Introduction
Hepatocellular carcinoma (HCC) is the third most
prevalent cause of cancer-associated mortality worldwide, and is
one of the most lethal malignancies (1). Currently, HCC is refractory to
conventional chemotherapies. Early-stage HCC with normal liver
function may be effectively treated with liver resection,
transplantation or percutaneously, with patients experiencing a
considerable 5-year survival rate. However, the majority of
patients are diagnosed at later or inoperable stages (2). Generally, the progression of HCC is
characterized by numerous aberrant cell behaviors, including the
loss of tissue-specific gene expression, a decrease in cell
differentiation, an increase in proliferation and the induction of
metastasis (3). Patients with HCC
frequently present with tumor cell invasion and migration prior to
a definite diagnosis (4,5). Therefore, it is imperative to clarify
the molecular mechanisms underlying HCC etiology, and develop novel
therapeutic agents to successfully treat the disease.
With the development of deep gene sequencing
technology, an increasing amount of research is being focused on
non-coding RNAs (ncRNAs). ncRNAs are transcribed from the human
genome, and include microRNAs (miRNAs), small interfering RNAs
(siRNAs), long ncRNAs (lncRNAs) and PIWI-interacting RNAs (6,7). A large
number of studies have concentrated on siRNAs and miRNAs, with
their functions and molecular mechanisms being elucidated in recent
years (8–10). However, the understanding of the
functions of lncRNAs in various diseases remains extremely limited.
Recent studies have reported that lncRNAs may also serve a key role
in transcription, chromatin modification and post-transcriptional
processing (11–13).
The number of studies investigating lncRNAs has
increased unexpectedly in recent years (14,15). The
nuclear-enriched abundant transcript 2, MALAT1, located at human
chromosome 11q13, was reported to exhibit high expression in
numerous species, and was demonstrated to regulate primary
transcripts either transcriptionally or post-transcriptionally
(16,17). Lai et al (18) reported that MALAT1 expression was
upregulated in HCC tissues and cell lines and functioned as an
independent prognostic factor for HCC recurrence. HOX transcript
antisense RNA (HOTAIR) was noted as an additional lncRNA involved
in cancer migration and metastasis. Ishibashi et al
(19) reported that a high expression
level of HOTAIR was detected in patients with primary HCC. Patients
with high HOTAIR expression consistently exhibit poor prognoses and
a low 5-year survival rate. Another extensively investigated lncRNA
is maternally expressed 3 (MEG3), which serves an important role in
regulating growth and cell development. Braconi et al
(20) reported that MEG3 was
downregulated in HCC tumor tissues. Restoration of MEG3 in HCC
cells significantly inhibited cell growth and induced
apoptosis.
Deep gene sequencing technology has aided the
investigation of various dysregulated lncRNAs in HCC. However, when
compared with miRNAs, the current understanding of lncRNAs is
limited. Previous studies have demonstrated that colon cancer
associated transcript 2 (CCAT2) was upregulated in lung and gastric
cancer (21,22), indicating that the lncRNA possesses
oncogenic characteristics. The present study aimed to verify the
expression level of CCAT2 in HCC tissues and cell lines, and assess
the impact of CCAT2 on cell proliferation, migration and apoptosis,
with the results demonstrating that CCAT2 functions as a oncogene
in HCC.
Materials and methods
Patients and tissue samples
A total of 50 HCC tissue samples were obtained from
50 different patients between July 2008 and June 2013 from The
Second Xiangya Hospital of Central South University (Changsha,
China). None of the patients received percutaneous ablation,
chemoembolization or radiotherapy prior to surgery. For all
patients, paired tumor and non-tumor liver tissue samples were
collected immediately following liver resection and were snapfrozen
at −80°C until use. The pathological diagnosis was confirmed in all
cases by the Department of Pathology, The Second Xiangya Hospital
of Central South University. The clinical pathological data of the
patients is presented in Table I. The
Ethics Committee of The Second Xiangya Hospital of Central South
University granted approval for the study prior to sample
collection, and the patients or a family member signed a consent
form permitting the collection and use of their samples for the
present study.
 | Table I.Clinical and pathological
characteristics of patients with hepatocellular carcinoma. |
Table I.
Clinical and pathological
characteristics of patients with hepatocellular carcinoma.
| Characteristics | Number of
patients |
|---|
| Age, years |
|
| ≤50 | 22 |
|
>50 | 28 |
| Gender |
|
| Male | 37 |
|
Female | 13 |
| Tumor size, cm |
|
| ≤5 | 27 |
|
>5 | 23 |
| Clinical TNM
stage |
|
| I | 7 |
| II | 28 |
|
III | 13 |
| IV | 2 |
Cell culture and transfection
HCC cell lines (HepG2, HEP3B, HCCLM3 and HuH7) and
one normal liver cell line (L02) were obtained from the American
Type Culture Collection (Mannasas, VA, USA). The cells were
cultured with Dulbecco's modified Eagle's medium (Invitrogen;
Thermo Fisher Scientific, Inc., Waltham, MA, USA) supplemented with
10% fetal bovine serum (Invitrogen; Thermo Fisher Scientific,
Inc.). All cells were maintained at 37°C in a humidified chamber
with 95% air and 5% CO2.
CCAT2 small hairpin RNA (shRNA;target sequence,
5′-UUAACCUCUUCCUAUCUCATT-3′) were purchased from GeneChem Co., Ltd.
(Shanghai, China). The CCAT2 overexpression plasmid,
CCAT2-pcDNA3.1(+), was synthesized by Life Technologies (Thermo
Fisher Scientific, Inc.). HepG2 and HuH7 cells were transfected
with 2 µg of CCAT2 shRNA or CCAT2-pcDNA3.1(+) vector using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The first-strand cDNA was synthesized using oligo-dT
primers from the RevertAid™ First Strand cDNA Synthesis kit
(catalog no., K1621; Thermo Fisher Scientific, Inc.). The primers
used were as follows: Human CCAT2, forward,
5′-CCCTGGTCAAATTGCTTAACCT-3′, and reverse,
5′-TTATTCGTCCCTCTGTTTTATGGAT-3′; and human glyceraldehyde
3-phosphate dehydrogenase (GAPDH), forward,
5′-CCACATCGCTCAGACACCAT-3′, and reverse 5′-ACCAGGCGCCCAATACG-3′.
GAPDH was utilized as an internal control. RT-qPCR was performed
using the SYBR® Premix Ex Taq™ II (Tli RNaseH Plus)
(catalog no., RR820A; Takara Biotechnology Co., Ltd., Dalian,
China) according to the manufacturer's protocol on the Light-cycler
480® II Real-Time PCR System (Roche Diagnostics, Basel,
Switzerland). The results were normalized using the
2−ΔΔCq method (23). The
conditions for PCR were as follows: 98°C for 5 min, followed by 45
cycles of 98°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec. The
experiments were performed in triplicate and repeated at least
three times.
Cell proliferation assay
Cell growth was analyzed using an MTT assay
performed according to the manufacturer's protocol. HepG2 and HuH7
cells (~1×104 cells) were seeded into a 96-well culture
plate 24 h prior to transfection with CCAT2 shRNA and shRNA
negative control, or CCAT2-pcDNA3.1(+) vector and pcDNA3.1(+) empty
vector. After 0, 24, 48 or 72 h transfection, 20 µl
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
5 mg/ml; Invitrogen; Thermo Fisher Scientific, Inc.) was added to
each plate and incubated for 4 h at 37°C. Subsequently, 150 µl
dimethyl sulfoxide was added to each plate and agitated for 10 min
at 37°C in order to solubilize the crystals. The number of cells
per plate were measured by the absorbance (540 nm) at the indicated
time points. Assays were repeated at least three times.
Cell migration assay
The 24-well Boyden chamber with 8-µm pore size
polycarbonate membrane (Corning Incorporated, Corning, NY, USA) was
used to evaluate cell motility. HepG2 and HuH7 cells were
transfected with CCAT2 shRNA and negative control, or
CCAT2-pcDNA3.1(+) vector and pcDNA3.1(+) empty vector using
Lipofectamine 2000. Matrigel (BD Biosciences, San Jose, CA, USA)
was used to simulate a matrix barrier for the migration assay.
Following 48 h of transfection, a total of 4×103 cells
were seeded in the upper chamber with 200-µl serum-free medium. A
total of 600 µl of medium containing 20% serum, which served as a
chemoattractant, was added into the lower chamber. Following 24 h
of incubation, the membranes were fixed with methanol and stained
with 0.1% crystal violet at 37°C. Three visual fields were randomly
selected from each membrane, and the number of cells were counted
using a light-microscope. Experiments were run in three independent
repeats in triplicate, and were examined in a double-blind manner
by at least two observers.
Cell apoptosis assay
At 48 h post-transfection, apoptosis induced by
CCAT2 shRNA and negative control, or CCAT2-pcDNA3.1(+) vector and
pcDNA3.1(+) empty vector, was evaluated by calculating the activity
of caspase 3 using the Human Caspase 3 ELISA kit (Cusabio, Wuhan,
China) and Hoechst 33258 staining (Invitrogen; Thermo Fisher
Scientific, Inc.). The results were observed by a Laser Scanning
Confocal Microscope (Zeiss AG, Oberkochen, Germany) and analyzed
using Image-Pro® Plus 5.1 software (Media Cybernetics
Inc, Bethesda, MD, USA). All experiments were performed three
times.
Statistical analysis
Statistical analysis was conducted using SPSS 17.0
(SPSS, Inc., Chicago, IL, USA). Statistical significance was
determined with Student's t-test. Data are presented as the mean ±
standard deviation. For the comparison of CCAT2 expression levels
in matched tumor vs. normal samples, a paired t-test was used.
P<0.05 was considered to indicate a statistically significant
difference.
Results
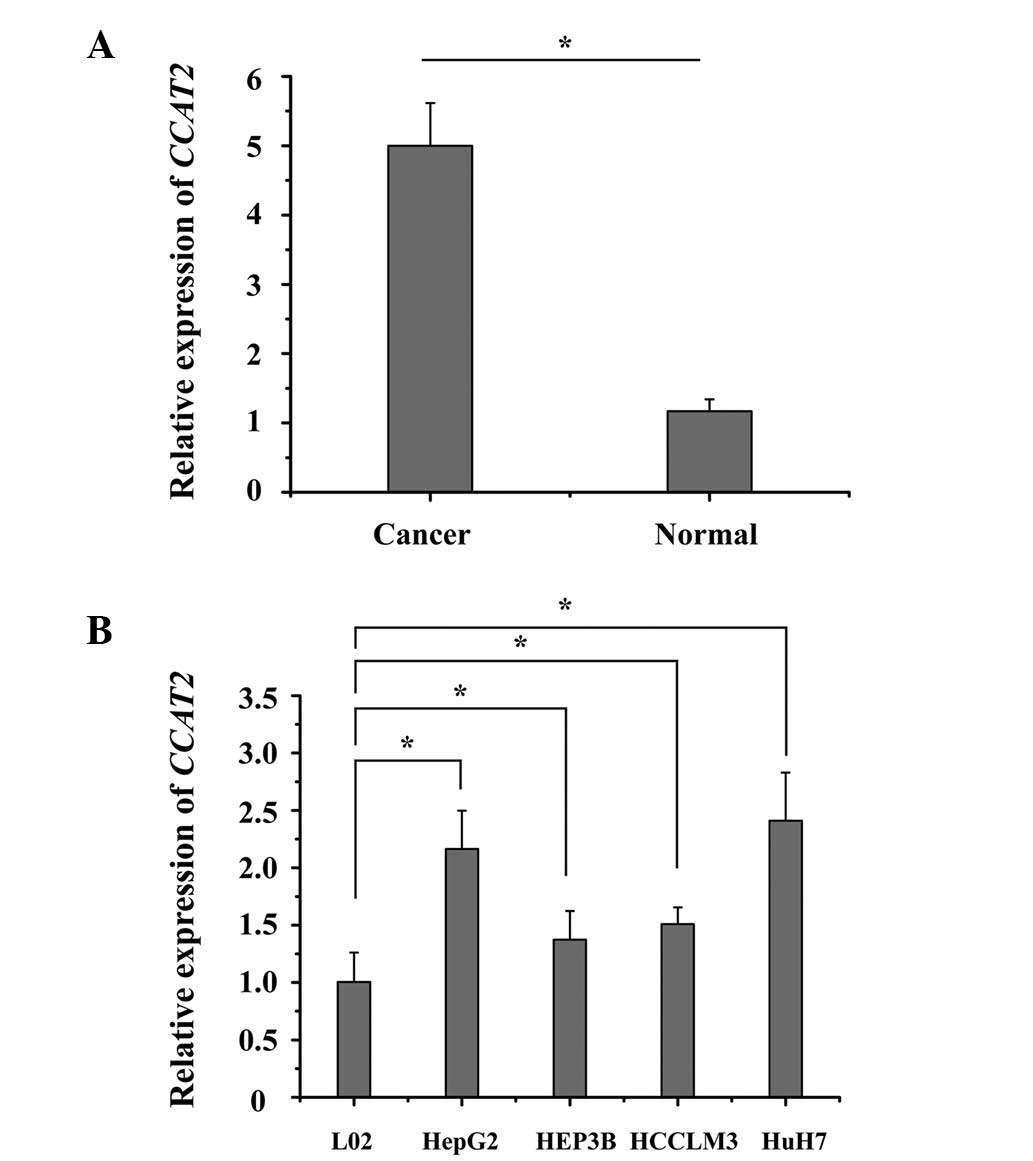
CCAT2 is upregulated in HCC tissues
and cell lines
The endogenous expression of CCAT2 in human HCC
tissues was compared with adjacent non-cancerous tissues by
RT-qPCR. It was observed that the expression of CCAT2 was
significantly upregulated in the HCC tissues when compared with the
matched normal liver tissue (P<0.05; Fig. 1A). Furthermore, it was demonstrated
that CCAT2 expression was significantly higher in all 4 HCC cell
lines when compared with that in the normal liver epithelial L02
cells (P<0.05; Fig. 1B).
Collectively, these results suggest that CCAT2 is upregulated in
HCC tissues and cell lines.
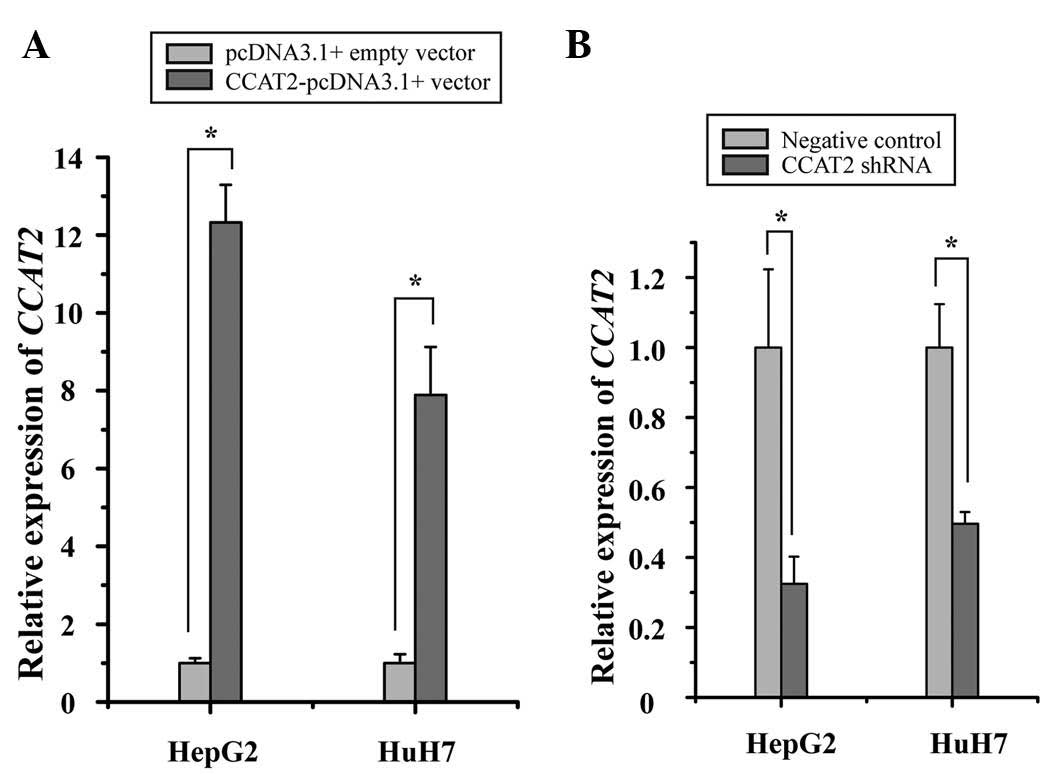
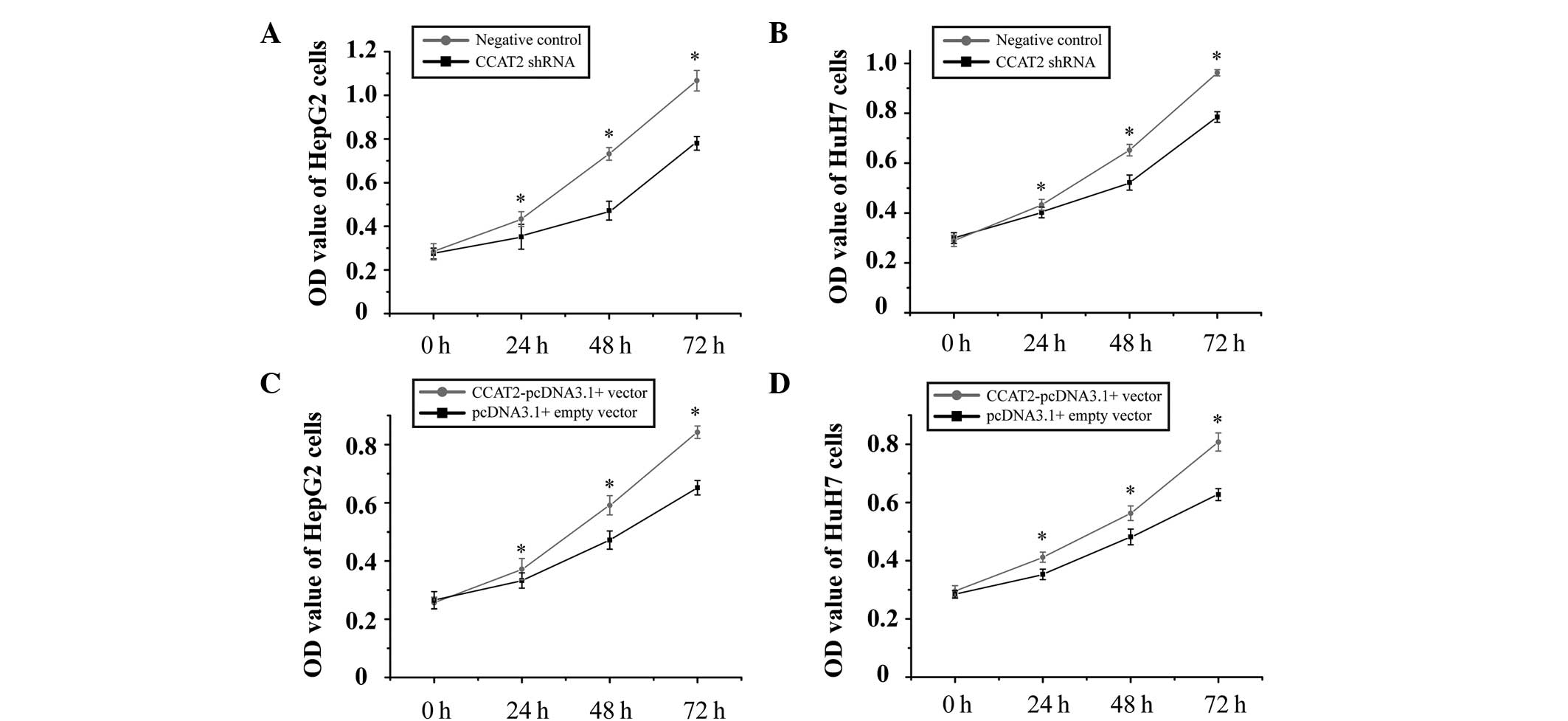
CCAT2 promotes proliferation of HCC
cells
To investigate the potential role of CCAT2 in HCC,
the inhibitory effect of CCAT2 on HCC cells was analyzed. HepG2 and
HuH7 cell proliferation was measured by MTT assay following
transfection with CCAT2 shRNA and negative control, or
CCAT2-pcDNA3.1(+) vector and pcDNA3.1(+) empty vector. As expected,
transfection of the CCAT2-pcDNA3.1(+) vector into the HepG2 and
HuH7 cells resulted in a significant increase in CCAT2 expression
when compared with the pcDNA3.1(+) empty vector (Fig. 2A), and the transfection of the CCAT2
shRNA into the HepG2 and HuH7 cells resulted in a decrease in CCAT2
expression when compared with negative control (Fig. 2B). Furthermore, it was observed that
the suppression of CCAT2 expression significantly inhibited the
proliferation of the HCC cells, while an increase in its expression
promoted cell proliferation (Fig. 3).
These results suggest that CCAT2 may function as an oncogene in the
development of HCC.
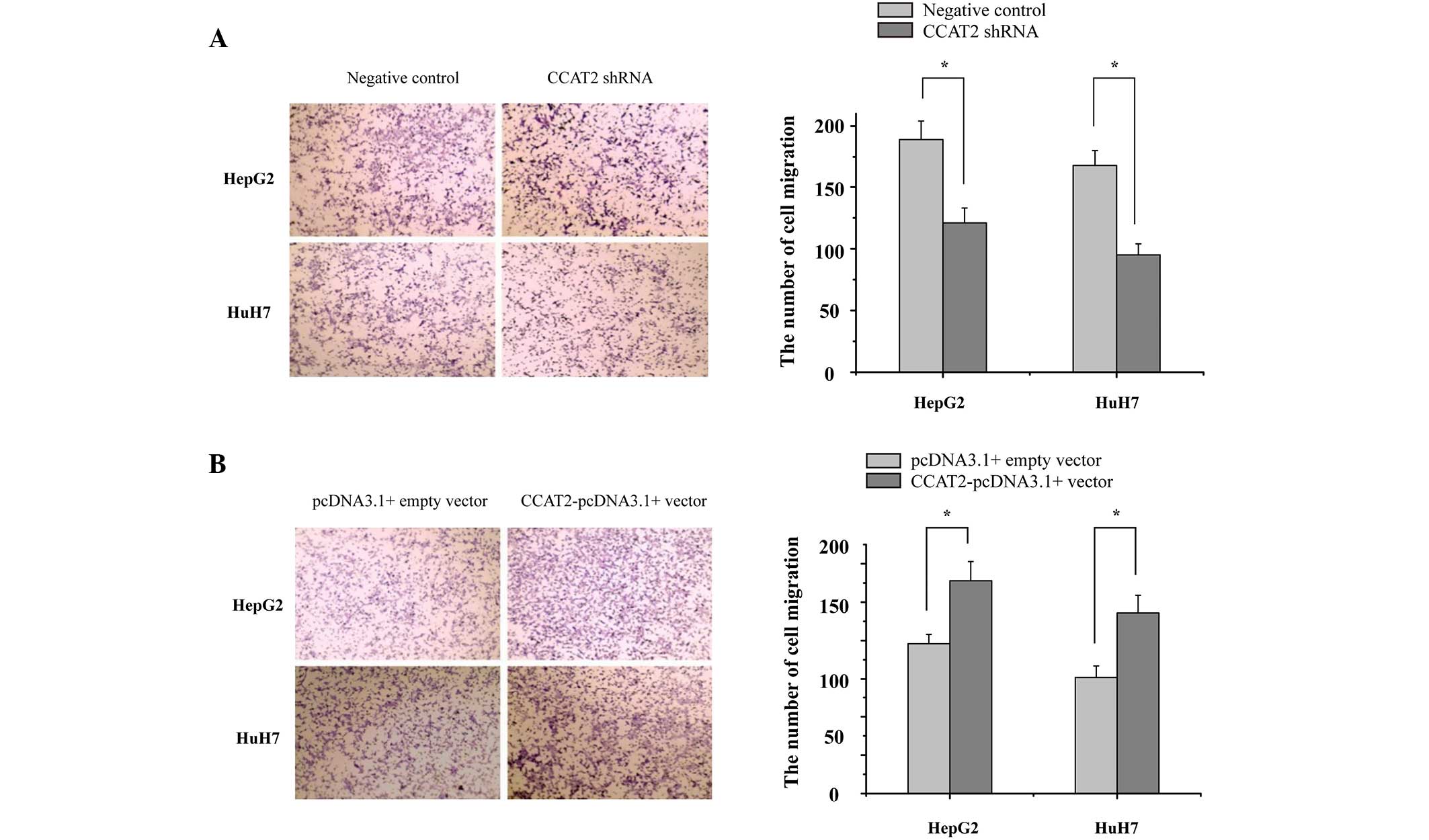
CCAT2 promotes HCC cell migration
In order to identify the potential role of CCAT2 in
HCC metastasis, the present study investigated the effect of CCAT2
on the migratory capacity of HCC cells. HepG2 and HuH7 cells were
transfected with CCAT2 shRNA and negative control or
CCAT2-pcDNA3.1(+) vector and pcDNA3.1(+) empty vector, and were
then evaluated by cell migration assays. The results demonstrated
that the suppression of CCAT2 expression significantly reduced the
migration rate of the HepG2 and HuH7 cells when compared with the
control (Fig. 4A). By contrast, the
overexpression of CCAT2 significantly increased the migration rate
of the HepG2 and HuH7 cells when compared with the control
(Fig. 4B). These results indicate
that CCAT2 functions as a oncogenic lncRNA and promotes the
migration of HCC cells.
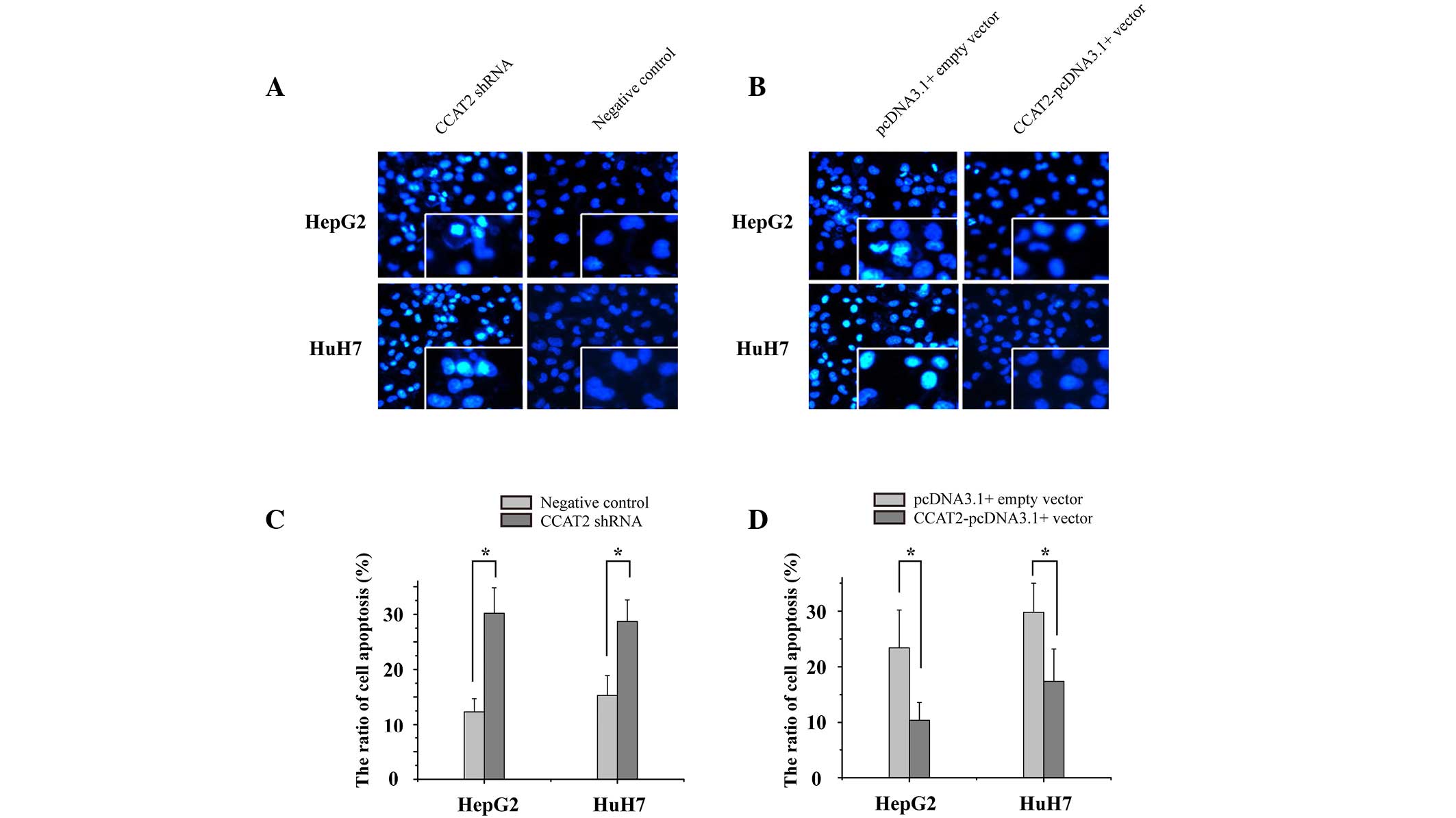
CCAT2 inhibits HCC cell apoptosis
It has been previously reported that lncRNAs serve a
crucial role in cell apoptosis, particularly in the avoidance of
apoptosis observed in cancer cells. To determine the impact of
CCAT2 on HCC cell apoptosis, an enzyme-linked immunosorbent assay
was utilized to measure the rate of apoptosis in the HCC cell
lines. As presented in Fig. 5, the
apoptosis rates of the HepG2 cells transfected with CCAT2 shRNA and
the CCAT2-pcDNA3.1(+) vector were 28.5 vs. 2.3%, respectively, and
the apoptosis rates of the HuH7 cells were 26.1 vs. 16.5%,
respectively; thus, demonstrating the upregulation of
CCAT2-inhibited HCC cell apoptosis. When combined, the results
provide strong evidence of CCAT2 functioning as a oncogene in HCC
development.
Discussion
An increasing number of studies have demonstrated
that the dysregulation of certain lncRNAs may contribute to
tumorigenesis (24,25). lncRNAs form a large family of
highly-conserved molecules that regulate a wide array of
cancer-associated genes, and may therefore be considered as a novel
family of tumor suppressor genes and oncogenes (24,25). The
lncRNA CCAT2 is located on chromosome 8q24.21, and is
differentially expressed in benign and malignant tumors (26–28).
Previous studies have reported that CCAT2 is overexpressed in
gastric and lung cancer (21,22); however, to the best of our knowledge,
the expression pattern and functional role of CCAT2 in HCC has not
yet been elucidated.
HCC is the fifth most prevalent malignancy and
accounts for 85–90% of all primary liver cancer cases (29). A number of studies have reported that
lncRNAs serve an important function in the regulation of gene
expression in cancer development (30–32). The
dysregulation of lncRNAs has been identified in various types of
cancer, including HOTAIR in ovarian and colorectal cancer, GAS5 in
breast cancer and MALAT1 in prostate cancer (33–35). The
dysregulation of lncRNAs has also been notably indicated in HCC,
with H19, hepatocellular carcinoma up-regulated EZH2-associated
lncRNA, microvascular invasion in HCC, hepatocellular carcinoma
up-regulated lncRNA and MEG3 demonstrating dysregulation in HCC
(36–38). Therefore, determination of the
expression profile and function of aberrant lncRNAs in HCC would be
particularly useful to further understand how lncRNAs assist the
development of the disease.
In the present study, the expression of CCAT2 was
analyzed in 50 HCC tissues samples, with the results demonstrating
that CCAT2 expression is higher in HCC tissues compared with paired
adjacent non-tumoral tissues. Recent studies have reported that
CCAT2 was upregulated in numerous types of cancer, which suggests
that CCAT2 may function as a oncogene in cancer (22,39).
Previous studies have also demonstrated that CCAT2, a regulator of
malignant cell progression, was associated with metastatic
processes in human cancer. To elucidate the role of CCAT2 in the
development of HCC, the present study performed HCC cell
transfection. Restoration of CCAT2 in the HCC cells, as a result of
transfection with the synthetic overexpression CCAT2-pcDNA3.1(+)
vector, was demonstrated to promote proliferation and migration. In
addition, the downregulation of CCAT2 in the HCC cells inhibited
the proliferation and migration of the HCC cells. The results of
the present study suggest that CCAT2 serves a crucial role in HCC
cell proliferation, migration and apoptosis.
In conclusion, the current study demonstrated that
CCAT2 was upregulated in HCC tissue, and indicated that this lncRNA
may function in HCC development through the dysregulation of
cellular migration, proliferation and apoptosis. CCAT2 is a
promising biomarker and/or a therapeutic target for HCC, with
further study required to elucidate the underlying molecular
mechanisms of CCAT2 in HCC development.
References
|
1
|
Lin L, Liang H, Wang Y, Yin X, Hu Y, Huang
J, Ren T, Xu H, Zheng L and Chen X: MicroRNA-141 inhibits cell
proliferation and invasion and promotes apoptosis by targeting
hepatocyte nuclear factor-3β in hepatocellular carcinoma cells. BMC
Cancer. 14:8792014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Block TM, Mehta AS, Fimmel CJ and Jordan
R: Molecular viral oncology of hepatocellular carcinoma. Oncogene.
22:5093–5107. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Braconi C, Valeri N, Kogure T, Gasparini
P, Huang N, Nuovo GJ, Terracciano L, Croce CM and Patel T:
Expression and functional role of a transcribed noncoding RNA with
an ultraconserved element in hepatocellular carcinoma. Proc Natl
Acad Sci USA. 108:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li G, Zhang H, Wan X, Yang X, Zhu C, Wang
A, He L, Miao R, Chen S and Zhao H: Long noncoding RNA plays a key
role in metastasis and prognosis of hepatocellular carcinoma.
Biomed Res Int. 2014:7805212014.PubMed/NCBI
|
|
5
|
Cui M, Zheng M, Sun B, Wang Y, Ye L and
Zhang X: A long noncoding RNA perturbs the circadian rhythm of
hepatoma cells to facilitate hepatocarcinogenesis. Neoplasia.
17:79–88. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Dong P, Yu F, Fan X, Lin Z, Chen Y and Li
J: Inhibition of ATIR by shRNA prevents collagen synthesis in
hepatic stellate cells. Mol Cell Biochem. 344:195–202. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gomes AQ, Nolasco S and Soares H:
Non-coding RNAs: Multi-tasking molecules in the cell. Int J Mol
Sci. 14:16010–16039. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao SM, Xing CY, Chen CQ, Lin SS, Dong PH
and Yu FJ: miR-15a and miR-16-1 inhibit the proliferation of
leukemic cells by down-regulating WT1 protein level. J Exp Clin
Cancer Res. 30:1102011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yu FJ, Dong PH, Fan XF, Lin Z, Chen YP and
Li J: Down-regulation of angiotensin II by shRNA reduces collagen
synthesis in hepatic stellate cells. Int J Mol Med. 25:801–806.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Chen SL, Zheng MH, Yang T, Song M and Chen
YP: Disparate profiles of dys-regulated miRNAs in activated hepatic
stellate cells. Hepatology. 57:1285–1286. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kitagawa M, Kotake Y and Ohhata T: Long
non-coding RNAs involved in cancer development and cell fate
determination. Curr Drug Targets. 13:1616–1621. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yoon JH, Abdelmohsen K, Srikantan S, Yang
X, Martindale JL, De S, Huarte M, Zhan M, Becker KG and Gorospe M:
LincRNA-p21 suppresses target mRNA translation. Mol Cell.
47:648–655. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Yoon JH, Abdelmohsen K and Gorospe M:
Posttranscriptional gene regulation by long noncoding RNA. J Mol
Biol. 425:3723–3730. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang Y, Yang L and Chen LL: Life without
A tail: New formats of long noncoding RNAs. Int J Biochem Cell
Biol. 54:338–349. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nadal-Ribelles M, Solé C, Xu Z, Steinmetz
LM, de Nadal E and Posas F: Control of Cdc28 CDK1 by a
stress-induced lncRNA. Mol Cell. 53:549–561. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang F, Yi F, Han X, Du Q and Liang Z:
MALAT-1 interacts with hnRNP C in cell cycle regulation. FEBS Lett.
587:3175–3181. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Shi X, Sun M, Liu H, Yao Y and Song Y:
Long non-coding RNAs: A new frontier in the study of human
diseases. Cancer Lett. 339:159–166. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lai MC, Yang Z, Zhou L, Zhu QQ, Xie HY,
Zhang F, Wu LM, Chen LM and Zheng SS: Long non-coding RNA MALAT-1
overexpression predicts tumor recurrence of hepatocellular
carcinoma after liver transplantation. Med Oncol. 29:1810–1816.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ishibashi M, Kogo R, Shibata K, Sawada G,
Takahashi Y, Kurashige J, Akiyoshi S, Sasaki S, Iwaya T, Sudo T, et
al: Clinical significance of the expression of long non-coding RNA
HOTAIR in primary hepatocellular carcinoma. Oncol Rep. 29:946–950.
2013.PubMed/NCBI
|
|
20
|
Braconi C, Kogure T, Valeri N, Huang N,
Nuovo G, Costinean S, Negrini M, Miotto E, Croce CM and Patel T:
microRNA-29 can regulate expression of the long non-coding RNA gene
MEG3 in hepatocellular cancer. Oncogene. 30:4750–4756. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang CY, Hua L, Yao KH, Chen JT, Zhang JJ
and Hu JH: Long non-coding RNA CCAT2 is up-regulated in gastric
cancer and associated with poor prognosis. Int J Clin Exp Pathol.
8:779–785. 2015.PubMed/NCBI
|
|
22
|
Qiu M, Xu Y, Yang X, Wang J, Hu J, Xu L
and Yin R: CCAT2 is a lung adenocarcinoma-specific long non-coding
RNA and promotes invasion of non-small cell lung cancer. Tumour
Biol. 35:5375–5380. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Deng R, Liu B, Wang Y, Yan F, Hu S, Wang
H, Wang T, Li B, Deng X, Xiang S, et al: High expression of the
newly found long noncoding RNA Z38 promotes cell proliferation and
oncogenic activity in breast cancer. J Cancer. 7:576–586. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng W, Wang J, Zhang J, Cai J, Bai Z and
Zhang Z: TET2 regulates LncRNA-ANRIL expression and inhibits the
growth of human gastric cancer cells. IUBMB Life. 68:355–364. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang J, Qiu M, Xu Y, Li M, Dong G, Mao Q,
Yin R and Xu L: Long noncoding RNA CCAT2 correlates with smoking in
esophageal squamous cell carcinoma. Tumour Biol. 36:5523–5528.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Redis RS, Sieuwerts AM, Look MP, Tudoran
O, Ivan C, Spizzo R, Zhang X, de Weerd V, Shimizu M, Ling H, et al:
CCAT2, a novel long non-coding RNA in breast cancer: Expression
study and clinical correlations. Oncotarget. 4:1748–1762. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu J, Liu S, Ye F, Shen Y, Tie Y, Zhu J,
Jin Y, Zheng X, Wu Y and Fu H: The long noncoding RNA expression
profile of hepatocellular carcinoma identified by microarray
analysis. PLoS One. 9:e1017072014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Pan YF, Feng L, Zhang XQ, Song LJ, Liang
HX, Li ZQ and Tao FB: Role of long non-coding RNAs in gene
regulation and oncogenesis. Chin Med J (Engl). 124:2378–2383.
2011.PubMed/NCBI
|
|
31
|
Li J, Meng H, Bai Y and Wang K: Regulation
of lncRNA and its role in cancer metastasis. Oncol Res. 23:205–217.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Adams BD, Anastasiadou E, Esteller M, He L
and Slack FJ: The inescapable influence of noncoding RNAs in
cancer. Cancer Res. 75:5206–5210. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Faghihi MA, Modarresi F, Khalil AM, Wood
DE, Sahagan BG, Morgan TE, Finch CE, St Laurent G III, Kenny PJ and
Wahlestedt C: Expression of a noncoding RNA is elevated in
Alzheimer's disease and drives rapid feed-forward regulation of
beta-secretase. Nat Med. 14:723–730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Matouk IJ, DeGroot N, Mezan S, Ayesh S,
Abu-lail R, Hochberg A and Galun E: The H19 non-coding RNA is
essential for human tumor growth. PLoS One. 2:e8452007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen X, Liu L and Zhu W: Up-regulation of
long non-coding RNA CCAT2 correlates with tumor metastasis and poor
prognosis in cervical squamous cell cancer patients. Int J Clin Exp
Pathol. 8:13261–13266. 2015.PubMed/NCBI
|