Introduction
Cervical cancer is the third most commonly diagnosed
cancer worldwide and the fourth leading cause of cancer-associated
mortality in women, with 529,800 new cases and 275,100 mortalities
among women in 2008 worldwide (1).
The global incidence and mortality of cervical cancer have
gradually decreased. In China, however, the incidence of cervical
cancer remains high, particularly in young women (2). The widespread use of cervical screening
programs and advances in various therapies, including surgery,
chemotherapy, radiotherapy and combined therapy, have dramatically
reduced the morbidity and mortality of cervical cancer. However,
the number of newly diagnosed cases worldwide remains large, with
~500,000 new cases each year (3).
Despite the high prevalence, understanding of the molecular
background in terms of genesis, growth and progress remain
insufficient. Unfortunately, due to the lack of specific
biomarkers, the gene diagnosis and biological treatment of cervical
caner were restricted.
The wings apart-like (WAPL) gene of Drosophila
melanogaster, which is located on X chromosome region 2D5-2D5,
encodes a protein that regulates heterochromatin structure.
Mutation of WAPL prevent the normal close apposition of sister
chromatids in heterochromatin regions, but does not appear to
affect either heterochromatin condensation or chromosomal
segregation. This suggests that WAPL is required to hold sister
chromatids together in mitotic heterochromatin. WAPL has also been
implicated in heterochromatin pairing during female meiosis and the
modulation of position effect variegation (4,5). The hWAPL
gene, which is 30,793 bp long and located on 10q23.2., is a human
homologue of the WAPL gene in Drosophila melanogaster. This
gene encodes a cohesin-binding protein that facilitates the timely
release of cohesin from chromosome arms during prophase. The
mechanism of action of the hWAPL gene is not clear. However,
overexpression of WAPL causes premature separation of sister
chromatids (6). hWAPL has the
characteristics of an oncogene and is associated with uterine
cervical cancer (7).
The present study aimed to investigate the
expression of the hWAPL gene in various tumor tissues. To identify
the expression of hWAPL in various cancer tissues, 9 common cancers
and CIN tissues were investigated, consisting of cervical, gastric
and lung cancers, and liver, bladder, esophageal, endometrial,
renal and rectal carcinoma. Furthermore, the expression of hWAPL
messenger (m)RNA was investigated in benign squamous epithelia and
cervical cancer tissues.
Materials and methods
Patients
The immunohistochemical analysis was conducted using
paraffin-embedded tissues obtained from 413 patients, consisting of
27 benign squamous epithelia, 47 cervical cancer, 30 cervical
intraepithelial neoplasia (CIN)I, 33 CINII, 38 CINIII, 29 gastric
cancer, 28 liver carcinoma, 26 bladder carcinoma, 35 esophageal
carcinoma, 25 endometrial carcinoma, 26 renal carcinoma, 36 rectal
carcinoma and 33 lung cancer tissues. The selection criteria were
as follows: i) Patients with no prior treatment; and ii) patients
whose cancer is primary. In total, 8 benign squamous epithelia and
11 cervical cancer tissues were analyzed by reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) for
the expression of hWAPL mRNA. All histopathological characteristics
were confirmed by pathologists. Written informed consent was
obtained from the patients for the publication of the present
study. The study was approved by the Ethics Committee of the Second
Affiliated Hosptial of Zhengzhou University (Zhengzhou, China).
Immunohistochemistry staining
Representative formalin fixed paraffin-embedded
tissue blocks were selected and 5-µm sections were cut,
deparaffinized with xylene (Tianjin Rgent Chemical Co., Ltd.,
Tianjin, China) and rehydrated through graded alcohols (anhydrous
alcohol, 95% alcohol, 85% alcohol, 75% alcohol; (Tianjin Rgent
Chemical Co., Ltd.). Antigen retrieval was performed by heating the
slides in citrate buffer (1 mol/l; ZSGB-Bio, Bejing, China) at 98°C
for 15 min in a water bath. Endogenous peroxidase was quenched for
10 min with peroxidase blocking reagent. Primary rabbit polyclonal
anti-hWAPL antibody (catalog no., NBP1-92579; dilution, 1:200;
Novus Biologicals Canada LLC, Oakville, ON, Canada) was incubated
for 12 h at room temperature. Antibody staining was visualized
using 2-step plus poly-HRP anti-mouse/rabbit IgG Detection System
(catalog no., PV-9000; ZSGB-Bio). The sections were counterstained
using Meyer's hematoxylin solution (ST Infinity HE Staining System;
Leica Biosystems GmbH, Wetzlar, Germany). Negative controls (PBS
instead of anti-hWAPL antibodies) were run simultaneously. The
hWAPL protein expression was scored on the following scales, taking
into account the intensity of staining and the proportion of cells
stained in those cells. The staining intensity was classified as
follows: 0, No cells stained; 1, weak staining; 2, yellow
cytoplasmic staining; and 3, deep brown cytoplasmic staining. The
rate of cells expressing hWAPL was calculated as follows: Rate (%)
= number of positive cells / total number of cells. The cells in 5
high-power fields were counted under a high-power lens
(magnification, ×400; BX43 Microscope; Olympus Corporation, Tokyo,
Japan) and scored as follows: 0, 0–5% hWAPL-positive cells; 1,
5–25% hWAPL-positive cells; 2, 26–50% hWAPL-positive cells; 3,
51–75% hWAPL-positive cells; and 4, ≥76% hWAPL-positive cells. The
product of the staining intensity and rate of cells expressing
hWAPL was the final score. A score of >3 was considered
positive. Two observers quantified the scores independently. The
staining score was expressed as mean ± standard deviation.
RT-qPCR analysis
Total RNA was isolated from cervical squamous cell
carcinoma tissues using the RNA Extraction kit (Beijing Dingguo
Changsheng Biotechnology Co., Ltd., Beijing China). Complementary
(c)DNA was synthesized in reaction mixture containing 3 µl total
RNA (1 µg/µl), 1 µl deoxynucleoside triphosphates (dNTPs; 10 mM),
0.5 µl forward primer (10 µM), 0.5 µl reverse primer (10 µM), 4 µl
5X first strand buffer, 0.5 µl RNase (40 U/µl), 0.5 µl Moloney
murine leukemia virus reverse transcriptase (200 U/µl) and 10 µl
diethylpyrocarbonate (DEPC)-treated water. The reactions were
incubated at 42°C for 1 h, followed by 95°C for 10 min to terminate
the reaction, and then quenched on ice for 5 min. The hWAPL mRNA
expression level was determined by RT-qPCR using an Applied
Biosystems 7700 Sequence Detection System (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and the following hWAPL
primers: Forward, primer, 5′-AATTGTCGAGCACTGATAGAG-3′; and reverse,
5′-TTAAGTCAGCCTCAAGTACCC-3′. To normalize the amount of cDNA in
each sample, the internal reference gene β-actin was quantified and
used as the control for the experiment, with the following primers:
Forward, 5′-ATCATGTTTGAGACCTTCAACA-3′; and reverse,
5′-CATCTCTTGCTCGAAGTCCA-3′. Each reaction contained 2 µl cDNA, 2.5
µl 10X PCR buffer, 0.5 µl dNTPs, 1 µl SYBR green, 1.5 µl
MgCl2 (25 mM), 0.5 µl forward primer, 0.5 µl reverse
primer, 0.5 µl Cap Taq polymerase (2 U/µl) and DEPC-treated water
was added to make a final volume of 25 µl. DNTPs, forward and
revers primers, first strand buffer, RNase, Moloney murine leukemia
virus reverse transcriptase and DEPC were all purchased from
Beijing Dingguo Changsheng Biotechnology Co., Ltd. The reaction
conditions were as follows: Initial denaturation for 2 min at 94°C;
35 cycles of denaturation for 30 sec at 94°C; renaturation for 30
sec at 58°C; and extension for 40 sec at 72°C. Following
amplification, the critical cycle number (cycle threshold, Cq)
(8), the amplification curve and the
copy number of the target gene were provided by the PCR
amplification instrument automatically. The relative quantification
of hWAPL mRNA = copies of hWAPL mRNA / copies of reference β-actin.
qPCR was repeated 3 times and the relative level of hWAPL mRNA was
expressed as the mean ± standard deviation.
Statistical analysis
Statistical analysis was performed using SPSS 10.0
software (SPSS, Inc., Chicago, IL, USA). The immunohistochemical
staining score was analyzed using one-way analysis of variance. The
percentage of tissues expressing hWAPL was analyzed using the
χ2 test. The mRNA level of hWAPL in cervical carcinoma
tissues and the normal tissues was analyzed by Student's t-test.
P<0.05 was considered to indicate a statistically significant
difference. The confidence intervals were evaluated, with a 0.05
level of significance.
Results
Protein level of hWAPL in cervical
carcinoma, CIN and benign squamous epithelial tissues
The expression of hWAPL, which was located in the
cytochylema and stained yellow or brown (Fig. 1), in terms of the staining scores and
the percentage of tissues expressing hWAPL, were significantly
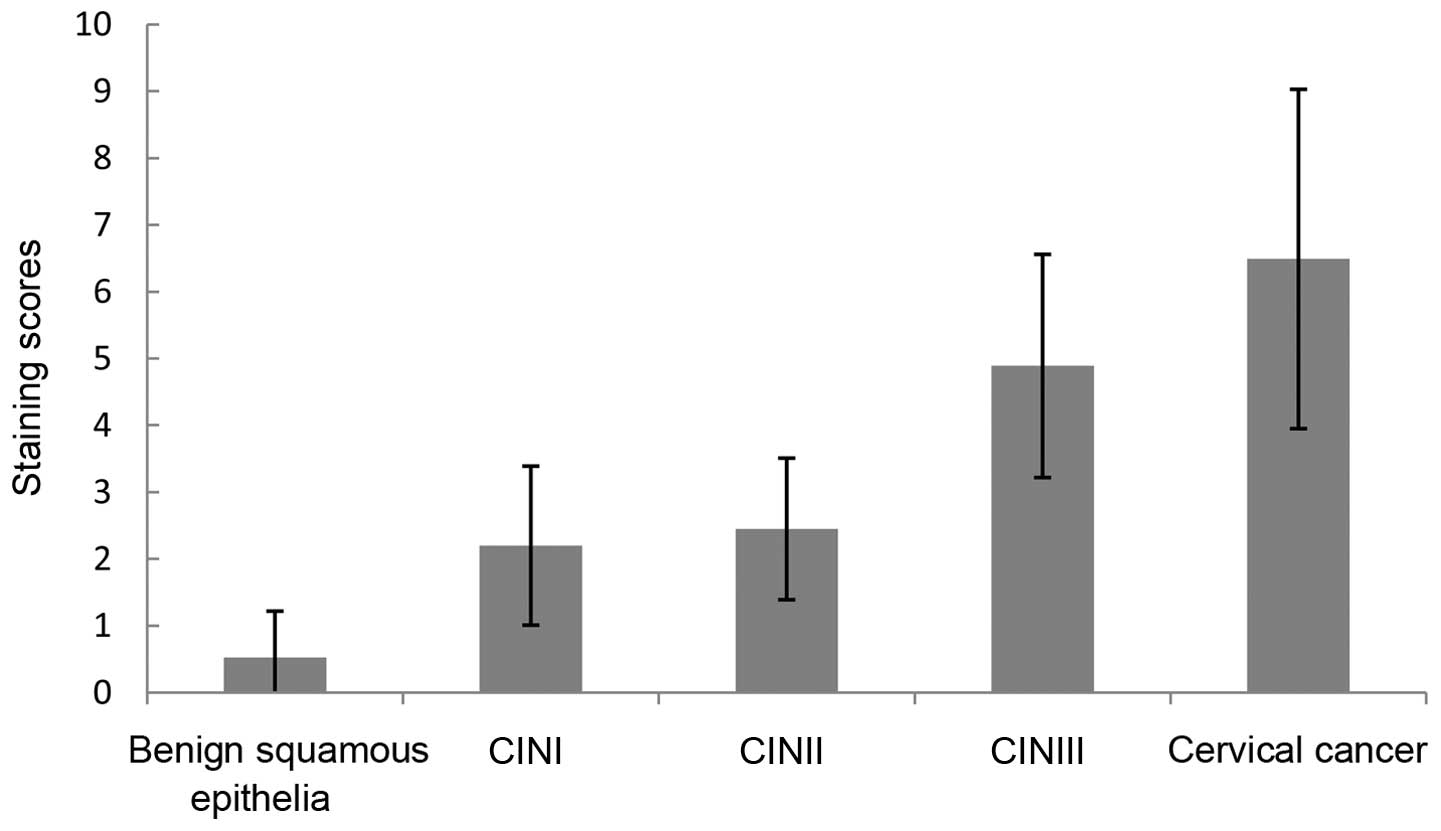
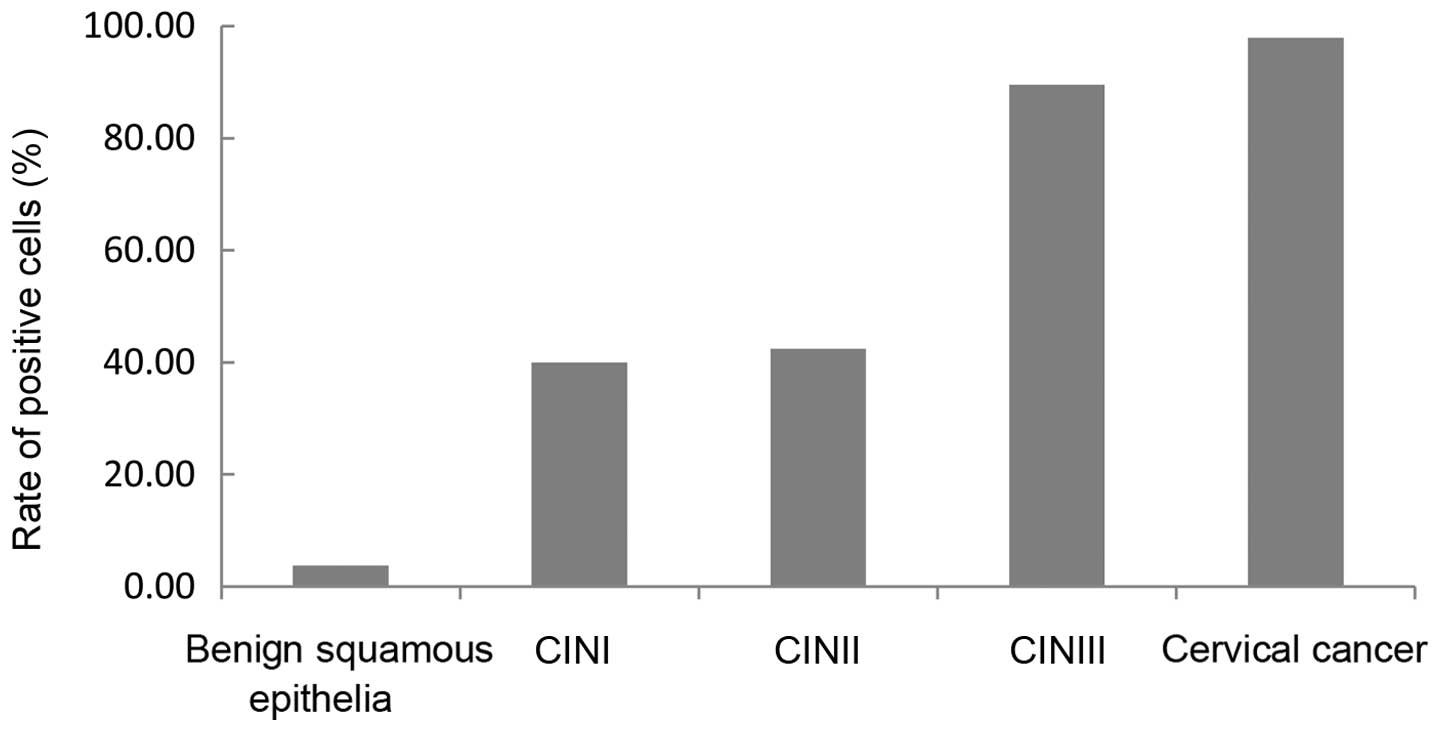
increased in cervical squamous cell carcinoma (staining score,
6.49±2.54; expression rate, 97.87%), whereas no or weak staining
was detected in benign squamous epithelial tissues (staining score,
0.52±0.70; expression rate, 3.70%). The staining scores and the
percentage of tissues expressing hWAPL increased gradually with the
development of CIN between CINI (staining score, 2.20±1.19;
expression rate, 40.00%), CINII (staining score, 2.45±1.06;
expression rate, 89.47%) and CINIII (staining score, 4.89±1.67;
expression rate, 42.42%). The staining scores and the percentage of
tissues expressing hWAPL in cervical cancer and CINIII were
significantly increased compared with CINI and CINII tissues
(P<0.001; Tables I and II; Figs. 2
and 3). Despite the difference in the
staining scores (P<0.001), no significant difference was
observed in the percentage of tissues expressing hWAPL (P=0.102)
between cervical cancer and CINIII. Furthermore, the differences
between the staining scores and percentage of tissues expressing
hWAPL in CINI and CINII were not significant. The development of
the cervical lesion was therefore associated with an increased
expression of hWAPL.
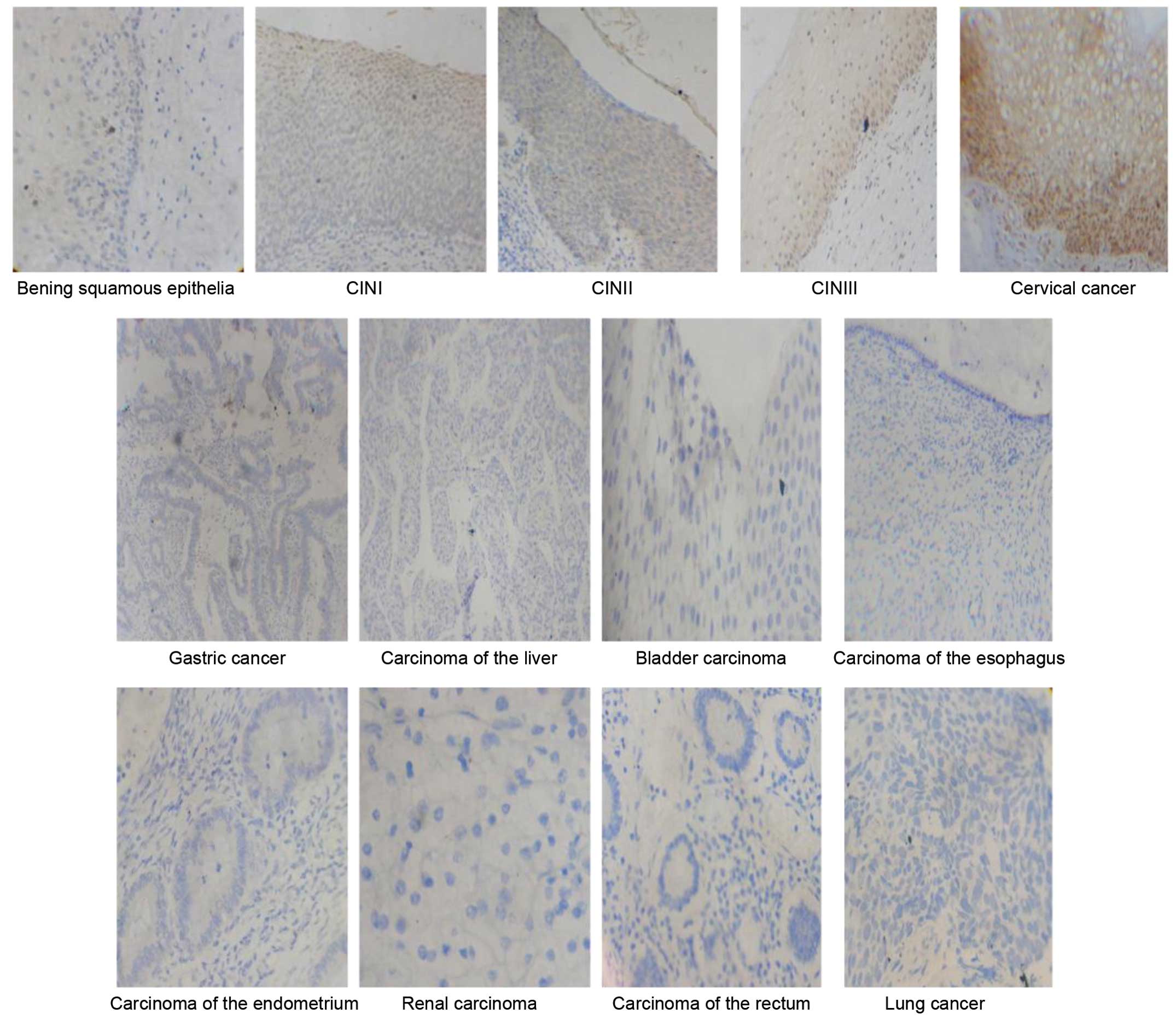 | Figure 1.Immunostaining for human wings
apart-like messenger RNA in benign squamous epithelia tissues,
CINI–III tissues and tissues from 9 types of cancer (magnification,
×400). Benign squamous epithelia, hWAPL-negative expression
0.52±0.70; CINI, hWAPL-positive expression 2.20±1.19; CINII,
hWAPL-positive expression 2.45±1.06; CINIII, hWAPL-positive
expression 4.89±1.67; cervical cancer, hWAPL-positive expression
6.49±2.54; gastric and lung cancer, carcinoma of the liver,
esophagus, endometrium and rectum and bladder and renal carcinoma,
hWAPL-negative expression. Staining scores presented as the mean ±
standard deviation. |
 | Table I.The staining scores of benign squamous
epithelia, CINI, CINII, CINIII and cervical cancer. |
Table I.
The staining scores of benign squamous
epithelia, CINI, CINII, CINIII and cervical cancer.
|
| Staining scores |
|
|
|---|
|
|
|
|
|
|---|
| Tissue type | Range | Mean ± standard
deviation | F | P-value |
|---|
| Benign squamous
epithelia | 0.0–3.1 |
0.52±0.70a | 70.26 | <0.001 |
| CINI | 1.2–4.3 |
2.20±1.19b |
|
|
| CINII | 1.3–4.2 |
2.45±1.06c |
|
|
| CINIII | 2.5–8.1 |
4.89±1.67d |
|
|
| Cervical cancer |
2.6–12.3 |
6.49±2.54e |
|
|
 | Table II.The rate of positive cell in benign
squamous epithelia, CINI, CINII, CINIII and cervical cancer. |
Table II.
The rate of positive cell in benign
squamous epithelia, CINI, CINII, CINIII and cervical cancer.
| Tissue type | Total, n | Tissues expressing
hWAPL, n (%) | χ2 | P-value |
|---|
| Benign squamous
epithelia | 27 | 1
(3.70)a | 87.531 | <0.001 |
| CINI | 30 | 12
(40.0)b |
|
| CINII | 33 | 14
(42.4)c |
|
| CINIII | 38 | 34
(89.5)d |
|
| Cervical cancer | 47 | 46
(97.9)e |
|
Protein level of hWAPL in other cancer
tissues
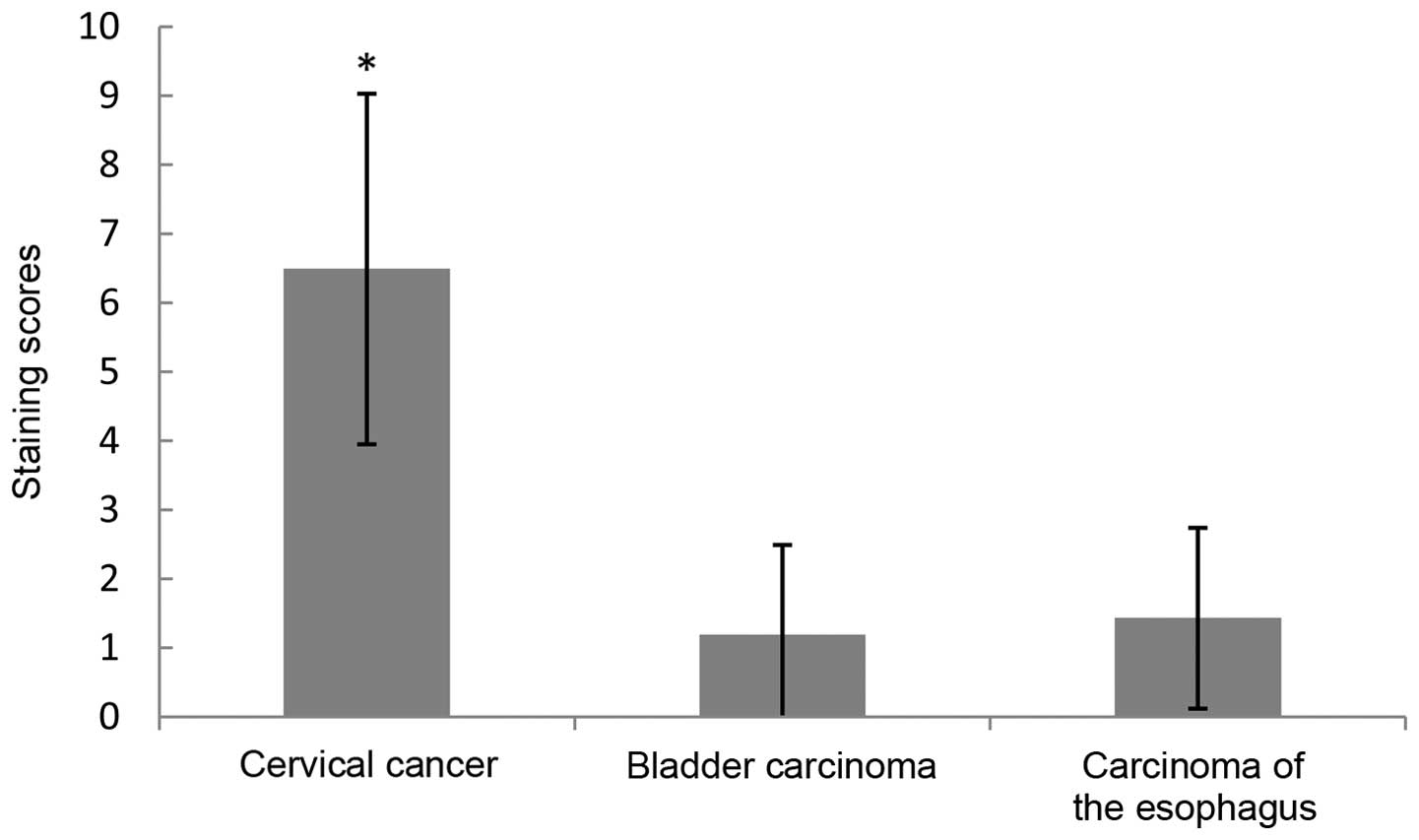
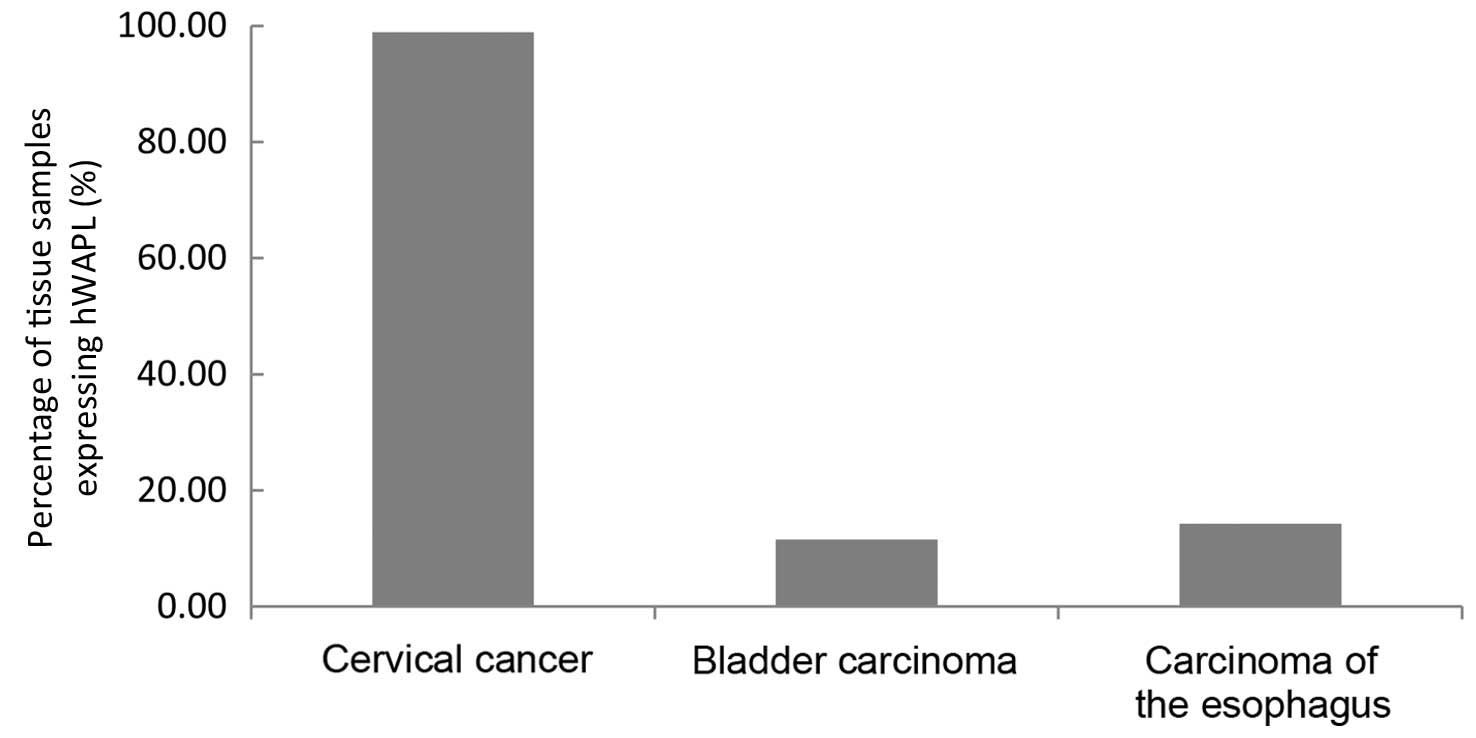
The expression of hWAPL was not present in gastric
cancer, liver, endometrial, renal or rectal carcinoma, or lung
cancer. In contrast to the staining scores and the percentage of
tissues expressing hWAPL in bladder carcinoma (staining score,
1.19±1.03; expression rate, 11.54%) and esophageal carcinoma
(staining score, 1.43±1.31; expression rate, 14.28%), the staining
scores and the percentage of tissues expressing hWAPL were
significantly increased in cervical cancer (P<0.001; Figs. 4 and 5).
The hWAPL gene was overexpressed only in cervical cancer.
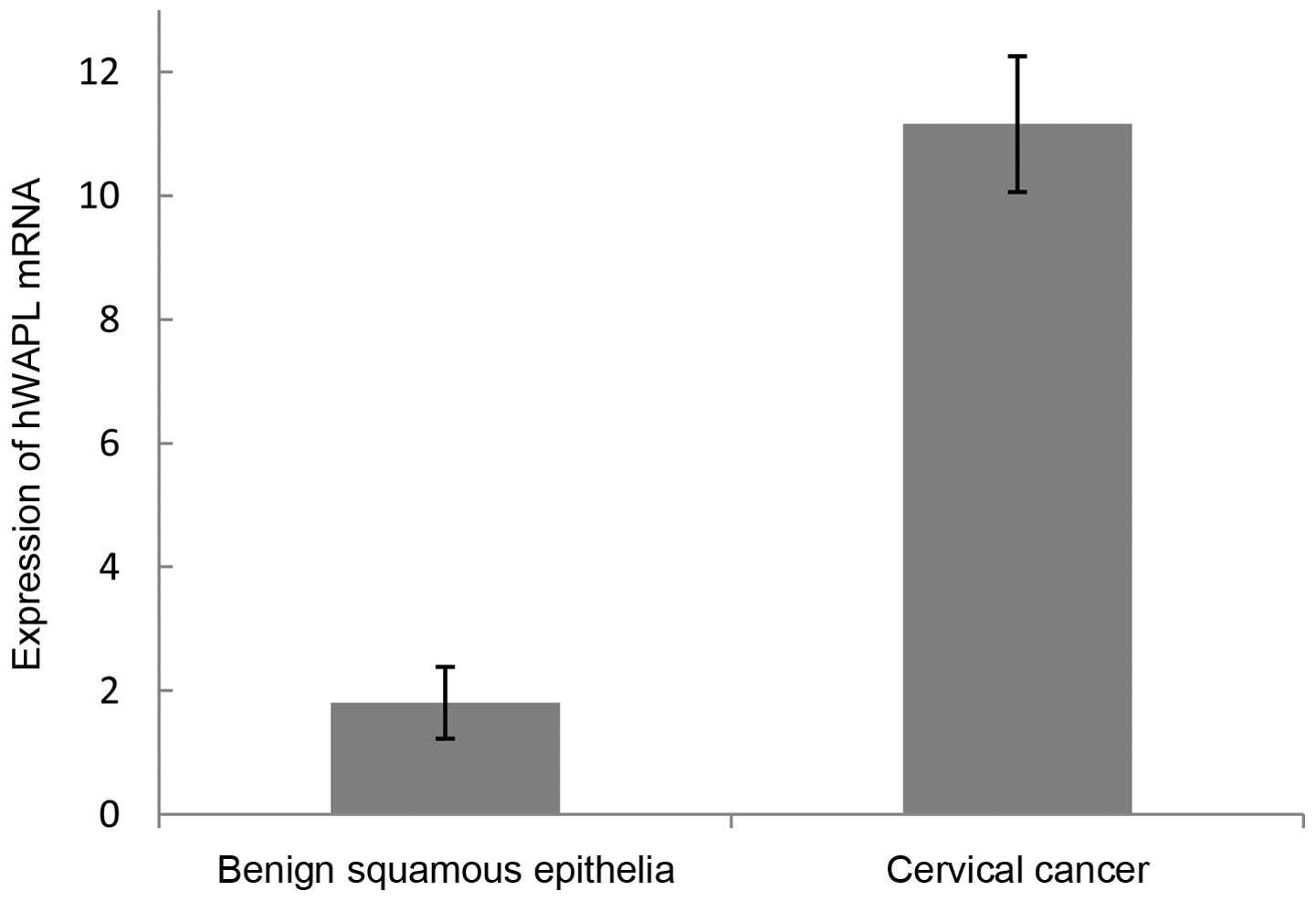
hWAPL mRNA level in cervical carcinoma
and normal cervical tissues
In total, 10 cervical carcinoma tissues and 7 normal
cervical tissues were examined for hWAPL gene expression by
RT-qPCR. The mean hWAPL mRNA level was 11.16±1.20 in cervical
cancer tissues and 1.81±0.58 in normal cervical tissues (Table III; Fig.
6). The difference between cervical tissues and normal cervical
tissues was significant (t=21.838; P<0.01). Thus, the
expression of the hWAPL gene was significantly increased in
cervical carcinoma tissues.
 | Table III.Expression of hWAPL mRNA in benign
squamous epithelia and cervical cancer. |
Table III.
Expression of hWAPL mRNA in benign
squamous epithelia and cervical cancer.
| Tissue type | Total, n | hWAPL mRNA level | t | P-value |
|---|
| Benign squamous
epithelia | 8 | 1.81±0.58 | 21.838 | <0.001 |
| Cervical cancer | 11 | 11.16±1.097 |
|
|
Discussion
The most important finding of the present study was
that the expression level of the hWAPL gene was significantly
increased in cervical cancer tissues compared with the 8 other
common cancers analyzed. Findings of previous study have suggested
that the hWAPL gene is specifically overexpressed in cervical
cancer (7). It is well known that a
pathological cervical lesion develops through step-by-step events,
progressing between normal benign squamous epithelia, cervical
intraepithelial neoplasia and cervical cancer. The present study
demonstrated that the staining scores and the percentage of tissues
expressing hWAPL increased gradually between normal benign squamous
epithelia, CINI, CINII, CINIII and cervical cancer. In normal
benign squamous epithelia, there was no expression of hWAPL or the
expression was limited to the basement membrane of the epithelium.
In addition, the expression of hWAPL was located in the bottom of
the epithelium. Notably, the expression of hWAPL was positive in
the total layer of epithelium in CINIII and cervical cancer. These
results indicated that the overexpression of hWAPL may play an
important role in the occurrence and development of cervical
cancer. This is in accordance with the analyses performed by Oikawa
et al (7).
Currently, the cause of hWAPL
specific-overexpression is not clear in cervical cancer. To the
best of our knowledge, it is well understood that, as a dominant
reason, HPV play a canonical role in occurrence and development of
cervical cancer (9,10). The expression of HPV E6 and E7
oncogenes is responsible for cervical neoplasia (11). An association has been revealed
between hWAPL overexpression and HPV infection in a previous study
(12). hWAPL expression was increased
by HPV E6 and E7 oncoproteins (12).
In the present study, expression of hWAPL, which was observed in
certain esophageal carcinoma tissues, may be due to HPV infection.
The pathway of hWAPL overexpression and the association between
hWAPL over-expression and HPV infection require additional
elucidation.
In conclusion, the hWAPL gene may be specifically
overexpressed in cervical cancer. The expression of hWAPL is
associated with the grade of the cervix lesion, and the hWAPL gene
may be a novel target for the diagnosis and therapy of cervical
cancer.
Acknowledgements
This work was supported by the Natural Science
Foundation of the Education Department of Henan Province, China
(grant no. 2006320012).
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pan Z, Li J, Pan X, Chen S, Wang Z, Li F,
Qu S and Shao R: Methylation of the RASSF1A gene promoter in Uigur
women with cervical squamous cell carcinoma. Tumori. 95:76–80.
2009.PubMed/NCBI
|
|
3
|
Dueñas-González A, Lizano M, Candelaria M,
Cetina L, Arce C and Cervera E: Epigenetics of cervical cancer. An
overview and therapeutic perspectives. Mol Cancer. 4:382005.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Verní F, Gandhi R, Goldberg ML and Gatti
M: Genetic and molecular analysis of wings apart-like (WAPL), a
gene controlling heterochromatin organization in drosophila
melanogaster. Genetics. 154:1693–1710. 2000.PubMed/NCBI
|
|
5
|
Dobie KW, Kennedy CD, Velasco VM, McGrath
TL, Weko J, Patterson RW and Karpen GH: Identification of
chromosome inheritance modifiers in drosophila melanogaster.
Genetics. 157:1623–1637. 2001.PubMed/NCBI
|
|
6
|
Gandhi R, Gillespie PJ and Hirano T: Human
Wapl is a cohesin-binding protein that promotes sister-chromatid
resolution in mitotic prophase. Curr Biol. 16:2406–2417. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oikawa K, Ohbayashi T, Kiyono T, Nishi H,
Isaka K, Umezawa A, Kuroda M and Mukai K: Expression of a novel
human gene, human wings apart-like (hWAPL), is associated with
cervical carcinogenesis an tumor progression. Cancer Res.
64:3545–3549. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Adersson S, Rylander E, Larsson B, Strand
A, Silfversvärd C and Wilander E: The role of human papillomavirus
in cervical adenocarcinoma carcinogenesis. Eur J Cancer.
37:246–250. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Muñoz N, Bosch FX, de Sanjosé S, Herrero
R, Castellsagué X, Shah KV, Snijders PJ and Meijer CJ:
International Agency for Research on Cancer Multicenter Cervical
Cancer Study Group: Epidemiologic classification of human
papillomavirus types associated with cervical caner. N Engl J Med.
348:518–527. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Doorbar J, Quint W, Banks L, Bravo IG,
Stoler M, Broker TR and Stanley MA: The biology and life-cycle of
human papillomaviruses. Vaccine. 30(Suppl 5): F55–F70. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kuroda M, Kiyono T, Oikawa K, Yoshida K
and Mukai K: The human papillomavirus E6 and E7 inducible oncogene,
hWAPL, exhibits potential as a therapeutic target. Br J Cancer.
92:290–293. 2005.PubMed/NCBI
|