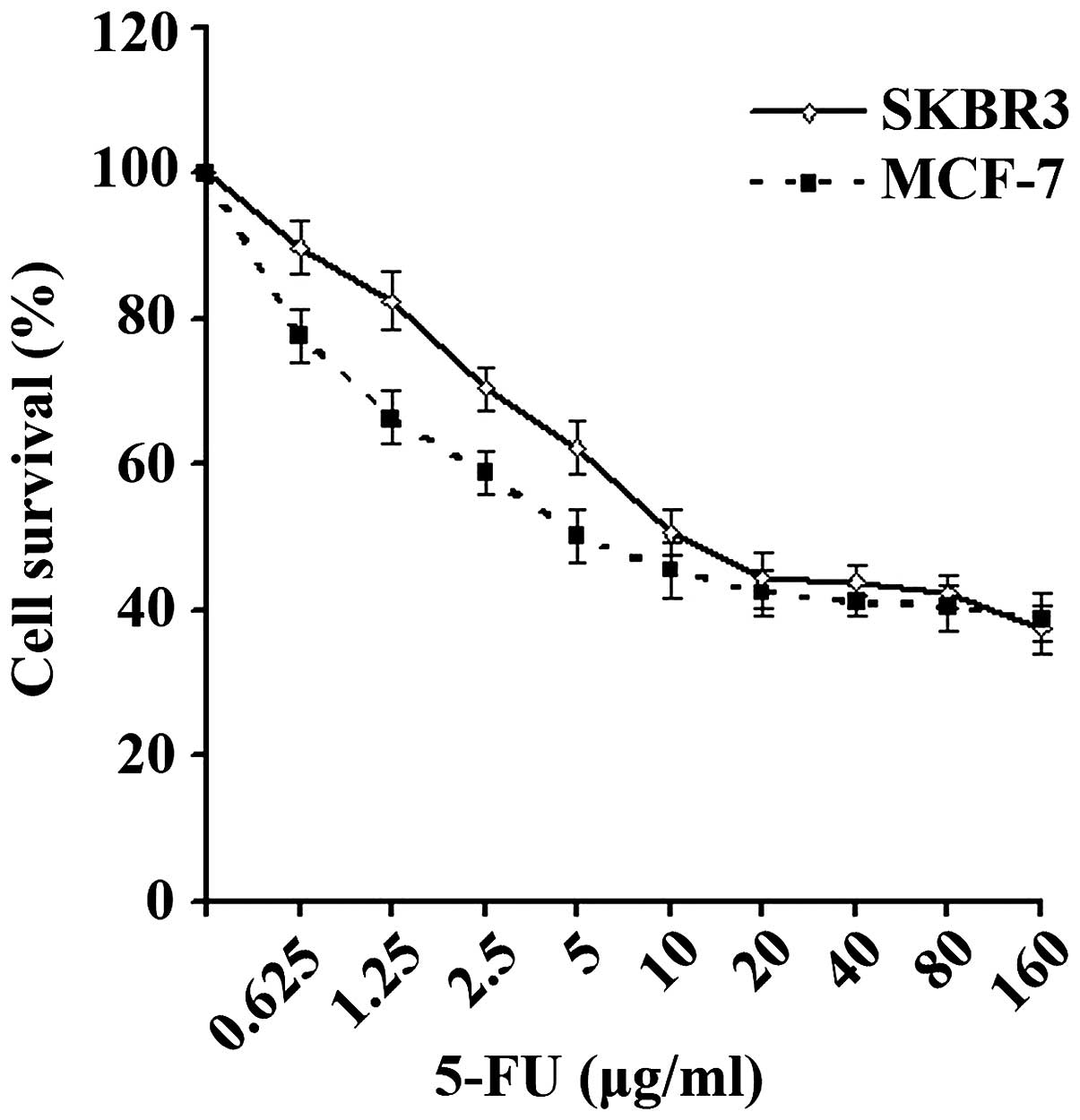
The survival curves of the SKBR3 and MCF-7 cells are
shown in Fig. 1. The IC50
values of 5-FU against the breast cancer SKBR3 and MCF-7 cell lines
were 8.77 and 7.73 µg/ml, respectively, which were then used in
subsequent experiments.
SKBR3 cells demonstrated increased
sensitivity to 5-FU following GEN1 gene suppression
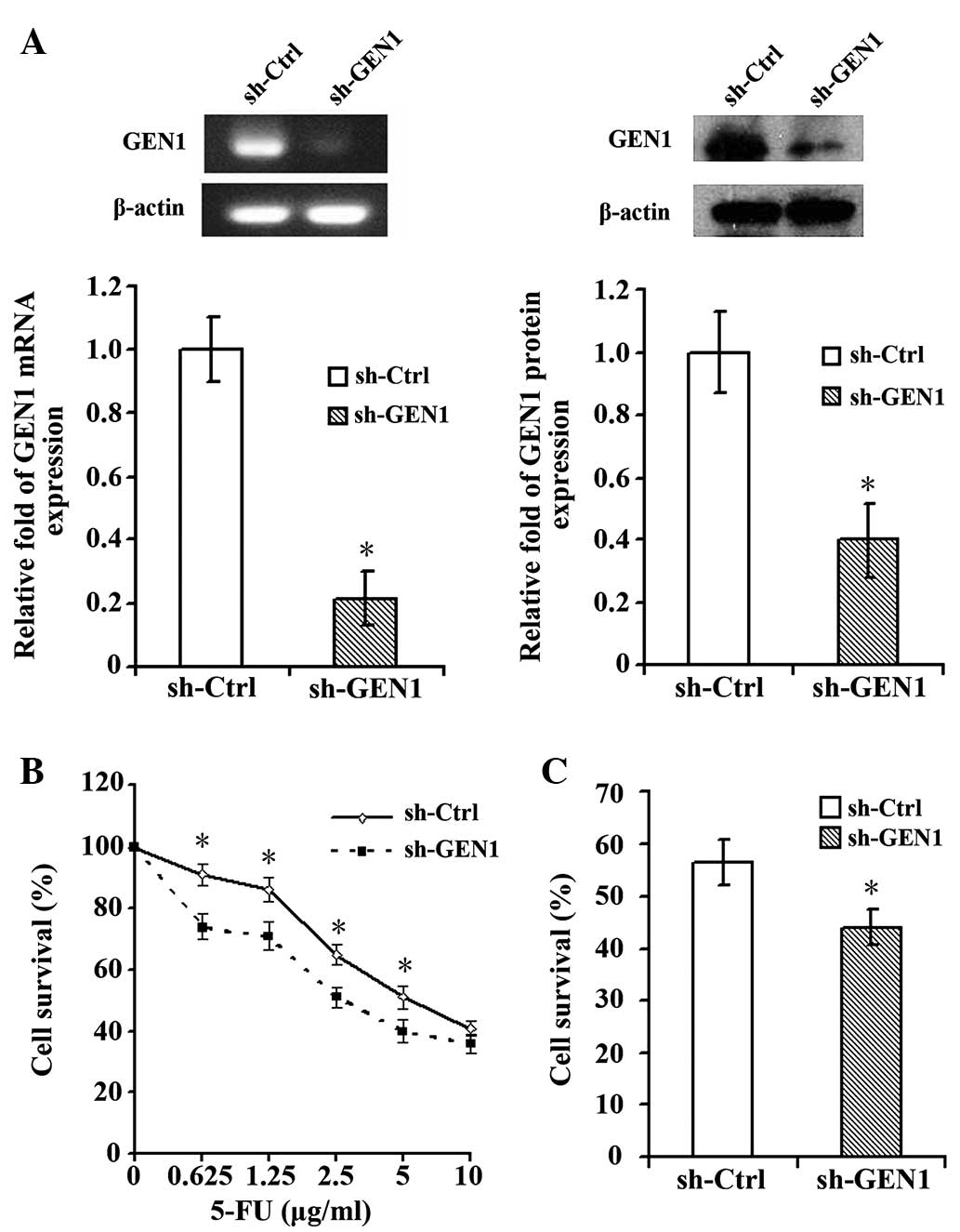
Silencing of the GEN1 gene was confirmed by RT-qPCR
and western blot analysis (Fig. 2A).
Following suppression of the GEN1 gene, the SKBR3 cells
demonstrated significantly enhanced sensitivity to 5-FU. At 5-FU
concentrations of 0.625, 1.25, 2.5 and 5.0 µg/ml, the survival of
cells in the experimental sh-GEN1 group was significantly decreased
compared with the control sh-Ctrl group (P<0.05; Fig. 2B). When treated with the
IC50 (8.77 µg/ml) of 5-FU, the survival of SKBR3 cells
in the sh-GEN1 group was significantly reduced (Fig. 2C).
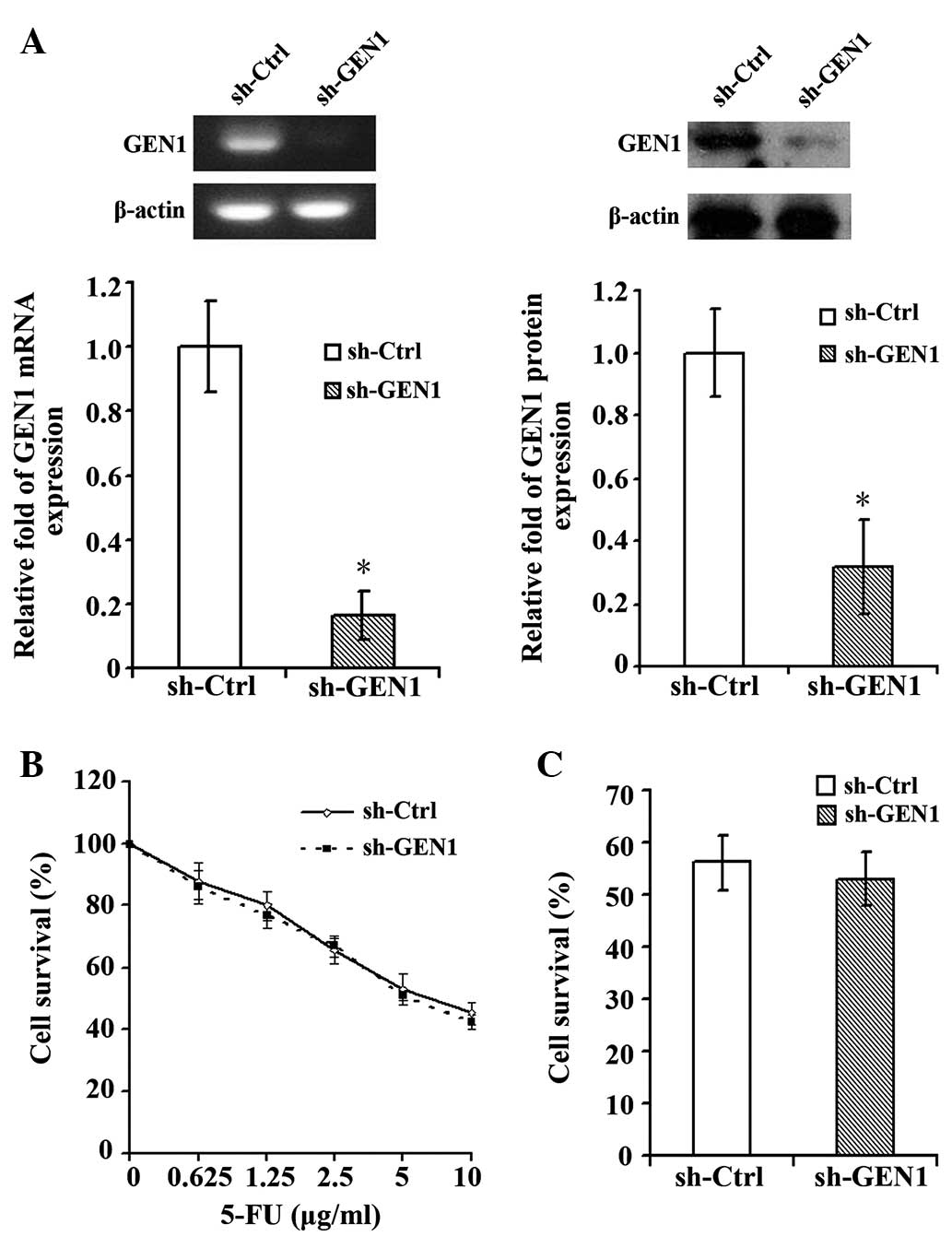
Silencing GEN1 in the MCF-7 cell line
had no significant effect on the sensitivity to 5-FU
The silencing of GEN1 was confirmed by RT-qPCR and
western blot (Fig. 3A). Subsequent to
suppression of GEN1, the MCF-7 cell line did not demonstrate a
significant change in the sensitivity to 5-FU. Under various
concentrations of 5-FU, the survival of the sh-GEN1 group was
similar with to the survival of the sh-Ctrl group (Fig. 3B). When treated with the
IC50 of 5-FU (7.73 µg/ml), the survival rates of the
sh-Ctrl and sh-GEN1 groups exhibited no evident difference in
sensitivity (Fig. 3C).
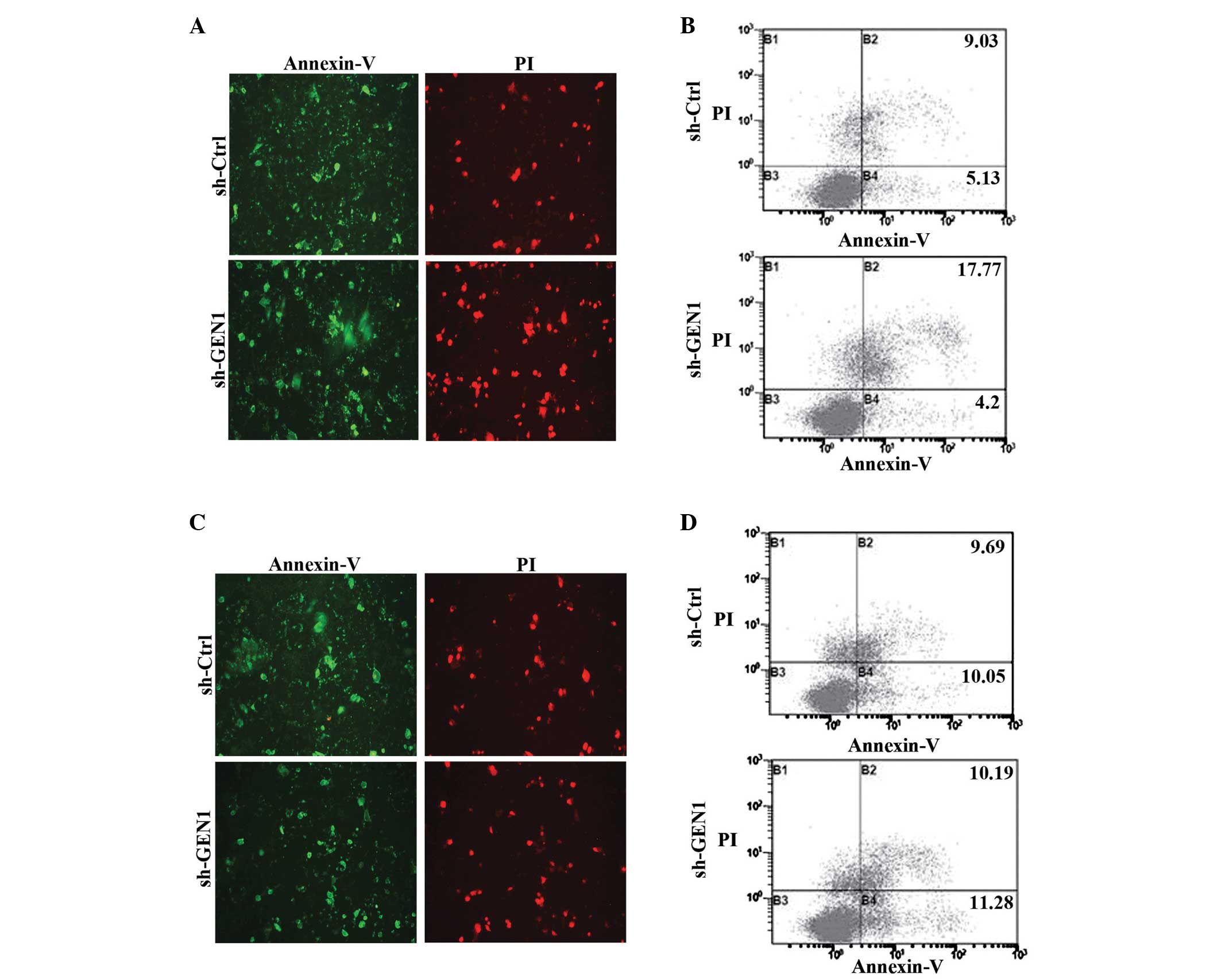
Effect of silencing GEN1 on the
apoptosis of SKBR3 and MCF-7 cell lines
Following the suppression of GEN1 for 24 h, the
cells were treated with 5-FU and the apoptosis was detected using
fluorescence microscopy and flow cytometry. Under the fluorescence
microscope, the fluorescence intensities of the SKBR3 cells of the
sh-GEN1 group were significantly stronger (P<0.05) compared with
the sh-Ctrl group (Fig. 4A),
particularly red fluorescence. The result of flow cytometry also
showed that the apoptosis of the sh-GEN1 group (21.98±3.23%) was
significantly increased (P<0.05) compared with the control group
(14.76±2.87%) (Fig. 4B). However, in
the MCF-7 cells, the results of each method did not demonstrate
statistical differences between the two groups (Fig. 4C and D).
Suppression of the GEN1 and Mus81
genes increased the sensitivity of MCF-7 cells to 5-FU
The expression of the GEN1 and Mus81 genes was
simultaneously suppressed to observe the effect on the sensitivity
of MCF-7 cells to 5-FU. The silencing of the genes was confirmed by
RT-qPCR and western blot analysis (Fig.
5A and B). The sh-Ctrl-1, sh-Ctrl-2, sh-GEN1, sh-Mus81 and
sh-GEN1+sh-Mus81 groups were transfected with 4 µg control plasmid,
8 µg control plasmid, 4 µg sh-GEN1 plasmid, 4 µg sh-Mus81 plasmid
and 4 µg sh-GEN1 plasmid + 4 µg sh-Mus81 plasmid, respectively. No
significant difference in cell survival was identified between the
sh-Ctrl-1 and sh-Ctrl-2 groups and the sh-GEN1 group. Unlike GEN1
suppression alone, MCF-7 cells demonstrated significantly enhanced
sensitivity to 5-FU when the GEN1 and Mus81 genes were
simultaneously suppressed (Fig.
5C).
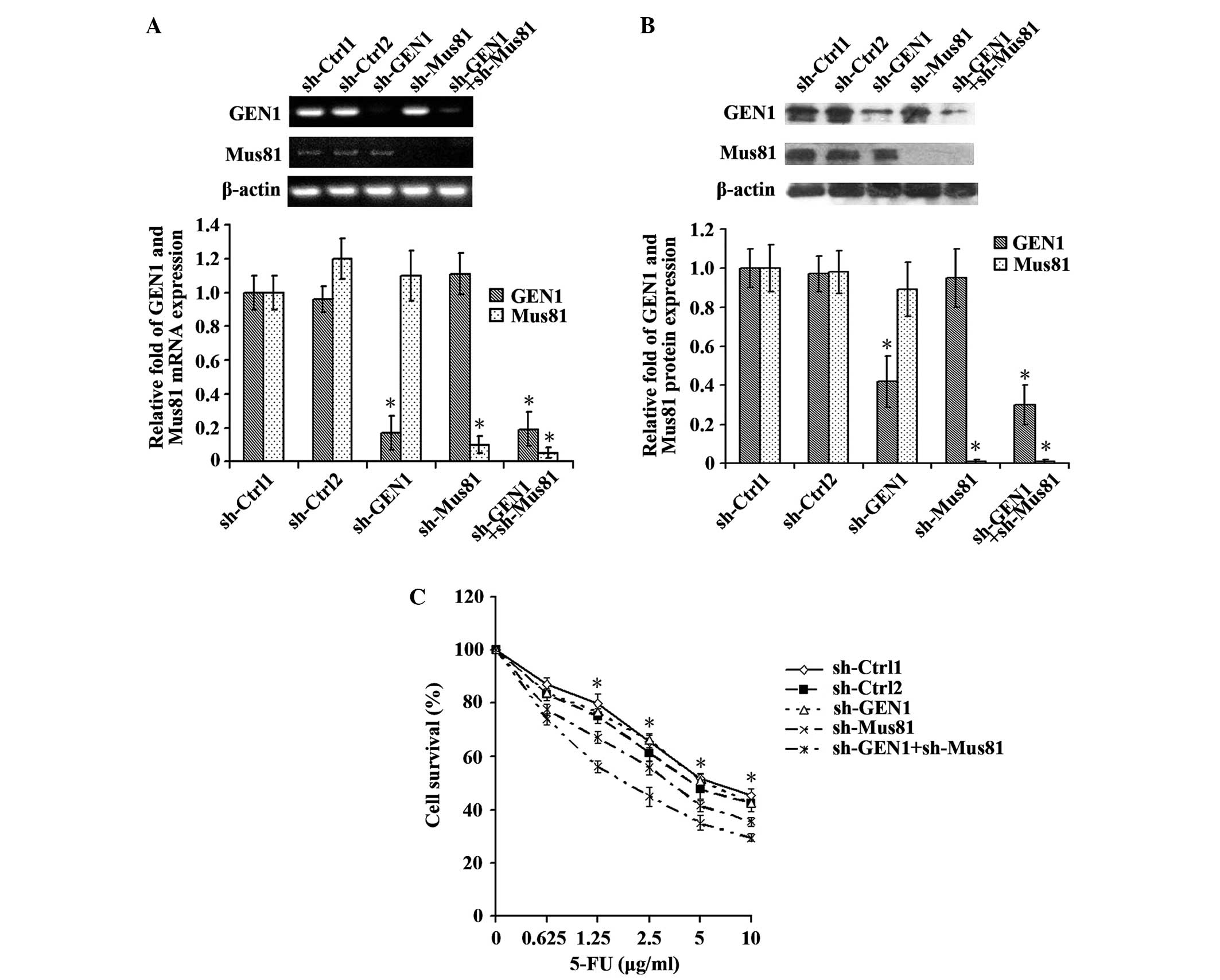 | Figure 5.The MCF-7 cell line demonstrated
increased sensitivity to 5-FU subsequent to the simultaneous
suppression of the GEN1 and Mus81 genes. Reverse
transcription-quantitative polymerase chain reaction and western
blot analysis confirmed the silencing of the GEN1 and Mus81 genes
by assessing the (A) mRNA and (B) protein expression. (C) The
sh-Ctrl-1, sh-Ctrl-2, sh-GEN1, sh-Mus81 and sh-GEN1+sh-Mus81 groups
were transfected with 4 µg control plasmid, 8 µg control plasmid, 4
µg sh-GEN1 plasmid, 4 µg sh-Mus81 plasmid and 4 µg sh-GEN1 plasmid
+ 4 µg sh-Mus81 plasmid, respectively. No significant difference in
cell survival was identified between the sh-Ctrl-1 and sh-Ctrl-2
groups and the sh-GEN1 group. In contrast to the suppression of
GEN1 alone, MCF-7 cells demonstrated significantly enhanced
sensitivity to 5-FU when the GEN1 and Mus81 genes were
simultaneously suppressed (P<0.05). *P<0.05. GEN1, flap
endonuclease GEN homolog 1; Mus81, MUS81 structure-specific
endonuclease subunit; sh-Ctrl; control short hairpin RNA; sh-GEN1,
GEN1 short hairpin RNA; sh-Mus81, Mus81 short hairpin RNA; 5-FU,
5-fluorouracil. |
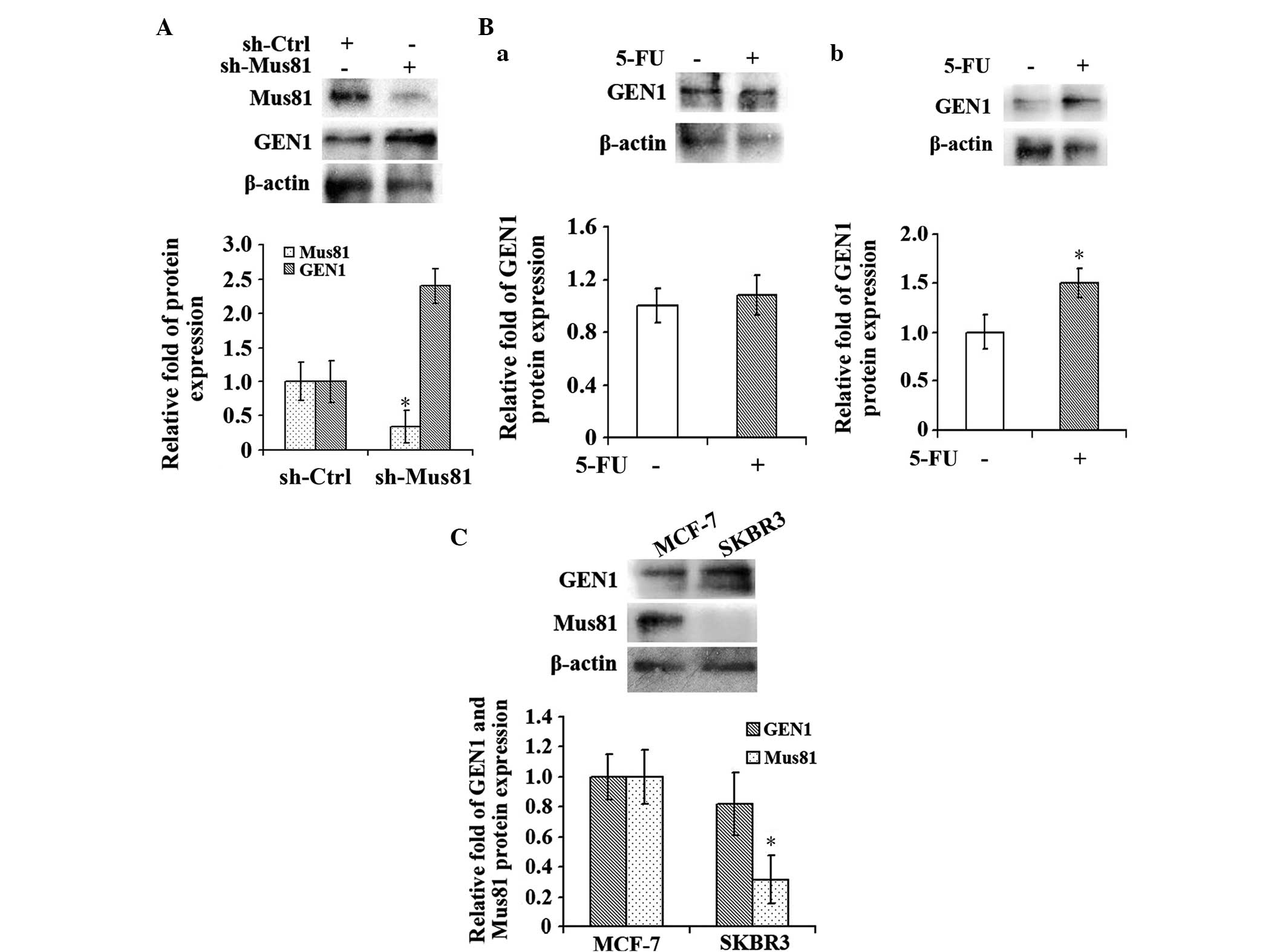
Under a low concentration of 5-FU, the
GEN1 expression level increased when the Mus81 level was low
Subsequent to suppression of Mus81 for 24 h, a low
concentration of 1 µg/ml 5-FU, which was approximately one-quarter
of the IC50 value, was added into the MCF-7 cells and
the cells were incubated for 96 h. Following incubation, the
expression levels of GEN1 and Mus81 were detected by RT-qPCR and
western blot analysis. The results showed that the level of GEN1 in
the experimental sh-Mus81 group was increased compared with the
control sh-Ctrl group (P<0.05; Fig.
6A). Additionally, the MCF-7 and SKBR3 cells of the
experimental groups were incubated with 1 µg/ml 5-FU for 96 h and
cells cultured in normal medium were considered the control group.
Subsequently, the expression level of GEN1 was detected in each
group by western blot analysis (Fig.
6B). The GEN1 expression level in the MCF-7 cells, which
demonstrated high Mus81 expression (Fig.
6C), did not change significantly when the cells were treated
with 5-FU (Fig. 6Ba). However,
compared with the control group, the level of GEN1 expression was
increased in the SKBR3 cells (P<0.05; Fig. 6Bb), which demonstrated a low Mus81
expression level (Fig. 6C).
Discussion
GEN1 was first identified in rice by Furukawa et
al (3). Ishikawa et al
classified GEN1 into a new class (class IV) of the Rad2/XPG
nuclease family (4). GEN1 contains an
N-terminal, internal conserved Rad2/XPG endonuclease region and a
helix-hairpin-helix region (17). Gao
et al found that GEN1 functioned as an HJ resolvase in
vivo as well as in vitro (18). In addition, a number of studies have
confirmed that the HJ may be broken down by GEN1 (19,20).
Similar to the Escherichia coli HJ resolvase RuvC, GEN1
specifically binds and resolves HJs by a dual incision mechanism
(17,21). GEN1 resolves HJ in a symmetrical
manner and exists in two forms, consisting of the monomeric form
and dimeric complexes. Efficient HJ resolution requires dimeric
complexes in order to provide the two active sites required for
near simultaneous dual incision (17). This monomer/dimer substrate-driven
switch distinguishes GEN1 from other HJ resolvases, such as Mus81,
which resolves HJ in an asymmetric manner (11).
HJs were first identified by Robin Holliday
(22). Numerous studies hypothesized
that HJ resolved properly is the key to correct DNA repair
(8,9).
DNA damaging drugs, such as 5-FU, may injure tumor cells by
damaging the DNA of the cells (23).
During this process, numerous HJs may be produced (24,25). It
has been confirmed that subsequent to suppressing HJ
resolvase-Slx4, the sensitivity to DNA damaging agents of tumor
cells increased significantly. Previous studies have also confirmed
that targeting Mus81 may increase sensitivity to DNA damaging
drugs, such as 5-FU, in breast cancer cells (13). Thus, the HJ resolution capacity of
tumor cells has a significant effect on the sensitivity to drugs.
However, it remains unknown whether GEN1 may have an impact on the
chemosensitivity of tumor cells. It has been proved that GEN1
interference may increase pharmaceutical sensitivity of yeast and
Drosophila alone or in combination with other genes
(11,15). Therefore, the present study was
designed to explore the effect of GEN1 interference on the
chemosensitivity of breast cancer cell lines MCF-7 and SKBR3.
In the present study, the functional studies of GEN1
were investigated in MCF-7 and SKBR3 cells. shRNA was used to
suppress the expression of GEN1 and the cells were incubated with
5-FU. Cell survival and apoptosis were detected to evaluate the
effect of GEN1 expression on the two cell lines. The results showed
that following suppression of GEN1, survival of SKBR3 cells in the
experimental group was significantly decreased compared with the
control group, while the level of apoptosis was significantly
increased. These results suggested that the SKBR3 cell line
increased the sensitivity to 5-FU following suppression of the GEN1
gene. By contrast, the MCF-7 cell line did not show significant
changes under the same conditions, which indicated that targeting
GEN1 had no evident effect on the chemosensitivity of MCF-7 cells.
The aforementioned results demonstrated that there was a marked
difference between the effect of GEN1 expression on the sensitivity
to injury drugs in the two breast cell lines, which requires
additional investigation. A previous study showed that the
expression of Mus81 in the SKBR3 cells was decreased compared with
MCF-7 cells (Fig. 6C). Since GEN1 and
Mus81 did not resolve HJ in the same manner, with GEN1 adopting a
symmetrical manner and Mus81 using an asymmetric mode, the present
study hypothesized that GEN1 acted as a collaborative gene of Mus81
and did not play a key role in breast cells with a high level of
Mus81. However, the activity of GEN1 increases when the expression
of Mus81 is low. To verify this hypothesis, the expression of GEN1
and Mus81 was suppressed simultaneously in the MCF-7 cell line. The
results showed that although silencing GEN1 alone did not increase
the sensitivity of MCF-7 cells to 5-FU, targeting GEN1 and Mus81
together significantly enhanced this sensitivity. Furthermore, the
enhancement is also stronger than the effect of silencing Mus81
alone. Therefore, the possibility that this change was merely due
to Mus81 interference was excluded, as the effect of GEN1 was also
involved in the sensitivity of cells to 5-FU. However, no
significant differences in the cell survival were identified
between the sh-Ctrl-1 and sh-GEN1 groups, which indicated that GEN1
did not play a significant role in cell survival. The
aforementioned results showed that GEN1 did not play a significant
role in the presence of Mus81 and its effect will be reflected when
Mus81 level was low. In addition, GEN1 interference enhanced the
effect of Mus81 on the chemotherapy of breast cancer cells.
To further confirm the aforementioned statement, the
MCF-7 cells were incubated with a low concentration of 5-FU for 96
h subsequent to Mus81 suppression. The level of GEN1 was determined
by western blot analysis. The result showed that the GEN1 level in
the experimental group was significantly increased compared with
the control group. Therefore, when the SKBR3 cell line, which
exhibited low Mus81 expression, was incubated with a low
concentration of 5-FU, the GEN1 level in the experimental group
also increased significantly. The aforementioned results indicated
that GEN1 demonstrates enhanced function when the Mus81 level is
low. By contrast, the normal MCF-7 cell line, which exhibited high
Mus81 expression, did not demonstrate an enhanced GEN1 level when
treated with 5-FU, which supports the present conclusion.
Overall, the present study showed that GEN1 may play
different roles in diverse breast cancer cell lines. The function
of GEN1 may be affected by the level of Mus81 in the cell line.
GEN1 acts as a collaborative gene of Mus81 and did not play a key
role in the breast cell line with a high Mus81 level. The activity
of GEN1 increases, however, in the cells with low expression of
Mus81. In addition, it is worth mentioning that GEN1 interference
improves the chemotherapy sensitizing effect brought by targeting
Mus81 alone. If Mus81 alone is targeted over the course of breast
cancer treatment, the expression level of GEN1 may increase
gradually thus to reduce the treatment effect. Therefore, the
present study hypothesizes that targeting Mus81 together with GEN1
may result in an improved effect of chemotherapy on breast
cancer.
Acknowledgements
The present study was supported by the Natural
Science Foundation of Zhejiang Province (grant no. Y14H200002),
Medicines Health Platform Key Project of Zhejiang Province (grant
no. 2013ZDA024) and Science and Technology Project of Shaoxing City
(grant no. 2013D10040).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kuczynski EA, Sargent DJ, Grothey A and
Kerbel RS: Drug rechallenge and treatment beyond progression -
implications for drug resistance. Nat Rev Clin Oncol. 10:571–587.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Furukawa T, Kimura S, Ishibashi T, Mori Y,
Hashimoto J and Sakaguchi K: OsSEND-1: A new RAD2 nuclease family
member in higher plants. Plant Mol Biol. 51:59–70. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kanai Y, Ishikawa G, Takeuchi R, Ruike T,
Nakamura R, Ihara A, Ohashi T, Takata K, Kimura S and Sakaguchi K:
DmGEN shows a flap endonuclease activity, cleaving the blocked-flap
structure and model replication fork. FEBS J. 274:3914–3927. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Muñoz-Galván S, Tous C, Blanco MG,
Schwartz EK, Ehmsen KT, West SC, Heyer WD and Aguilera A: Distinct
roles of Mus81, Yen1, Slx1-Slx4 and Rad1 nucleases in the repair of
replication-born double-strand breaks by sister chromatid exchange.
Mol Cell Biol. 32:1592–1603. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Agmon N, Yovel M, Harari Y, Liefshitz B
and Kupiec M: The role of Holliday junction resolvases in the
repair of spontaneous and induced DNA damage. Nucleic Acids Res.
39:7009–7019. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ip SC, Rass U, Blanco MG, Flynn HR, Skehel
JM and West SC: Identification of Holliday junction resolvases from
humans and yeast. Nature. 456:357–361. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ashton TM, Mankouri HW, Heidenblut A,
McHugh PJ and Hickson ID: Pathways for Holliday junction processing
during homologous recombination in saccharomyces cerevisiae. Mol
Cell Biol. 31:1921–1933. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Machwe A, Karale R, Xu X, Liu Y and Orren
DK: The Werner and Bloom syndrome proteins help resolve replication
blockage by converting (regressed) holliday junctions to functional
replication forks. Biochemistry. 50:6774–6788. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saito TT, Mohideen F, Meyer K, Harper JW
and Colaiácovo MP: SLX-1 is required for maintaining genomic
integrity and promoting meiotic noncrossovers in the Caenorhabditis
elegans germline. PLoS Genet. 8:e10028882012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Andersen SL, Kuo HK, Savukoski D, Brodsky
MH and Sekelsky J: Three structure-selective endonucleases are
essential in the absence of BLM helicase in Drosophila. PLoS
Genet. 7:e10023152011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Svendsen JM, Smogorzewska A, Sowa ME,
O'Connell BC, Gygi SP, Elledge SJ and Harper JW: Mammalian
BTBD12/SLX4 assembles a Holliday junction resolvase and is required
for DNA repair. Cell. 138:63–77. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Qian Y, Liu Y, Yan Q, Lv J, Ni X, Wu Y and
Dong X: Inhibition of Mus81 by siRNA enhances sensitivity to 5-FU
in breast carcinoma cell lines. Onco Targets Ther. 7:1883–1890.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vizeacoumar FJ, Arnold R, Vizeacoumar FS,
Chandrashekhar M, Buzina A, Young JT, Kwan JH, Sayad A, Mero P,
Lawo S, et al: A negative genetic interaction map in isogenic
cancer cell lines reveals cancer cell vulnerabilities. Mol Syst
Biol. 9:6962013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ho CK, Mazón G, Lam AF and Symington LS:
Mus81 and Yen1 promote reciprocal exchange during mitotic
recombination to maintain genome integrity in budding yeast. Mol
Cell. 40:988–1000. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wechsler T, Newman S and West SC: Aberrant
chromosome morphology in human cells defective for Holliday
junction resolution. Nature. 471:642–646. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Rass U, Compton SA, Matos J, Singleton MR,
Ip SC, Blanco MG, Griffith JD and West SC: Mechanism of Holliday
junction resolution by the human GEN1 protein. Genes Dev.
24:1559–1569. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gao M, Rendtlew Danielsen J, Wei LZ, Zhou
DP, Xu Q, Li MM, Wang ZQ, Tong WM and Yang YG: A novel role of
human holliday junction resolvase GEN1 in the maintenance of
centrosome integrity. PLoS One. 7:e496872012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bailly AP, Freeman A, Hall J, Déclais AC,
Alpi A, Lilley DM, Ahmed S and Gartner A: The caenorhabditis
elegans homolog of Gen1/Yen1 resolvases links DNA damage signaling
to DNA double-strand break repair. PLoS Genet. 6:e10010252010.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tay YD and Wu L: Overlapping roles for
Yen1 and Mus81 in cellular Holliday junction processing. J Biol
Chem. 285:11427–11432. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Górecka KM, Komorowska W and Nowotny M:
Crystal structure of RuvC resolvase in complex with Holliday
junction substrate. Nucleic Acids Res. 41:9945–9955. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Holliday R: A mechanism for gene
conversion in fungi. Genet Res. 5:282–304. 1964. View Article : Google Scholar
|
|
23
|
Tetu SG, Johnson DA, Varkey D, Phillippy
K, Stuart RK, Dupont CL, Hassan KA, Palenik B and Paulsen IT:
Impact of DNA damaging agents on genome-wide transcriptional
profiles in two marine Synechococcus species. Front Microbiol.
16:2322013.
|
|
24
|
Lorenz A, West SC and Whitby MC: The human
Holliday junction resolvase GEN1 rescues the meiotic phenotype of a
Schizosaccharomyces pombe mus81 mutant. Nucleic Acids Res.
38:1866–1873. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mankouri HW, Ashton TM and Hickson ID:
Holliday junction-containing DNA structures persist in cells
lacking Sgs1 or Top3 following exposure to DNA damage. Proc Natl
Acad Sci USA. 108:4944–4949. 2011. View Article : Google Scholar : PubMed/NCBI
|