Introduction
T-cell acute lymphoblastic leukemia (T-ALL)
comprises an aggressive malignancy, in which multiple genetic
abnormalities collaborate in the transformation of T-cell
progenitors. Abundant genetic alterations in T-ALL have been
identified by whole-genome sequencing and DNA copy number analysis
of candidate genes, including deletions and/or sequence mutations
of the cyclin-dependent kinase inhibitor 2A/B, lymphoid enhancer
binding factor 1, Notch homolog 1, translocation-associated
(Drosophila) (NOTCH1), F-box and WD repeat domain containing 7
(FBXW7), phosphatase and tensin homolog (PTEN), neuroblastoma RAS
viral oncogene homolog, Wilms tumor 1, plant homeodomain finger
protein 6 (PHF6), interleukin 7 receptor and runt related
transcription factor 1 genes (1–4).
PHF6 encodes a PHD factor containing four
nuclear localization signals and two PHD zinc finger domains, and
has a proposed role in the control of gene expression (5). Inactivating mutations of the PHF6
gene were originally found to be associated with a form of
syndromic X-linked mental retardation, Börjeson-Forssman-Lehmann
syndrome (6). In addition,
PHF6 has been identified as a novel key tumor suppressor
gene in T-ALL. PHF6 mutations were detected in adult and
pediatric patients with T-ALL, as well as also in adults with acute
myeloid leukemia (7,8); however, the association between
PHF6 mutations and clinical outcome in these patients has
not been fully determined.
NOTCH1 mutations have been shown to play an
important role in the pathogenesis of T-ALL (9); however, the clinical significance of the
co-existence of NOTCH1 and PHF6 mutations
(PHF6mutNOTCH1mut) has not been
sufficiently explored. The present study demonstrated the
characteristics of PHF6 mutations in adult Chinese patients
with T-ALL, with 10/16 of the detected mutations being reported for
the first time. In addition, the correlation between
PHF6mutNOTCH1mut co-existence
in T-ALL and event-free survival (EFS) was explored and found to be
significant in this cohort of adult T-ALL patients.
Materials and methods
Patients and samples
Bone marrow (BM) samples were collected from 79
adult patients (age, 14–62 years) with ALL (57 males and 22
females; 59 T-ALL and 20 B-ALL samples) at the First Affiliated
Hospital of Nanjing Medical University (Nanjing, China) between May
2008 and 2014. The diagnosis of ALL was made based on the
molecular, immunophenotypic, morphologic and cytogenetic criteria
established by the World Health Organization Diagnosis and
Classification of ALL (2008) (10).
Informed consent was obtained from all individuals prior to their
participation to the study, in accordance with the Declaration of
Helsinki. The study was approved by the Institutional Review Board
of Nanjing Medical University.
Mutation analysis of PHF6, NOTCH1, FBXW7, PTEN
and Janus kinase 1 (JAK1). Mutation analysis was performed
for PHF6 exons 2–10. Genomic DNA was isolated with a QIAamp DNA
Blood Mini kit (Qiagen Inc., Valencia, CA, USA) following the
manufacturer's instructions. DNA fragments spanning the above
PHF6 exons were amplified by polymerase chain reaction (PCR)
using AmpliTaq Gold (Applied Biosystems, Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and exon-specific primers (5). DNA sequencing was performed on the
purified PCR products. In addition, exon 26/N-terminal region of
the heterodimerization domain (HD-N), exon 27/C-terminal region of
the heterodimerization domain (HD-C), exon 28 and exon
34/proline-glutamic acid-serine-threonine (PEST) domain of
NOTCH1 were amplified for mutation screening, as previously
described (9). Exons in FBXW7,
PTEN and JAK1 were also screened as previously reported
(11,12).
Cytogenetic and molecular
analysis
Conventional cytogenetic analysis was performed
using unstimulated short-term cultures at the time of diagnosis,
according to the International System for Human Cytogenetic
Nomenclature recommendations (13).
For each sample, ≥20 BM metaphase cells were analyzed.
Immunophenotypic analysis was performed by flow
cytometry on fresh BM samples. The cell-surface antigen was
considered positive when fluorescence intensity of ≥20% of cells
exceeded the fluorescence of negative control.
Statistical analysis
Patients were divided into high or low PHF6
expression groups [the median (0.0020925) was used as cut-off value
based on SPSS 17.0]. For quantitative parameters, the overall
differences between the cohorts were evaluated using the
Mann-Whitney U-test. For qualitative parameters, the overall group
differences were analyzed using the χ2 test. Survival
analysis was calculated using the Kaplan-Meier method. Experimental
data are presented as the mean ± standard error. Determinations of
statistical significance were performed using a Student
t-test for comparisons of two groups or using analysis of
variance (ANOVA) for comparing multiple groups. Statistical
analysis was performed using the SPSS 17.0 software (SPSS Inc.,
Chicago, IL, USA). P<0.05 was considered to indicate a
statistically significant difference.
Results
PHF6 mutations in adult T-ALL
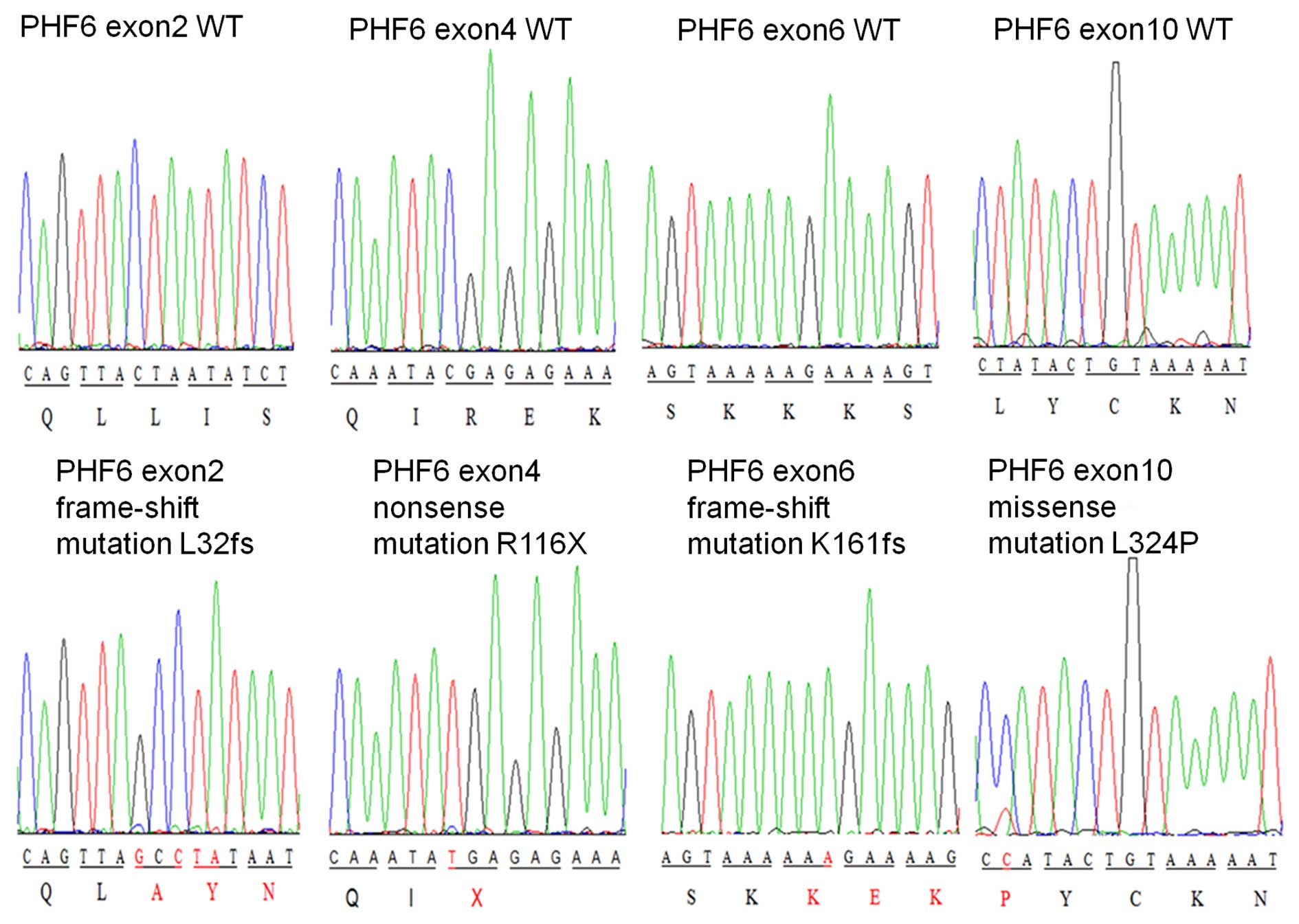
PHF6 mutations were detected in 27.1% of the Chinese
adults with T-ALL (16/59). The identified mutations located in
exons 2 and 4–10. The most common locations for mutations were in
exon 9 and exon 8, in which the mutation rate reached 25.0 and
18.8%, respectively.
The majority of the mutations detected were nonsense
mutations (7/16, 43.8%), followed by insertion (4/16, 25.0%) and
missense mutations (3/16, 18.75%), a deletion (1/16, 6.3%), and
insertion/deletion mutations (1/16, 6.3%) (Fig. 1 and Table
I). No PHF6 mutations were identified in the 20 DNA
samples from patients with B-ALL (data not shown), suggesting that
PHF6 mutations in lymphoid tumors could be restricted to
T-ALL. Of note, 10/16 (62.5%) PHF6 mutations identified in
the present study were novel mutations (Table I). In addition, 6/16 (37.5%) mutations
were frame-shift mutations, which may result in the deletion of the
gene.
 | Table I.Plant homeodomain finger protein 6
mutations in adult T-cell acute lymphoblastic leukemia. |
Table I.
Plant homeodomain finger protein 6
mutations in adult T-cell acute lymphoblastic leukemia.
| Patient ID | Mutation
(nucleotide) | Exon | Type of
mutation | Mutation (amino
acid) | Previously
reported |
|---|
| PHF6mu 1# | c.631A>T | 7 | Nonsense | p.K211X | N |
| PHF6mu 2# | c.479_480insA | 6 | Frame-shift | p.K161fs | N |
| PHF6mu 3# | c.820C>T | 8 | Nonsense | p.R274X | Y |
| PHF6mu 4# |
c.93_94insG+94_95insCTA | 2 | Frame-shift | p.L32fs | N |
| PHF6mu 5# | c.517A>T | 6 | Nonsense | p.K173X | N |
| PHF6mu 6# | c.821G>A | 8 | Missense | p.R274Q | Y |
| PHF6mu 7# | c.731_732delTG | 8 | Frame-shift | p.L244fs | N |
| PHF6mu 8# | c.955C>T | 9 | Nonsense | p.R319X | Y |
| PHF6mu 9# | c.385C>T | 5 | Nonsense | p.R129X | Y |
| PHF6mu 10# | c.346C>T | 4 | Nonsense | p.R116X | Y |
| PHF6mu 11# |
c.134delG+insCC | 2 | Frame-shift | p.C45fs | N |
| PHF6mu 12# | c.586_587insA | 7 | Frame-shift | p.R196 K | N |
| PHF6mu 13# | c.971T>C | 10 | Missense | p.L324P | N |
| PHF6mu 14# | c.957_958insGT | 9 | Frame-shift | p.G320fs | N |
| PHF6mu 15# | c.903C>A | 9 | Nonsense | p.Y301X | Y |
| PHF6mu 16# | c.905A>C | 9 | Missense | p.H302P | N |
PHF6 expression and its association
with mutations
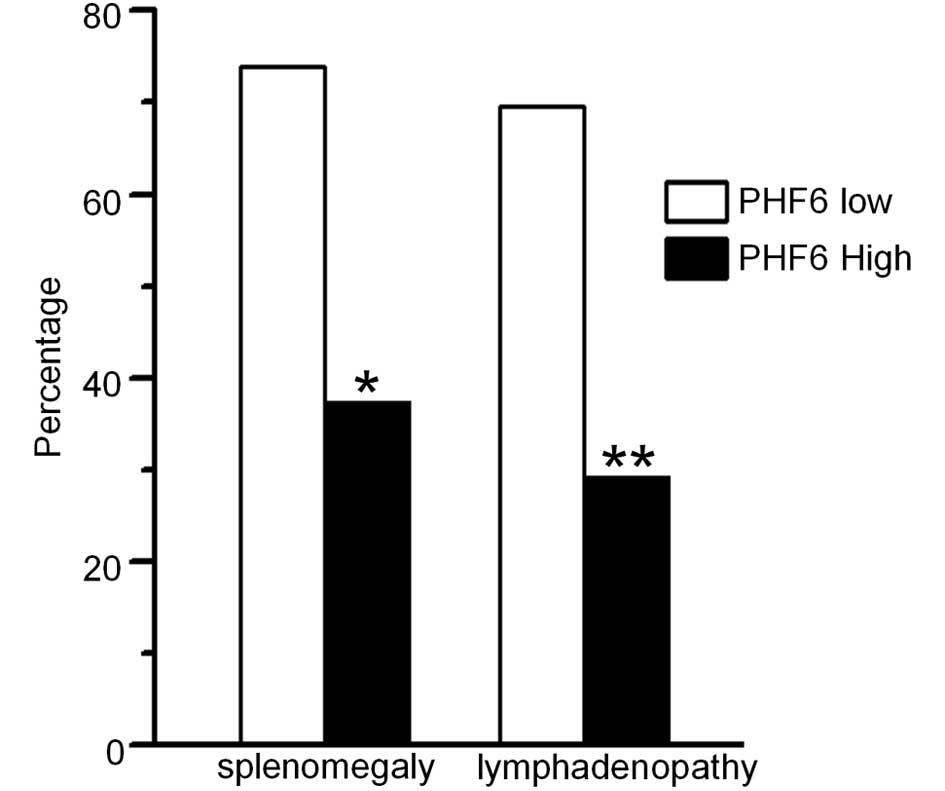
PHF6 mRNA expression was detected in 46 patients
whose cDNA samples were available. The PHF6 expression was
divided into high and low expression groups (G1–2 vs. G3–4). A
significant correlation was observed between low PHF6
expression levels and high frequency of splenomegaly and
lymphadenopathy in adult T-ALL [73.9 (17/23) vs. 37.5% (9/24)
(P=0.012) and 69.6 (16/23) vs. 29.2% (7/24) (P=0.006),
respectively] (Fig. 2), suggesting
that low PHF6 expression levels may be associated with the
markers of leukemic cell proliferation, which involved
extramedullary infiltration in T-ALL. Furthermore, it was observed
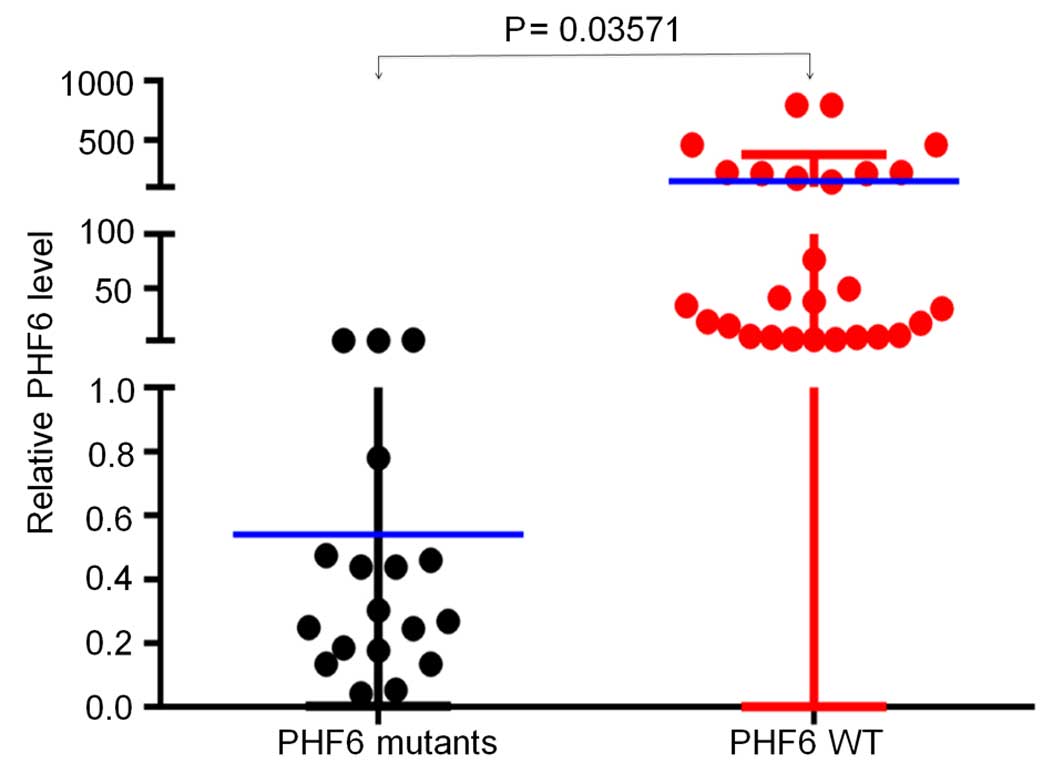
that PHF6 expression was significantly lower in patients
with T-ALL with PHF6 mutations, as compared with those with
PHF6 wild-type (WT) (0.00423 vs. 0.06464; P=0.035) (Fig. 3), which further indicated PHF6
mutations could result in loss of function mutations.
Cooperative genetic lesions of PHF6
mutations in adult T-ALL
The mutations in NOTCH1, FBXW7, PTEN and
JAK1 were also screened in this cohort of patients with
T-ALL. It was found that the frequency of
PHF6mutNOTCH1mut co-existence
was significantly higher in patients with PHF6 mutations
than in those with PHF6 WT (75.0 vs. 44.2; P=0.035),
suggesting an association between PHF6 and NOTCH1
mutations in this cohort (Table II).
No significant associations were observed between PHF6 and
FBXW7, PTEN or JAK1 mutations (Table II).
 | Table II.Association of PHF6 mutation with
clinical feature in adult T-cell acute lymphoblastic leukemia. |
Table II.
Association of PHF6 mutation with
clinical feature in adult T-cell acute lymphoblastic leukemia.
|
| PHF6 |
|---|
|
|
|
|---|
| Clinical
features | WT (n=43) | Mutation
(n=16) | P-value |
|---|
| Male, % | 83.7 | 68.8 | 0.365 |
| Age, median
(range) | 26 (14–62)
years | 39 (14–60)
years | 0.043 |
| WBC, median
(range) | 47.9 (1.2–546.0)
×109/l | 19.0 (2.7–175.3)
×109/l | 0.060 |
| Platelet, median
(range) | 56.0 (17.0–267.0)
×109/l | 53.0 (25.0–223.0)
×109/l | 0.871 |
| Hemoglobin, median
(range) | 119.0 (56.0–171.0)
g/l | 99.5 (56.0–153.0)
g/l | 0.044 |
| LDH, median
(range) | 1038.0
(131.0–8601.0) U/l | 811.5
(294.0–5353.0) U/l | 0.747 |
| Marrow blasts,
median (range) | 75.0
(22.0–98.0)% | 80.0
(27.0–99.0)% | 0.615 |
| Peripheral blasts,
median (range) | 80.0 (0–90.9)% | 28.0
(5.0–94.0)% | 0.080 |
| Immunophenotype
(≥20%) |
|
|
|
|
CD13 | 24.2 | 71.4 | 0.002 |
|
CD33 | 35.3 | 54.5 | 0.436 |
|
CD34 | 48.6 | 66.7 | 0.278 |
| Other genetic
abnormalities, % |
|
|
|
| Complex
Karyotype | 11.8 | 7.7 | 1.000 |
|
NOTCH1 mutation | 44.2 | 75.0 | 0.035 |
|
FBXW7 mutation | 11.6 | 6.3 | 0.902 |
|
PTEN mutation | 11.6 | 6.3 | 0.902 |
|
JAK1 mutation | 7.0 | 12.5 | 0.880 |
| Extramedullary
infiltration, % |
|
|
|
|
Hepatomegaly | 16.3 | 18.8 | 1.000 |
|
Splenomegaly | 39.5 | 68.8 | 0.046 |
|
Lymphadenopathy | 44.2 | 81.3 | 0.011 |
The domains involving
PHF6mutNOTCH1mut co-existence
were further analyzed. The most commonly mutated domains in
NOTCH1 co-existing with PHF6 mutations in same
patient were the HD-N domain (6/12, 50.0%), followed by HD-C (2/12,
16.7%), PEST (2/12, 16.7%), HD-C + PEST (1/12, 8.3%) and HD-N +
HD-C 1/12 (8.3%) (Table III). These
data indicated that the HD domain (particularly the HD-N) of
NOTCH1 may contribute to the synergistic oncogenic effect of
the two genes.
 | Table III.Correlation of PHF6 mutations
with NOTCH1 mutations in adult T-cell acute lymphoblastic
leukemia. |
Table III.
Correlation of PHF6 mutations
with NOTCH1 mutations in adult T-cell acute lymphoblastic
leukemia.
|
| PHF6
mutations | NOTCH1
mutations |
|---|
|
|
|
|
|---|
| Patient ID | Nucleotide | Exon | Amino acid | Nucleotide | Exon | Amino acid | Domain |
|---|
| PHF6mu 1# | c.631A>T | 7 | p.K211X | c.7355C>A | 34 | p.A2452E | PEST |
| PHF6mu 2# | c.479_480insA | 6 | p.K161fs |
c.4732_4734delGTG | 26 | p.V1578delV | HD-N |
| PHF6mu 3# | c.820C>T | 8 | p.R274X |
c.4732_4734delGTG | 26 | p.V1578delV |
|
|
|
|
|
| c.5094C>T |
| p.D1698D | HD-N |
| PHF6mu 4# | c.93_94ins | 2 | p.L32fs | c.5126T>C | 27 | p.L1709P | HD-C |
|
| G+94_95insCTA |
|
|
|
|
|
|
| PHF6mu 6# | c.821G>A | 8 | p.R274Q |
c.4815_4817delinsAGC | 26 | p.F1606AGGD | HD-N |
|
|
|
|
| CGGGGGGGA |
| p.E2515fs*1 |
|
|
|
|
|
|
c.7541_7542insC |
|
|
|
| PHF6mu 8# | c.955C>T | 9 | p.R319X | c.4799T>A | 26 | p.L1600Q | HD-N |
| PHF6mu 9# | c.385C>T | 5 | p.R129X | c.4721T>C | 26 | p.L1574P | HD-N |
| PHF6mu 10# | c.346C>T | 4 | p.R116X | c.5033T>C | 27 | p.L1678P | HD-C |
|
|
|
|
| c.7400C>A | 34 | p.S2467* | PEST |
| PHF6mu 13# | c.971T>C | 10 | p.L324P |
C.4845_4846insCCT | 26 |
p.1615_1616insP | HD-N |
|
|
|
|
| c.5114C>T | 27 | p.A1705 V | HD-C |
| PHF6mu 14# | c.957_958insGT | 9 | p.G320fs |
c.7368_7369insTA | 34 | p.L2457fs*21 | PEST |
| PHF6mu 15# | c.903C>A | 9 | p.Y301X |
c.4776_4777insGAA | 26 | p.F1592LNPTLP | HD-N |
|
|
|
|
| TCCAACCCTCCC |
|
|
|
| PHF6mu 16# | c.905A>C | 9 | p.H302P | c.5033T>C | 27 | p.L1678P | HD-C |
Correlation of PHF6 mutations with
clinical features
The association between PHF6 mutations and
clinical characteristics was analyzed in the patients with T-ALL.
No gender differences were observed in the incidence of PHF6
mutations in this cohort of adult Chinese patients with T-ALL.
It was found that the PHF6 mutations were
significantly associated with older age, lower hemoglobin levels,
higher frequency of CD13 positivity, and higher incidence of
splenomegaly or lymphadenopathy, as compared with PHF6 WT
patients (Table II).
Since an association was found between PHF6
and NOTCH1,
PHF6mutNOTCH1mut co-existence
was further analyzed in relation to the clinical features of the
cohort. It was found that the patients with
PHF6mutNOTCH1mut co-existence
had lower hemoglobin levels, along with a higher incidence of
splenomegaly or lymphadenopathy, as compared with patients without
such co-existence (Table III).
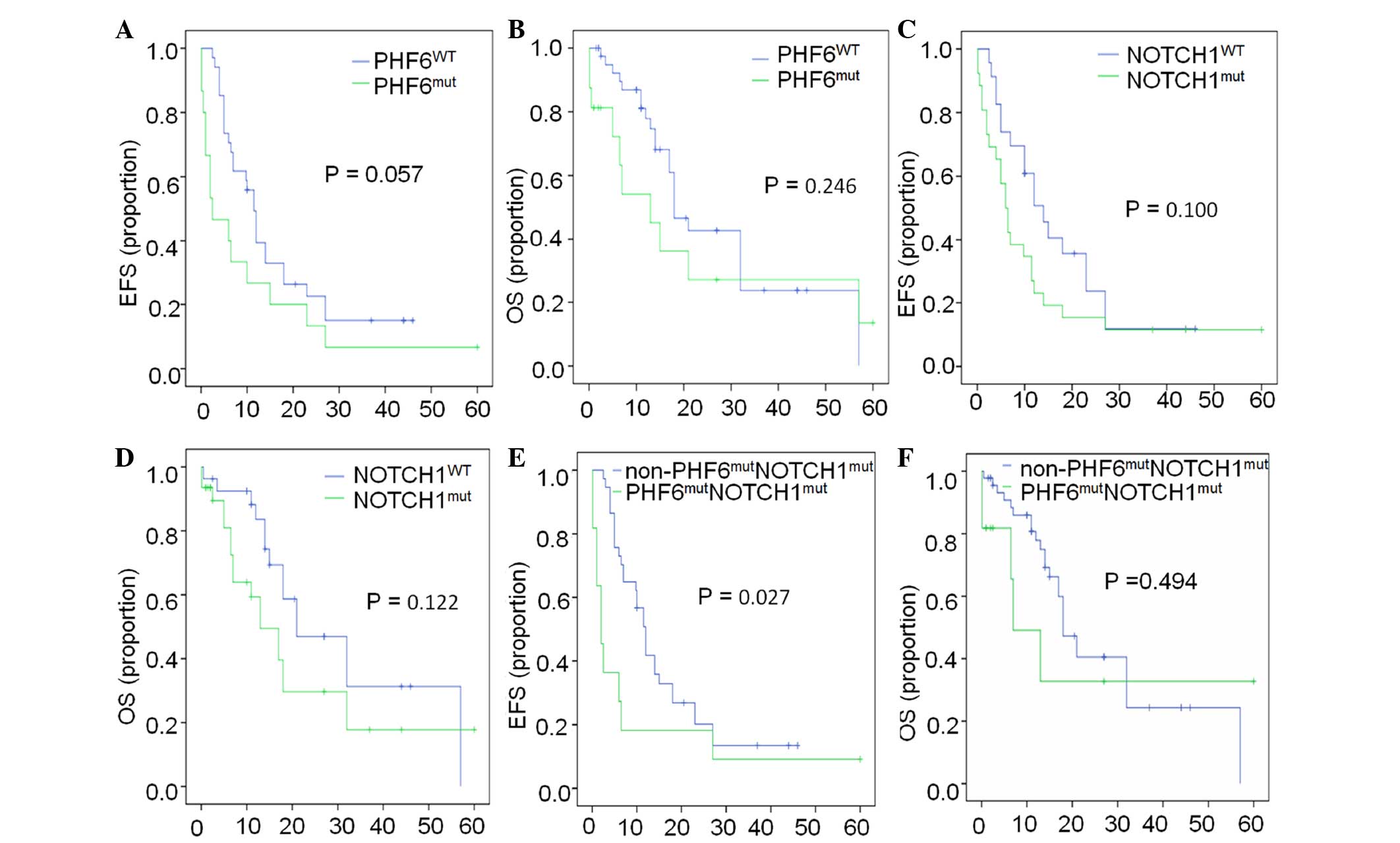
In addition, the survival status of this population
was analyzed. The EFS and overall survival (OS) in patients with
PHF6 mutations vs. PHF6 WT were 2.5 vs. 11.5 months
(P=0.057) and 13.0 vs. 18.0 months (P=0.246), respectively
(Fig. 4A and B), while those in
patients with NOTCH1 mutations vs. NOTCH1 WT were 6.0
vs. 14.0 months (P=0.100) and 13.0 vs. 21.0 months (P=0.122),
respectively (Fig. 4C and D). These
results indicated that there were no significant differences in the
EFS and OS between patients with PHF6 or NOTCH1
mutations and patients with PHF6 WT or NOTCH1 WT;
however, it was found that patients with
PHF6mutNOTCH1mut co-existence
had a significantly shorter EFS compared with that of patients
without such co-existence (2.0 vs. 12.0 months, respectively;
P=0.027). No differences in the OS were observed between patients
with PHF6mutNOTCH1mut
co-existence and those without (7.0 vs. 18.0 months, respectively;
P=0.494) (Fig. 4E and F). The
patients with PHF6mutNOTCH1mut
co-existence also exhibited a higher rate of splenomegaly and
lymphadenopathy compared with those without this co-existence
(Table IV).
 | Table IV.Association of
PHF6mutNOTCH1mut co-existence with
clinical feature in adult T-cell acute lymphoblastic leukemia. |
Table IV.
Association of
PHF6mutNOTCH1mut co-existence with
clinical feature in adult T-cell acute lymphoblastic leukemia.
|
|
PHF6mutNOTCH1mut
co-existence |
|---|
|
|
|
|---|
| Clinical
features | WT (n=47) | Mutation
(n=11) | P-value |
|---|
| Male, % | 83.0 | 63.6 | 0.311 |
| Age, median
(range) | 26 (14–62)
years | 27 (14–60)
years | 0.456 |
| WBC, median
(range) | 47.9 (1.2–546.0)
×10-9/l | 20.5 (2.9–175.3)
×10-9/l | 0.144 |
| Platelet, median
(range) | 57.0 (17.0–267.0)
×10-9/l | 46.0 (25.0–223.0)
×10-9/l | 0.979 |
| Hemoglobin, median
(range) | 122.0 (56.0–171.0)
g/l | 95.0 (56.0–117.0)
g/l | 0.007 |
| LDH, median
(range) | 1038.0
(131.0–8601.0) U/l | 861.0
(294.0–5353.0) U/l | 0.921 |
| Marrow blasts,
median (range) | 0.75
(0.22–0.98)% | 0.85
(0.27–0.99)% | 0.585 |
| Peripheral blasts,
median (range) | 0.70
(0.00–0.99)% | 0.40
(0.06–0.94)% | 0.310 |
| Immunophenotype
(≥20%) |
|
|
|
|
CD13 | 29.7 | 66.7 | 0.094 |
|
CD33 | 35.1 | 57.1 | 0.501 |
|
CD34 | 50.0 | 62.5 | 0.800 |
| Other genetic
abnormalities, % |
|
|
|
| Complex
karyotype | 10.8 | 10.0 | 1.000 |
|
FBXW7 mutation | 12.8 | 0.0 | 0.483 |
|
PTEN mutation | 12.8 | 0.0 | 0.483 |
|
JAK1 mutation | 6.4 | 9.1 | 1.000 |
| Extramedullary
infiltration, % |
|
|
|
|
Hepatomegaly | 14.9 | 18.2 | 1.000 |
|
Splenomegaly | 38.3 | 81.8 | 0.009 |
|
Lymphadenopathy | 44.7 | 90.9 | 0.006 |
Discussion
To the best of our knowledge, this study is the
second report of PHF6 mutations in an Asian adult population
with T-ALL. Of note, a higher incidence of PHF6 mutations
was observed in Asian adults with T-ALL in the present study
(27.1%), compared with the previous one (18.6%) (8). A different study showed that the
frequency of PHF6 mutations in pediatric patients with T-ALL
in the USA was 16% (5), which was
lower than the frequency found in the present study (27.1%). This
variance was possibly attributable to differences in age, area and
race of the observed populations.
PHF6 has been reported to be a novel tumor
suppressor in T-ALL (5). Of note, out
of the 16 PHF6 mutations identified in this cohort of adult
patients with T-ALL, 10 were novel mutations. Consistent with other
reports (5,7,9), no
PHF6 mutations were detected in B-ALL samples, suggesting
that PHF6 inactivating mutations are restricted to lymphoid
tumors of the T-cell lineage.
It has been reported that PHF6 is an X-linked
gene, and that PHF6 mutations were almost exclusively found
in T-ALL samples from male subjects (5); however, no significant differences were
observed in PHF6 mutations in relation to gender in this
cohort of Chinese adults with T-ALL, which is consistent with
another study conducted on a Chinese population (8,9), which
suggests that ethnic factors may contribute to gender differences
in the risk of PHF6 mutations, and this requires further
investigation in larger cohorts.
In the present study, it was observed that patients
with T-ALL exhibited significantly lower PHF6 expression,
and that low PHF6 expression in T-ALL is associated with
leukemic cell proliferation. In addition, 6 of the 16 mutations
were found to induce a frame-shift, which may result in the
deletion of the PHF6 protein and its eventual dysfunction. These
data indicated that PHF6 inactivation in T-ALL is a result
of genetic abnormalities and/or low PHF6 expression.
No associations were observed between PHF6
and NOTCH1 mutations in either pediatric (n=65) or adult
(n=34) cohorts with T-ALL in the previous study (5). However, the significant correlation
found between PHF6 and NOTCH1 mutations in the
present study is consistent with another study on Chinese patients
with T-ALL (8). PHF6 serves a
potential role in transcriptional regulation, but its effects on
genomics are not fully understood. Both PHF6 and
NOTCH1 mutations have a high incidence in T-ALL. Whether
PHF6 inactivating mutations could induce the genomic
alterations of other genes in T-ALL, such as NOTCH1,
requires further research.
In order to further explore the effect of
PHF6 mutations on clinical outcomes, the OS and EFS of
patients with T-ALL were analyzed. Despite the fact that no
significant differences were identified in the OS and EFS of
patients with PHF6 mutations compared with those with
PHF6 WT, a significantly shorter EFS was observed in
patients with PHF6mutNOTCH1mut
co-existence. This result further indicated the synergistic effects
of PHF6 and NOTCH1 mutations on the oncogenesis of
T-ALL; therefore,
PHF6mutNOTCH1mut co-existence
could serve as a prognostic marker for the disease and should be
integrated into future prognostic models of adult T-ALL.
In conclusion, a high incidence of PHF6
mutations was observed in Chinese adults with T-ALL. The low
expression of PHF6 was found to be associated with the
markers of leukemic cell proliferation. A
PHF6mutNOTCH1mut co-existence
was observed and shown to be correlated with a shorter EFS in
patients with T-ALL. The present results indicated a synergistic
effect of PHF6 and NOTCH1 mutations on the
oncogenesis of adult T-ALL, suggesting that their co-existence
could serve as a prognostic marker for the disease.
Acknowledgements
This study was funded by The National Natural
Science Foundation of China (grant nos. 81270613 and 30973376),
Jiangsu Province Key Medical Talents (grant no. RC2011077),
Scientific Research Foundation for the Returned Overseas Chinese
Scholars, State Education Ministry (39th), China Postdoctoral
Science Foundation (grant no. 20090461134), Special grade of the
financial support from China Postdoctoral Science Foundation (grant
no. 201003598), Six Great Talent Peak Plan of Jiangsu Province
(grant nos. 2010-WS-024 and 2014-WSN-049), Scientific Research
Foundation for the Returned Overseas Chinese Scholars, Nanjing
Municipal Bureau of Personnel (2009) and The Key Project supported
by the Medical Science and Technology Development Foundation,
Nanjing Department of Health (grant no. ZKX14015).
References
|
1
|
Graux C, Cools J, Michaux L, Vandenberghe
P and Hagemeijer A: Cytogenetics and molecular genetics of T-cell
acute lymphoblastic leukemia: From thymocyte to lymphoblast.
Leukemia. 20:1496–1510. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mullighan CG and Downing JR: Genome-wide
profiling of genetic alterations in acute lymphoblastic leukemia:
Recent insights and future directions. Leukemia. 23:1209–1218.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang J, Ding L, Holmfeldt L, Wu G,
Heatley SL, Payne-Turner D, Easton J, Chen X, Wang J, Rusch M, et
al: The genetic basis of early T-cell precursor acute lymphoblastic
leukaemia. Nature. 481:157–163. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Della Gatta G, Palomero T, Perez-Garcia A,
Ambesi-Impiombato A, Bansal M, Carpenter ZW, De Keersmaecker K,
Sole X, Xu L, Paietta E, et al: Reverse engineering of TLX
oncogenic transcriptional networks identifies RUNX1 as tumor
suppressor in T-ALL. Nat Med. 18:436–440. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Van Vlierberghe P, Palomero T, Khiabanian
H, Van der Meulen J, Castillo M, Van Roy N, De Moerloose B,
Philippé J, González-García S, Toribio ML, et al: PHF6 mutations in
T-cell acute lymphoblastic leukemia. Nat Genet. 42:338–342. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lower KM, Turner G, Kerr BA, Mathews KD,
Shaw MA, Gedeon AK, Schelley S, Hoyme HE, White SM, Delatycki MB,
et al: Mutations in PHF6 are associated with
Börjeson-Forssman-Lehmann syndrome. Nat Genet. 32:661–665. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Van Vlierberghe P, Patel J, Abdel-Wahab O,
Lobry C, Hedvat CV, Balbin M, Nicolas C, Payer AR, Fernandez HF,
Tallman MS, et al: PHF6 mutations in adult acute myeloid leukemia.
Leukemia. 25:130–134. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wang Q, Qiu H, Jiang H, Wu L, Dong S, Pan
J, Wang W, Ping N, Xia J, Sun A, et al: Mutations of PHF6 are
associated with mutations of NOTCH1, JAK1 and rearrangement of
SET-NUP214 in T-cell acute lymphoblastic leukemia. Haematologica.
96:1808–1814. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lin ZK, Zhang R, Ge Z, Liu J, Guo X, Qiao
C, Wu YJ, Qiu HR, Zhang JF and Li JY: Characteristics of NOTCH1
mutation in adult T-cell acute lymphoblastic leukemia. Zhongguo Shi
Yan Xue Ye Xue Za Zhi. 21:1403–1408. 2013.(In Chinese). PubMed/NCBI
|
|
10
|
Swerdlow SH, Campo E, Harris NL, Jaffe ES,
Pileri SA, Stein H, Thiele J and Vardiman JW: WHO Classification of
Tumours of Haematopoietic and Lymphoid Tissues (4th). IARC press.
Lyon: 2008.
|
|
11
|
Guo X, Zhang R, Liu J, Li M, Song C, Dovat
S, Li J and Ge Z: Characterization of LEF1 high expression and
novel mutations in adult acute lymphoblastic leukemia. PLoS One.
10:e01254292015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo X, Zhang R, Ge Z, Xu JY, Li M, Qiao C,
Qiu HR and Li JY: Mutations of FBXW7 in adult T-Cell acute
lymphocytic leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi.
23:612–618. 2015.(In Chinese). PubMed/NCBI
|
|
13
|
Shaffer LG, Slovak ML and Campbell LJ: An
International System for Human Cytogenetic Nomenclature.
Recommendations of the International Standing Committee on Human
Cytogenetic Nomenclature. Karger S.AG: (Basel, Switzerland).
2009.
|