Introduction
Ovarian cancer is a malignant tumour posing a
serious threat to women's health. As the main type of ovarian
cancer, ovarian epithelial carcinoma accounts for 85–90% of all
ovarian cancers. The mortality rate of ovarian epithelial carcinoma
ranks first among all female reproductive tract malignancies.
Approximately 70% of ovarian cancer patients are in the late stage
when diagnosed. Most of these tumours easily develop drug
resistance in the course of post-surgery chemotherapy; therefore,
the therapeutic effect is greatly reduced, leading to a survival
rate of just 30% for ovarian cancer (1). Therefore, multidrug resistance is the
main cause of ovarian cancer chemotherapy failure. Studies have
demonstrated that multidrug resistance is the result of multiple
genes or proteins and a multistep process, or cross-reactivity of
multiple factors. Multidrug resistance involves several different
regulatory mechanisms, and epigenetic regulation is one of the
significant regulatory mechanisms in the development of ovarian
cancer multidrug resistance (2).
Epigenetic modification is a heritable change in gene expression
without a DNA sequence change (3).
Epigenetic modification includes DNA methylation, histone
modification, chromatin modification and microRNA modification
(4), and plays an essential role in
gene transcription regulation. As one of the main pathways of
epigenetic regulation, DNA methylation is closely related to the
multidrug resistance, development, progression, clinical diagnosis
and prognosis of ovarian cancer (5).
Therefore, a comprehensive study of the mechanisms of ovarian
cancer has crucial diagnostic and therapeutic significance.
DNA methylation is the process by which a methyl
group from the donor S-adenosyl-L-methionine is added to the
5-carbon position of cytosine to form 5-methylcytosine under the
catalysis of DNA methyltransferase (DNMT). DNA methylation includes
whole-genome hypomethylation and CpG island hypermethylation of a
promoter region. CpG island hypermethylation may lead to decreased
or even silenced gene expression levels and eventually plays a
significant role in the regulation of cancers, including ovarian
cancer (6). It has been proven that
DNA methylation plays a critical role in the regulation of
multidrug resistance in ovarian cancer. Therefore, in this study,
we systematically analysed 26 methylated genes that have
significant regulatory roles in multidrug-resistant ovarian cancer.
We conducted an integrated analysis of their correlation with
ovarian cancer clinical factors including malignant behaviour,
prognosis and staging. Additionally, we investigated the relevance
and integrity of these 26 genes through bioinformatics. This study
has overall reference value and significance in the understanding
of the regulatory mechanisms of ovarian cancer multidrug resistance
by DNA methylation.
Materials and methods
Gene search
Using ‘ovarian cancer’ or ‘ovarian carcinoma’, ‘DNA
methylation’ or ‘methylation’, ‘resistant’ or ‘resistance’ or
‘chemoresistance’ as key words, we screened methylated genes
associated with the regulation of drug resistance in ovarian cancer
from an advanced search in the PubMed database (http://www.ncbi.nlm.nih.gov/pubmed/).
Bioinformatics analysis
Biological process annotation and enrichment
We screened biological processes with significance
(P<0.05) and involved genes through biological process gene
annotation and enrichment analysis using the Database for
Visualization and Integrated Discovery (DAVID) software (http://david.abcc.ncifcrf.gov/) (7) and Coremine medical software (http://www.coremine.com/).
Protein interactions
Protein interaction analysis was conducted using the
search tool for the retrieval of interacting genes/proteins
(STRING) and online software (http://string-db.org/) (8). The protein interaction reliability score
was 0.400 (medium confidence).
Results
Comprehensive analysis of the
regulation of ovarian cancer multidrug resistance by methylated
genes
By fully integrating publication references for the
association of DNA methylation with ovarian cancer multidrug
resistance, we screened 26 methylated genes that are significantly
related to ovarian cancer multidrug resistance; namely MLH1, BRCA1,
FBXO32, DNAJC15, CSAG2, PROM1, ASS1, RASSF1, PTEN, TNFRSF10A,
ABCG2, ZMYND10, MDR1, TGFBI, RGS10, UCHL1, Sulf-1, SFRP, MAL,
TUBB3, L1TD1, CLDN4, HOXA10, HOXA9, HOXA11 and FANCF. With the
exception of CSAG2, MDR1, TUBB3, L1TD1, FANCF and HOXA10, all of
the genes were hypermethylated with low expression in
drug-resistant tissues and cells of ovarian cancer. This result
indicated that DNA high/hypermethylation is the main pathway for
the regulation of ovarian cancer multidrug resistance compared with
hypomethylation. However, there were a number of genes for which
the methylation status and expression level were uncertain. Eyre
et al (9) observed that ABCB1
(MDR1) gene expression presents hypomethylation in
paclitaxel-resistant ovarian cancer cells and is involved in
metastasis and resistance to chemotherapy in ovarian cancer through
cancer stem cells and side population cells. However, there are
also studies indicating that this gene has undergone DNA
hypermethylation in drug-resistant ovarian cancer cells, and that
its downregulation is involved in the development, progression and
multidrug resistance of ovarian cancer by the c-Jun/JNK signalling
pathway (10). Lee et al
(11) observed that the
downregulation of the MAL gene leads to tumourigenesis, while its
upregulation increases the drug resistance of ovarian cancer.
DNA methylation is an essential mechanism for the
regulation of the development of drug resistance in ovarian cancer,
but the regulatory mechanism varies with different genes. The 26
methylated genes associated with ovarian cancer drug resistance in
this study are involved in paclitaxel and cisplatin resistance of
ovarian cancer through various mechanisms, including gene mismatch
repair, gene microsatellite instability, cell repair defects,
diminished DNA recognition capacity of cells, abnormal cell
proliferation, apoptosis, cell growth, cell invasion and
metastasis, prevention of intracellular drug accumulation, and
associated signalling pathways (see Table
I) (10–52). In all of the regulatory mechanisms,
apoptosis may be a notable way for methylated genes to be involved
in the regulation of ovarian cancer drug resistance since there are
at least eight genes, including MLH1, PTEN and ABCG2, that
participate in regulating ovarian cancer drug resistance through
the cell apoptosis signalling pathway. In addition, five genes,
including PTEN and BLU, are directly or indirectly involved in the
regulation of the AKT signalling pathway, which has been reported
to be critical in the regulation of apoptosis, cell proliferation,
cell growth, metabolism and multidrug resistance (53). These results indicated that among the
26 methylated genes that are associated with the regulation of
ovarian cancer drug resistance, at least half respond to ovarian
cancer drug resistance by directly or indirectly involving an
apoptosis signalling pathway.
 | Table I.Comprehensive analysis of the
correlation between 26 methylated genes and ovarian cancer
multidrug resistance. |
Table I.
Comprehensive analysis of the
correlation between 26 methylated genes and ovarian cancer
multidrug resistance.
| Gene | Methylation status
in drug-resistant tissue/cell | Expression of
gene | Drugs | Regulation manner
of drug resistance | Drug resistance
after adding demethylation inhibitor | Refs. |
|---|
| MLH1 |
Hypermethylation | Silenced
expression | Platinum (13) | DNA mismatch
repair, cell proliferation, apoptosis | Yes | (14,15) |
| BRCA1 |
Hypomethylation | Upregulation | Cisplatin | DNA mismatch
repair | No | (16,17) |
| FBXO32 |
Hypermethylation | Silenced
expression | Platinum | Apoptosis, cell
invasion, TGF-β/SMAD4 signalling pathway | Yes | (18) |
| DNAJC15 |
Hypermethylation | Downregulation | Platinum,
taxol | Prevents
intracellular drug accumulation (10) | Yes | (19,20) |
| CSAG2 |
Hypomethylation | Upregulation | Taxol | – | Yes | (21) |
| PROM1 |
Hypomethylation | Upregulation | Platinum | – | No | (22) |
| ASS1 |
Hypermethylation | Silenced
expression | Platinum | – | – | (23) |
| RASSF1 |
Hypermethylation | Silenced
expression | Taxol | Apoptosis, RAS
signalling pathway | Yes | (24,25) |
| PTEN |
Hypermethylation | Silenced
expression | Taxol | Apoptosis, PI3K/AKT
signalling pathway (26) | Yes | (27) |
| TNFRSF10A |
Hypermethylation | Silenced
expression | Platinum | Apoptosis, cell
invasion, TRAIL signalling pathway (28) | Yes | (29) |
| ABCG2 |
Hypomethylation | Upregulation | Multidrug
resistance | – | No | (30) |
| ZMYND10 |
Hypermethylation | – | – | Cell growth,
apoptosis, AKT signalling pathway | – | (31,32) |
| MDR1 | Hypomethylation
(12), Hypermethylation (10) | Upregulation
(12), Silenced expression (10) | Taxol (12), Multidrug resistance (10) | c-Jun/JNK
signalling pathway (10) | – | – |
| TGFBI |
Hypermethylation | Silenced
expression | Taxol | Cell
proliferation | Yes | (33) |
| RGS10 |
Hypermethylation | Downregulation | Platinum | Apoptosis | Yes | (34) |
| UCHL1 |
Hypermethylation | Downregulation | Cisplatin | Apoptosis, cell
proliferation, AKT signalling pathway | Yes | (35,36) |
| Sulf-1 |
Hypermethylation | Silenced
expression | Platinum | By regulating the
expression of Bim | Yes | (37,38) |
| SFRP |
Hypermethylation | Silenced
expression | Cisplatin | Cell growth, cell
proliferation, cell invision, Wnt signalling pathway, through
TWIST-mediated EMT and AKT2 signalling | – | (39) |
| MAL |
Hypomethylation | Upregulation | Cisplatin | – | No | (11) |
| TUBB3 |
Hypomethylation | Upregulation | Taxol | – | – | (40,41) |
| L1TD1 |
Hypomethylation | Upregulation | Platinum | Regulating genomic
stability | Yes | (42) |
| CLDN4 |
Hypermethylation | Silenced
expression | Cisplatin | Cell invision, cell
migration, β-catenin signalling pathway, P13K/AKT signalling
pathway | Yes | (43–46) |
| HOXA10 | Hypomethylation
(47) | Upregulation
(47) | Platinum (48) | Cell proliferation,
cell migration, cell invision (49) | – | – |
| HOXA9, HOXA11 |
Hypermethylation | Downregulation | – | – | – | (50,51) |
| FANCF |
Hypomethylation | Upregulation | Alkylating agent,
cisplatin | Gene mismatch
repair, FA/BRCA signalling pathway | No | (52) |
Gene expression downregulation caused by DNA
hypermethylation plays a notable role in the development of drug
resistance in ovarian cancer. Therefore, therapies including the
use of DNA methylation inhibitors to reverse the expression of DNA
methylation may be a key trend for ovarian cancer treatment and for
the mitigation of multidrug resistance. Studies have revealed that
after adding the demethylation inhibitor 5-azacytidine or 5-aza-2
deoxycytidine to 11 genes, including MLH1, FBXO32 and TRAG-3, most
of the sensitivity of ovarian cancer cells to chemotherapy drugs
increased with varying degrees (see Table
I). Hence, demethylation inhibitors have been increasingly used
in clinical practice. However, certain genes, including BRCA1,
PROM1, ABCG2, MAL, L1TD1 and FANCF, presented hypomethylation in
ovarian cancer tissues, and the use of demethylation inhibitors may
decrease ovarian cancer cell sensitivity to chemotherapy drugs or
increase drug resistance (see Table
I).
Biological processes enrichment
We conducted a biological process analysis on the 26
methylated genes that are associated with the regulation of ovarian
cancer drug resistance (7). As shown
in Table II, among the clusters of
genes for the biological processes that have the highest score in
enrichment, cluster 1 consists of six genes primarily associated
with apoptosis-related biological processes, and cluster 2 consists
of four genes that are mainly related to cytoskeletal and cell
cycle processes. These results suggest that DNA methylation is
involved in the development of multidrug resistance in ovarian
cancer mainly through apoptosis and cell cycle regulation. In
addition, DAVID also enriched four methylated genes associated with
drug response.
 | Table II.Results of biological process
enrichment using DAVID. |
Table II.
Results of biological process
enrichment using DAVID.
| Enriched biological
processes | Enriched genes | P-value |
|---|
|
Apoptosis-related | BRCA1, MAL, MLH1,
PTEN, TNFRSF10, SFRP |
|
|
Induction of apoptosis |
| 0.0016 |
|
Positive regulation of
apoptosis |
| 0.0047 |
|
Regulation of apoptosis |
| 0.0083 |
| Cell
cycle-related | BRCA1, MLH1, TUBB3,
UCHL1 |
|
|
Microtubule-based process |
| 0.0077 |
| Cell
cycle |
| 0.0560 |
| Drug-related | ABCB1, ABCG2,
CSAG2, PTEN |
|
| Response to
drug |
| 0.0490 |
Protein interaction analysis for 26
methylated genes
To explain the correlation between all of the genes
and ovarian cancer drug resistance, we conducted a comprehensive
analysis of protein interactions for the 26 genes using the STRING
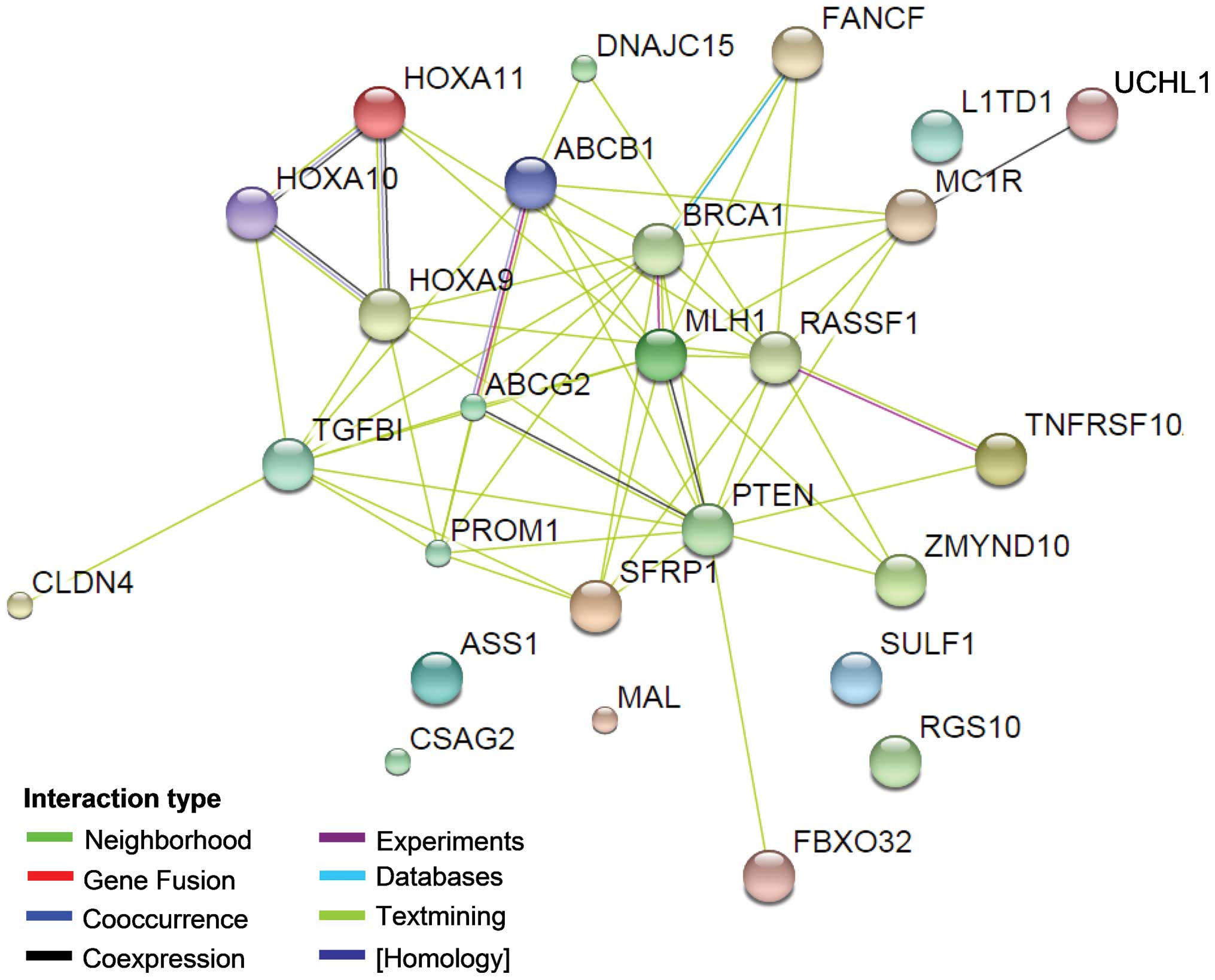
(8) tool. As shown in Fig. 1, with the exception of the ASS1, MAL,
CSAG2, Sulf-1, RGS10 and LITD1 genes/proteins, direct or indirect
interactions were identified for the all genes. PTEN demonstrated a
direct interaction with 13 genes, indicating that it may play a
central role in these ovarian cancer drug resistance-related genes.
PTEN has been proven to be a tumour suppressor gene, and it is
involved in the cell cycle, cell division and apoptosis through
negative regulation of the PTEN/P13K/AKT pathway (53,54).
Previous studies suggest that PTEN is also involved in drug
resistance via the PI3K/AKT pathway. Reduction in PTEN expression
was noted to result in the development of drug resistance in
OVCAR-3 cells and the alterations conferred resistance to cisplatin
through the activation of PI3K/AKT (55). Further research indicates that
overexpression of PTEN reverses chemoresistance to cisplatin in
human ovarian cancer cells through inactivation of the PI3K/AKT
cell survival pathway (56). In
addition to PTEN, ABCB1 and ABCG2 also have direct interactions
with several other proteins in the network. It is indicated that
these two genes may also play a key role in the ovarian cancer drug
resistance regulated by DNA methylation. ABCB1 and ABCG2 are genes
of the ABC transporter protein family and are associated with drug
transport. The P-gp protein encoded by ABCB1 regulates the drug
cumulative effect in tumour cells and eventually participates in
the multidrug resistance process by being involved in drug
transport across the membrane and in membrane stability maintenance
(57); ABCG2 possesses ATP activity
and is involved in biological processes, including drug transport
and cell membrane stability (58).
Chen et al (59) observed that
ABCB1 and ABCG2 are involved in ovarian cancer drug resistance
through the transcription factor Gli of the Hh signalling pathway.
Gli expression inhibition may reduce ABCB1 and ABCG2 gene
expression levels and strengthen the sensitivity of ovarian cancer
to chemotherapy. The underlying mechanism is mainly the result of
the joint action of Gli with its similar sequence in the ABCB1 and
ABCG2 promoter regions. In addition, Hatle et al (10) observed that the reduced expression of
DNAJC15 prevented the drug cumulative effect in cells by increasing
the expression of ABCB1, thus participating in the process of
chemotherapy resistance in ovarian cancer.
As noted above, among the 26 methylated genes that
are associated with ovarian cancer multidrug resistance, direct or
indirect interactions were identified among 20 genes and their
proteins (Fig. 1), indicating that
these genes have significant correlations in their function.
Therefore, to further analyse the for the regulation of ovarian
cancer drug resistance by these 20 functionally correlated genes,
we conducted an annotation analysis of the correlations among the
genes (MLH1, BRCA1, FBXO32, DNAJC15, PROM1, RASSF1, PTEN,
TNFRSF10A, ABCG2, ZMYND10, MDR1, TGFBI, UCHL1, SFRP, TUBB3, CLDN4,
HOXA10, HOXA9, HOXA11 and FANCF) and ovarian cancer (ovarian
neoplasms) and tumour resistance (drug resistance, neoplasms) using
Coremine software (Fig. 1). As shown
in Table III, a comprehensive
analysis annotated 12 biological processes that were significantly
correlated with the 20 genes, ovarian cancer and tumour drug
resistance (P<0.05), indicating that the 20 genes may be
involved in the regulation of ovarian cancer and multidrug
resistance by acting on these 12 biological processes. Among the 12
annotated biological processes, DNA methylation had the highest
score (DNA methylation, P=9.5E-4), fully illustrating a significant
correlation between DNA methylation and these 20 genes, ovarian
cancer and drug resistance. This result is consistent with the
results summarised in Table I and
they are considered to form a mutual basis for each other.
Moreover, in addition to the biological processes of DNA
methylation, the annotated biological processes also included
apoptosis, cell proliferation, cell cycle and signal
transduction.
 | Table III.Biological process annotation
analysis of the interaction among 20 methylated genes and ovarian
cancer (ovarian neoplasms) and tumour drug resistance (drug
resistance, neoplasms) using Coremine medical software. |
Table III.
Biological process annotation
analysis of the interaction among 20 methylated genes and ovarian
cancer (ovarian neoplasms) and tumour drug resistance (drug
resistance, neoplasms) using Coremine medical software.
| Input terms | Annotated
biological process | P-value |
|---|
| 20 genes (MLH1,
BRCA1, FBXO32, | DNA
methylation | 0.00094 |
| DNAJC15, PROM1,
RASSF1, PTEN, | Gene
expression | 0.00133 |
| TNFRSF10A, ABCG2,
ZMYND10, | Methylation | 0.00133 |
| MDR1, TGFBI, UCHL1,
SFRP, | Apoptotic
process | 0.00221 |
| TUBB3, CLDN4,
HOXA10, HOXA9, | Cell
proliferation | 0.00361 |
| HOXA11, FANCF) AND
ovarian neoplasms | Cell cycle | 0.00562 |
| AND drug
resistance, neoplasms | Signal
transduction | 0.00594 |
|
| RNA
interference | 0.00865 |
|
|
Phosphorylation | 0.01780 |
|
| Cell death | 0.02020 |
|
| Pathogenesis | 0.02610 |
Correlation analysis of ovarian cancer
drug resistance-related methylated genes with malignant behaviour
and prognosis
An integrated analysis of the correlation between
the integrated 26 methylated genes and ovarian cancer malignant
clinical behaviour and prognosis factors was also conducted. As
shown in Table IV (10,11,14,16,18–27,29–36,38,39,41–45,47–52,60–83),
we observed that a considerable number of DNA methylated genes,
including MLH1, FBXO32, PROM1, RASSF1, PTEN, SFRP, TUBB3, L1TD1 and
CLDN4, were associated with the invasion of ovarian cancer. Each
gene was hypermethylated, resulting in the silencing of gene
expression in ovarian cancer and drug-resistant tissue or cells and
ultimately regulating tumour invasion. MLH1, FBXO32 and TUBB3 are
associated with ovarian cancer lymph node metastasis, while HOXA10
hypomethylation is associated with ovarian cancer metastasis
behaviour (50). In addition, a
number of the 26 methylated genes revealed a significant
correlation with the type, pathological grade and clinical stage of
ovarian cancer. For example, methylation changes in MLH1, BRCA1,
PTEN, UCHL1, L1TD1 and HOXA10 were associated with the histological
type and pathological grade. Studies have revealed that MLH1
methylation changes are related to serous pathologically
well-differentiated ovarian cancer, with statistical significance,
while methylation changes in PTEN are related to mucinous ovarian
cancer, with statistical significance (61). In addition, the methylation of nine
genes (FBXO32, RASSF1, PTEN, UCHL1, SFRP, MAL, TUBB3, L1TD1 and
FANCF) was correlated with the clinical stage of ovarian cancer,
and the DNA methylation of four genes [MLH1 (14), BRCA1 (62), ASS1 (23) and SFRP (39)] was correlated with recurrence
(Table IV).
 | Table IV.Correlation analysis of the
methylation levels of 26 ovarian cancer drug resistance-related
genes with DNA methylation and clinical factors of ovarian
cancer. |
Table IV.
Correlation analysis of the
methylation levels of 26 ovarian cancer drug resistance-related
genes with DNA methylation and clinical factors of ovarian
cancer.
| Gene | Methylation status
of ovarian cancer | Invasion | Histological
type | Grade | Stage | Relapse | Prognosis | Plasma | Refs. |
|---|
| hMLH1 | Hypermethylation
(66) | Yes (66), LM (66) | Yes (66) | Yes (61) | – | Yes | Poor (PFS)
(48) | Yes | (14) |
| BRCA1 |
Hypermethylation | – | Yes (61) | Yes (61) | – | Yes (62) | Good (PFS)
(64) | Yes (65,67) | (16) |
| FBXO32 |
Hypermethylation | Yes | – | – | Yes | – | Poor (PFS) | – | (18) |
| MCJ |
Hypermethylation | – | – | – | – | – | Poor (OS) (20) | – | (19,20) |
| TRAG-3 |
Hypomethylation | – | – | – | – | – | Good (OS, PFS)
(9) | – | (21) |
| CD133 |
Hypermethylation | Yes | – | – | – | – | – | – | (22) |
| ASS1 |
Hypermethylation | – | – | – | – | Yes | Poor (OS, RFS) | – | (23) |
| RASSF1A |
Hypermethylation | Yes (66,68,69) | No (70) | Yes (70) | Yes (68,70,71) | – | Poor (PFS)
(48) | Yes (65,67) | (24,25) |
| PTEN |
Hypermethylation | Yes (72), LM (72) | Yes (61), No (73,74) | Yes (73) | Yes (73) | – | Poor (OS, PFS)
(12) | – | (27) |
| DR4 |
Hypermethylation | – | – | – | – | – | Poor (OS, PFS)
(75) | Yes (76) | (29) |
| ABCG2 |
Hypermethylation | – | – | – | – | – | – | – | (30) |
| BLU |
Hypermethylation | – | – | – | – | – | Poor (OS, PFS) | – | (31,32) |
| MDR1 | Hypomethylation
(11); Hypermethylation (10) | – | – | – | – | – | Poor (60) | – | – |
| TGFBI |
Hypermethylation | – | No association
(63) | No association
(63) | No association
(63) | – | No association
(63) | – | (26,33,63) |
| RGS10 |
Hypermethylation | – | – | – | – | – | No association | – | (34) |
| UCHL1 |
Hypermethylation | – | Yes (77) | Yes (77) | Yes (77) | – | Yes (77) | – | (35,36) |
| HSulf-1 |
Hypermethylation | – | – | – | – | – | – | – | (38) |
| SFRP |
Hypermethylation | Yes | – | – | Yes (78,79) | Yes (39) | Poor (OS) (39) | – | (39) |
| MAL |
Hypermethylation | – | – | – | Yes (80) | – | Poor (80) | – | (11) |
| TUBB3 |
Hypomethylation | Yes, LM | – | – | Yes | – | Poor (OS and PFS)
(81) | – | (41) |
| LINE-1 |
Hypomethylation | Yes | Yes | Yes | Yes | – | Poor (OS and
PFS) | Yes (67) | (42) |
| CLDN4 |
Hypermethylation | Yes (44,45) | – | – | – | – | – | – | (43) |
| HOXA10 | Hypomethylation
(47) | Yes (49) | Yes (82) | – | – | – | Poor (49) | – | – |
| HOXA9, HOXA11 |
Hypermethylation | – | – | Yes | – | – | Poor (PFS)
(81) | – | (50,51) |
| FANCF |
Hypomethylation | – | – | – | Yes (83) | – | – | – | (52) |
DNA methylated genes associated with drug resistance
regulation have a significant correlation with ovarian cancer
prognosis (Table IV). Among the 26
methylated genes, the methylation of ABCG2, Sulf-1, FANCF, CLDN4,
TGFBI (63) and RGS10 (34) has been reported not to be associated
with the prognosis of ovarian cancer. However, the methylation or
abnormal expression of the remaining 20 genes are all statistically
relevant to ovarian cancer prognosis. As shown in Table IV, most gene methylation and the
subsequent expression changes are relevant to the poor prognosis of
ovarian cancer, shortening overall survival (OS) and
progression-free survival (PFS), while methylation changes in the
BRCA1 and CSAG2 genes are correlated with longer PFS and OS
(84,64). In addition, studies have indicated
that the methylation of SOX1 and LMX1A is related to a patient's
long-term survival rate (85). Given
that ovarian cancer drug resistance-related methylated genes are
significantly related to ovarian cancer prognosis, the joint
detection of the DNA methylation levels in plasma may be an
significant approach for ovarian carcinoma clinical prediction and
prognostic analysis. In fact, there have been studies demonstrating
that plasma DNA methylation may be a biomarker for the early
diagnosis of ovarian cancer. As shown in Table IV, the methylation status and levels
of MLH1, BRCA1, RASSF1, TNFRSF10A and L1TD1 were significantly
changed in the plasma of patients with early ovarian cancer.
Further studies have revealed that in the plasma DNA in 50 ovarian
cancer patients, at least one promoter of BRCAl and RASSF1 was in a
state of hypermethylation in 68% of the patients. Methylation
testing combining these two genes with four other genes (APC, DAPK,
p14ARF and p16INK4a) increases the diagnosis susceptibility of
ovarian cancer to 100%. The methylation of all six genes was
negative in the plasma DNA of the peripheral blood of 20 normal
females; i.e., the diagnostic specificity was 100% (65). Thus, DNA methylation status detection,
particularly DNA methylation status detection in plasma, has broad
application prospects for the diagnosis of ovarian cancer.
Discussion
Ovarian cancer is a gynaecological cancer with a
high mortality rate. The main reason for chemotherapy failure is
the development of multidrug resistance. DNA methylation is an
epigenetic mechanism that plays a crucial role in the development,
progression and drug resistance of ovarian cancer. To date, there
has been extensive research into the correlation between ovarian
cancer drug resistance and DNA methylation, and notable relevant
research results have been reported. However, most of the current
studies in this area are focused on DNA methylation level assays in
ovarian cancer tissue or cells, or on the regulation of the
development and drug resistance in ovarian cancer by methylation of
one or several genes. Research is relatively dispersed, and there
are few studies concerning the comprehensive analysis of gene
methylation mechanisms related to ovarian cancer drug resistance
and their association with clinical factors. In the present study,
we conducted an integrated analysis of the correlation between the
26 methylated genes and ovarian cancer, multidrug resistance and a
number of other clinical factors. Focusing on the effects of these
genes on the drug resistance mechanisms, prognosis and malignant
behaviour of ovarian carcinoma, this study provides guidance on the
choice of chemotherapy drugs used in clinical settings, early
detection and prognosis. At the same time, we carried out
bioinformatics analyses, including biological process annotation
and protein-protein interaction, providing evidence for integrated
research of DNA methylation genes that play a role in ovarian
cancer and drug resistance.
Through DAVID bioprocess enrichment analysis, we
observed that the 26 ovarian cancer-related methylated genes were
significantly correlated with apoptosis (see Table IV); Coremine medical analysis of the
20 methylated genes that have the closest interaction among each
other (Fig. 1) demonstrated that
apoptosis is one of the biological processes that is significantly
correlated with these 20 genes, ovarian cancer and drug resistance
(Table III). These results indicate
that DNA methylation involves the regulation of multidrug
resistance in ovarian cancer through apoptosis. The latter finding
is in good agreement with that of previous studies (86,87). As
shown in Table I, there are at least
seven genes, including PTEN, BLU and UCHL1, that participate in the
regulation of multidrug resistance in ovarian cancer through cell
apoptosis. Apoptosis is autonomous programmed cell death for the
maintenance of homeostasis. Unlike necrosis, apoptosis is not a
passive process but an active process that involves a series of
functions, including gene activation, expression and regulation.
Apoptosis is not self-injury under pathological conditions but an
initiative death process to better adapt to the living environment
(88). Studies have revealed that a
number of genes are involved in the regulation of ovarian cancer
multidrug resistance through apoptosis. For example, the reduction
of the expression of the tumour suppressor gene PTEN was
accompanied by increased expression of Bcl-2, which is widely
recognised as an anti-apoptotic gene. The presence of Bcl-2 helps
to maintain normal cell proliferation and prevents
cisplatin-mediated apoptosis by blocking the release of cytochrome
c from mitochondria, thereby participating in the regulation
of ovarian cancer cisplatin resistance (89). Similarly, the tumour suppressor gene
BLU acts through the downregulation of Bcl-2 and the upregulation
of Bax, P21 and P53, causing apoptosis and responding to paclitaxel
resistance in ovarian cancer (31).
Another example is that UCHL1 promotes sensitivity to cisplatin in
ovarian cancer cells by promoting apoptosis (36). In summary, these results demonstrate
that apoptosis may be an essential mechanism by which methylated
genes regulate drug resistance in ovarian cancer. In-depth research
of the role of methylated genes in the regulation of apoptosis in
ovarian cancer is likely to provide further opportunities to
overcome multidrug resistance.
Platinum- and paclitaxel-based drugs are currently
the main chemotherapy drugs used in the clinical treatment of
ovarian cancer. Based on our overall analysis (Table I), gene methylation is the primary
reason for resistance to these types of drugs in ovarian cancer
cells. Thus, the reversal of gene methylation status may be the key
to overcoming resistance to platinum or paclitaxel. It has been
proven that the reversal of the methylation status of certain genes
effectively reverses the resistance to chemotherapeutic drugs in
ovarian cancer cells. Strathdee et al (90) demonstrated that the methylation level
of the hMLHl gene in cisplatin-resistant A2780 cells was
significantly higher than that in cisplatin-sensitive cells.
Following treatment with the DNA methylation inhibitor
5-azacytidine, A2780 drug-resistant cells restored the sensitivity
to cisplatin, while the DNA methylation levels in hMLHl were also
significantly decreased, indicating that increased sensitivity to
the drug in ovarian cancer is, at least in part, due to the
decreased DNA methylation level in the hMLHl gene. Additionally,
Kassler et al (25) noted that
the hypermethylation status in the RASSF1A gene promoter in
paclitaxel-resistant ovarian cancer cells and the forced increase
of RASSF1A gene expression enhanced the sensitivity of ovarian
cancer to paclitaxel. This result indicates that the expression
silencing caused by RASSF1A methylation causes paclitaxel
resistance of ovarian cancer cells and that the stimulation of the
demethylation status by forcing the re-expression of RASSF1A
effectively increases the sensitivity of the cells to the drugs.
These results indicate that the demethylation of methylated genes
reverses drug resistance in ovarian cancer cells to some extent.
Therefore, demethylating drugs have been increasingly used in
clinical settings. For example, DNMT inhibitors, including
5-azacytidine and its deoxyribose analogues (5-azacytidine) or
5-aza-2 deoxycytidine, decitabine and other demethylating drugs,
have been used in cancer chemotherapy. In summary, the
comprehensive analysis and detection of ovarian cancer drug
resistance-related methylated genes may guide clinical drug choice
to a certain extent, providing significant clinical value.
This study also performed an integrated analysis of
the correlation between ovarian cancer drug resistance-related
methylated genes and ovarian cancer malignant behaviour. Overall,
there are relatively few studies concerning ovarian cancer drug
resistance-related methylated genes and ovarian cancer malignant
behaviour. As shown in Table II,
among all 26 genes, 9 genes (including PTEN) are related to ovarian
cancer staging, 7 genes (including MLH1) are related to ovarian
cancer invasion, 6 genes (including BRCA1) are related to the
histological type of ovarian cancer, and 6 genes (including
RASSF1A) are related to ovarian cancer grading. However, the
correlation between the remaining methylated genes and ovarian
cancer malignant behaviour is unclear. Furthermore, methylation
status changes in drug resistance-related methylated genes in
ovarian cancer were significantly associated with the prognosis of
ovarian cancer. As shown in Table
IV, 19 of the 26 methylated genes, including hMLH1, are
associated with ovarian cancer prognosis, of which hypermethylation
of at least 13 genes, including ASS1, is significantly associated
with poor prognosis. The latter findings were mainly characterised
by a significant shortening of OS and PFS. These results suggest
that the methylation level of ovarian cancer resistance-related
genes is a good prognostic marker and that an integrated analysis
of multiple methylated genes may better predict the prognosis.
Conclusion
An integrated analysis of the correlation between 26
methylated genes and drug resistance in ovarian cancer and a
relevant bioinformatics analysis were conducted in this study.
Protein/gene interactions revealed that at least 20 of the 26 genes
interact with each other (Fig. 1) and
that PTEN, ABCB1 and ABCG2 have direct interaction with most of the
other genes in the network. This result suggests that overall,
these methylated genes may participate in the regulation of ovarian
cancer drug resistance and that genes including PTEN may be key
regulators. Annotations on biological processes using DAVID and
Coremine (Tables II and III) indicated that apoptosis may be a
significant mechanism of drug resistance by methylated genes. In
addition, the integrated analysis revealed that re-expression
caused by demethylation reverses tolerance to chemotherapeutic
drugs in ovarian cancer to a certain extent. This observation
indicates that the application prospect of demethylating drugs in
clinical practice may be broader than previously considered. In
addition, this study demonstrated that the methylation levels of
ovarian cancer drug-resistant methylated genes were significantly
associated with poor prognosis in ovarian cancer (Table IV). In summary, this study explains
the potential correlation between methylated genes and drug
resistance in ovarian cancer with the potential to guide our
understanding of the regulation of ovarian cancer drug resistance
by gene methylation, treatment and improvement of the prognosis of
ovarian cancer.
Acknowledgements
The study was supported by the Natural Science
Foundation of Guangxi (grant no. 2014jjAA40637) and the Key Health
Science Foundation of Guangxi (grant no. 14124004-1-24).
References
|
1
|
Cho KR and Shih IeM: Ovarian cancer. Annu
Rev Pathol. 4:287–313. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Balch C, Huang TH, Brown R and Nephew KP:
The epigenetics of ovarian cancer drug resistance and
resensitization. Am J Obstet Gynecol. 191:1552–1572. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Novik KL, Nimmrich I, Genc B, Maier S,
Piepenbrock C, Olek A and Beck S: Epigenomics: genome-wide study of
methylation phenomena. Curr Issues Mol Biol. 4:111–28.
2002.PubMed/NCBI
|
|
4
|
Lund AH and van Lohuizen M: Epigenetics
and cancer. Genes Dev. 18:2315–2335. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Seeber LM and van Diest PJ: Epigenetics in
ovarian cancer. Methods Mol Biol. 863:253–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Widschwendter M, Jiang G, Woods C, Müller
HM, Fiegl H, Goebel G, Marth C, Müller-Holzner E, Zeimet AG, Laird
PW and Ehrlich M: DNA hypomethylation and ovarian cancer biology.
Cancer Res. 64:4472–4480. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang B, Kirov S and Snoddy J: WebGestalt:
an integrated system for exploring gene sets in various biological
contexts. Nucleic Acids Res. 33(Web Server issue): W741–W748. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Franceschini A, Szklarczyk D, Frankild S,
Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C
and Jensen LJ: STRING v9.1: Protein-protein interaction networks,
with increased coverage and integration. Nucleic Acids Res.
41(Database issue): D808–D815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Eyre R, Harvey I, Stemke-Hale K, Lennard
TW, Tyson-Capper A and Meeson AP: Reversing paclitaxel resistance
in ovarian cancer cells via inhibition of the ABCB1 expressing side
population. Tumour Biol. 35:9879–9892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hatle KM, Neveu W, Dienz O, Rymarchyk S,
Barrantes R, Hale S, Farley N, Lounsbury KM, Bond JP, Taatjes D and
Rincón M: Methylation-controlled J protein promotes c-Jun
degradation to prevent ABCB1 transporter expression. Mol Cell Biol.
27:2952–66. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Lee PS, Teaberry VS, Bland AE, et al:
Elevated MAL expression is accompanied by promoter hypomethylation
and platinum resistance in epithelial ovarian cancer. Int J Cancer.
126:1378–1389. 2010.PubMed/NCBI
|
|
12
|
Harvey I, Stemke-Hale K, Lennard TW,
Tyson-Capper A and Meeson AP: Reversing paclitaxel resistance in
ovarian cancer cells via inhibition of the ABCB1 expressing side
population. Tumour Biol. 35:9879–9892. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Watanabe Y, Ueda H, Etoh T, Koike E,
Fujinami N, Mitsuhashi A and Hoshiai H: A change in promoter
methylation of hMLH1 is a cause of acquired resistance to
platinum-based chemotherapy in epithelial ovarian cancer.
Anticancer Res. 27:1449–1452. 2007.PubMed/NCBI
|
|
14
|
Gifford G, Paul J, Vasey PA, Kaye SB and
Brown R: The acquisition of hMLH1 methylation in plasma DNA after
chemotherapy predicts poor survival for ovarian cancer patients.
Clin Cancer Res. 10:4420–4426. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Calin GA, Sevignani C, Dumitru CD, Hyslop
T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M
and Croce CM: Human microRNA genes are frequently located at
fragile sites and genomic regions involved in cancers. Proc Natl
Acad Sci USA. 101:2999–3004. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Stordal B, Timms K, Farrelly A, Gallagher
D, Busschots S, Renaud M, Thery J, Williams D, Potter J, Tran T, et
al: BRCA1/2 mutation analysis in 41 ovarian cell lines reveals only
one functionally deleterious BRCA1 mutation. Mol Oncol. 7:567–579.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang YQ, Zhang JR, Li SD, He YY, Yang YX,
Liu XL and Wan XP: Aberrant methylation of breast and ovarian
cancer susceptibility gene 1 in chemosensitive human ovarian cancer
cells does not involve the phosphatidylinositol 3′-kinase-Akt
pathway. Cancer Sci. 101:1618–1623. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chou JL, Su HY, Chen LY, Liao YP,
Hartman-Frey C, Lai YH, Yang HW, Deatherage DE, Kuo CT, Huang YW,
et al: Promoter hypermethylation of FBXO32, a novel TGF-beta/SMAD4
target gene and tumor suppressor, is associated with poor prognosis
in human ovarian cancer. Lab Invest. 90:414–25. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Strathdee G, Vass JK, Oien KA, Siddiqui N,
Curto-Garcia J and Brown R: Demethylation of the MCJ gene in stage
III/IV epithelial ovarian cancer and response to chemotherapy.
Gynecol Oncol. 97:898–903. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Strathdee G, Davies BR, Vass JK, Siddiqui
N and Brown R: Cell type-specific methylation of an intronic CpG
island controls expression of the MCJ gene. Carcinogenesis.
25:693–701. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao X, Hu JF, Li T, Yang Y, Sun Z, Ulaner
GA, Vu TH and Hoffman AR: Epigenetic regulation of the taxol
resistance-associated gene TRAG-3 in human tumors. Cancer Genet
Cytogenet. 151:1–13. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Baba T, Convery PA, Matsumura N, Whitaker
RS, Kondoh E, Perry T, Huang Z, Bentley RC, Mori S, Fujii S, et al:
Epigenetic regulation of CD133 and tumorigenicity of CD133+ ovarian
cancer cells. Oncogene. 28:209–218. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nicholson LJ, Smith PR, Hiller L,
Szlosarek PW, Kimberley C, Sehouli J, Koensgen D, Mustea A, Schmid
P and Crook T: Epigenetic silencing of argininosuccinate synthetase
confers resistance to platinum-induced cell death but collateral
sensitivity to arginine auxotrophy in ovarian cancer. Int J Cancer.
125:1454–1463. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vos MD, Martinez A, Elam C, Dallol A,
Taylor BJ, Latif F and Clark GJ: A role for the RASSF1A tumor
suppressor in the regulation of tubulin polymerization and genomic
stability. Cancer Res. 64:4244–4250. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kassler S, Donninger H, Birrer MJ and
Clark GJ: RASSF1A and the Taxol response in ovarian cancer. Mol
Biol Int. 2012:2632672012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Blanco-Aparicio C, Renner O, Leal JF and
Carnero A: PTEN, more than the AKT pathway. Carcinogenesis.
28:1379–1386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Dai F, Zhang Y, Zhu X, Shan N and Chen Y:
Anticancer role of MUC1 aptamer-miR-29b chimera in epithelial
ovarian carcinoma cells through regulation of PTEN methylation.
Target Oncol. 7:217–225. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Horak P, Pils D, Haller G, Pribill I,
Roessler M, Tomek S, Horvat R, Zeillinger R, Zielinski C and
Krainer M: Contribution of epigenetic silencing of tumor necrosis
factor-related apoptosis inducing ligand receptor 1 (DR4) to TRAIL
resistance and ovarian cancer. Mol Cancer Res. 3:335–343. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Y, Hu W, Shen DY, Kavanagh JJ and Fu S:
Azacitidine enhances sensitivity of platinum-resistant ovarian
cancer cells to carboplatin through induction of apoptosis. Am J
Obstet Gynecol. 200(177): e1–e9. 2009.
|
|
30
|
Bram EE, Hu W, Shen DY, Kavanagh JJ and Fu
S: Chemotherapeutic drug-induced ABCG2 promoter demethylation as a
novel mechanism of acquired multidrug resistance. Neoplasia.
11:1359–1370. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Park ST, Byun HJ, Kim BR, Dong SM, Park
SH, Jang PR and Rho S: Tumor suppressor BLU promotes paclitaxel
antitumor activity by inducing apoptosis through the
down-regulation of Bcl-2 expression in tumorigenesis. Biochem
Biophys Res Commun. 435:153–159. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chiang YC, Chang MC, Chen PJ, Wu MM, Hsieh
CY, Cheng WF and Chen CA: Epigenetic silencing of BLU through
interfering apoptosis results in chemoresistance and poor prognosis
of ovarian serous carcinoma patients. Endocr Relat Cancer.
20:213–227. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wang N, Zhang H, Yao Q, Wang Y, Dai S and
Yang X: TGFBI promoter hypermethylation correlating with paclitaxel
chemoresistance in ovarian cancer. J Exp Clin Cancer Res. 31:62012.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ali MW, Cacan E, Liu Y, Pierce JY,
Creasman WT, Murph MM, Govindarajan R, Eblen ST, Greer SF and Hooks
SB: Transcriptional suppression, DNA methylation and histone
deacetylation of the regulator of G-protein signaling 10 (RGS10)
gene in ovarian cancer cells. PLoS One. 8:e601852013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okochi-Takada E, Nakazawa K, Wakabayashi
M, Mori A, Ichimura S, Yasugi T and Ushijima T: Silencing of the
UCHL1 gene in human colorectal and ovarian cancers. Int J Cancer.
119:1338–1344. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Jin C, Yu W, Lou X, Zhou F, Han X, Zhao N
and Lin B: UCHL1 is a putative tumor suppressor in ovarian cancer
cells and contributes to cisplatin resistance. J Cancer. 4:662–670.
2013. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
He X, Khurana A, Roy D, Kaufmann S and
Shridhar V: Loss of HSulf-1 expression enhances tumorigenicity by
inhibiting Bim expression in ovarian cancer. Int J Cancer.
135:1783–1789. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lai J, Chien J, Staub J, Avula R, Greene
EL, Matthews TA, Smith DI, Kaufmann SH, Roberts LR and Shridhar V:
Loss of HSulf-1 up-regulates heparin-binding growth factor
signaling in cancer. J Biol Chem. 278:23107–23117. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Su HY, Lai HC, Lin YW, Liu CY, Chen CK,
Chou YC, Lin SP, Lin WC, Lee HY and Yu MH: Epigenetic silencing of
SFRP5 is related to malignant phenotype and chemoresistance of
ovarian cancer through Wnt signaling pathway. Int J Cancer.
127:555–567. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gao S, Zhao X, Lin B, Hu Z, Yan L and Gao
J: Clinical implications of REST and TUBB3 in ovarian cancer and
its relationship to paclitaxel resistance. Tumour Biol.
33:1759–1765. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Izutsu N, Maesawa C, Shibazaki M, Oikawa
H, Shoji T, Sugiyama T and Masuda T: Epigenetic modification is
involved in aberrant expression of class III beta-tubulin, TUBB3,
in ovarian cancer cells. Int J Oncol. 32:1227–1235. 2008.PubMed/NCBI
|
|
42
|
Pattamadilok J, Huapai N, Rattanatanyong
P, Vasurattana A, Triratanachat S, Tresukosol D and Mutirangura A:
LINE-1 hypomethylation level as a potential prognostic factor for
epithelial ovarian cancer. Int J Gynecol Cancer. 18:711–717. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Litkouhi B, Kwong J, Lo CM, Smedley JG
III, McClane BA, Aponte M, Gao Z, Sarno JL, Hinners J, Welch WR, et
al: Claudin-4 overexpression in epithelial ovarian cancer is
associated with hypomethylation and is a potential target for
modulation of tight junction barrier function using a C-terminal
fragment of Clostridium perfringens enterotoxin. Neoplasia.
9:304–314. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Lin X, Shang X, Manorek G and Howell SB:
Regulation of the Epithelial-Mesenchymal Transition by Claudin-3
and Claudin-4. PLoS One. 8:e674962013. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shang X, Lin X, Alvarez E, Manorek G and
Howell SB: Tight junction proteins claudin-3 and claudin-4 control
tumor growth and metastases. Neoplasia. 14:974–985. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Shang X, Lin X, Manorek G and Howell SB:
Claudin-3 and claudin-4 regulate sensitivity to cisplatin by
controlling expression of the copper and cisplatin influx
transporter CTR1. Mol Pharmacol. 83:85–94. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Jiang Y, Chu Y, Tang W, Wan Y, Zhang L and
Cheng W: Transcription factor WT1 and promoter CpG hypomethylation
coactivate HOXA10 expression in ovarian cancer. Curr Pharm Des.
20:1647–1654. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Matei D, Fang F, Shen C, Schilder J,
Arnold A, Zeng Y, Berry WA, Huang T and Nephew KP: Epigenetic
resensitization to platinum in ovarian cancer. Cancer Res.
72:2197–2205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Li B, Jin H, Yu Y, Gu C, Zhou X, Zhao N
and Feng Y: HOXA10 is overexpressed in human ovarian clear cell
adenocarcinoma and correlates with poor survival. Int J Gynecol
Cancer. 19:1347–1352. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Widschwendter M, Apostolidou S, Jones AA,
Fourkala EO, Arora R, Pearce CL, Frasco MA, Ayhan A, Zikan M,
Cibula D, et al: HOXA methylation in normal endometrium from
premenopausal women is associated with the presence of ovarian
cancer: a proof of principle study. Int J Cancer. 125:2214–2218.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Fiegl H, Windbichler G, Mueller-Holzner E,
Goebel G, Lechner M, Jacobs IJ and Widschwendter M: HOXA11 DNA
methylation - a novel prognostic biomarker in ovarian cancer. Int J
Cancer. 123:725–729. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Taniguchi T, Tischkowitz M, Ameziane N,
Hodgson SV, Mathew CG, Joenje H, Mok SC and D'Andrea AD: Disruption
of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian
tumors. Nat Med. 9:568–574. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
53
|
Cai J, Xu L, Tang H, Yang Q, Yi X, Fang Y,
Zhu Y and Wang Z: The role of the PTEN/PI3K/Akt pathway on
prognosis in epithelial ovarian cancer: a meta-analysis.
Oncologist. 19:528–535. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chu EC and Tarnawski AS: PTEN regulatory
functions in tumor suppression and cell biology. Med Sci Monit.
10:RA235–RA241. 2004.PubMed/NCBI
|
|
55
|
Lee S, Choi EJ, Jin C and Kim DH:
Activation of PI3K/Akt pathway by PTEN reduction and PIK3CA mRNA
amplification contributes to cisplatin resistance in an ovarian
cancer cell line. Gynecol Oncol. 97:26–34. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Wu H, Cao Y, Weng D, Xing H, Song X, Zhou
J, Xu G, Lu Y, Wang S and Ma D: Effect of tumor suppressor gene
PTEN on the resistance to cisplatin in human ovarian cancer cell
lines and related mechanisms. Cancer Lett. 271:260–271. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sui H, Fan ZZ and Li Q: Signal
transduction pathways and transcriptional mechanisms of
ABCB1/Pgp-mediated multiple drug resistance in human cancer cells.
J Int Med Res. 40:426–435. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Krishnamurthy P and Schuetz JD: Role of
ABCG2/BCRP in biology and medicine. Annu Rev Pharmacol Toxicol.
46:381–410. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chen Y, Bieber MM and Teng NN: Hedgehog
signaling regulates drug sensitivity by targeting ABC transporters
ABCB1 and ABCG2 in epithelial ovarian cancer. Mol Carcinog.
53:625–634. 2014.PubMed/NCBI
|
|
60
|
Schneider J, Jimenez E, Marenbach K, Marx
D and Meden H: Co-expression of the MDR1 gene and HSP27 in human
ovarian cancer. Anticancer Res. 18:2967–2971. 1998.PubMed/NCBI
|
|
61
|
Yang HJ, Liu VW, Wang Y, Tsang PC and Ngan
HY: Differential DNA methylation profiles in gynecological cancers
and correlation with clinico-pathological data. BMC Cancer.
6:2122006. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Swisher EM, Gonzalez RM, Taniguchi T,
Garcia RL, Walsh T, Goff BA and Welcsh P: Methylation and protein
expression of DNA repair genes: association with chemotherapy
exposure and survival in sporadic ovarian and peritoneal
carcinomas. Mol Cancer. 8:482009. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Kang S, Dong SM and Park NH: Frequent
promoter hypermethylation of TGFBI in epithelial ovarian cancer.
Gynecol Oncol. 118:58–63. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ignatov T, Eggemann H, Costa SD, Roessner
A, Kalinski T and Ignatov A: BRCA1 promoter methylation is a marker
of better response to platinum-taxane-based therapy in sporadic
epithelial ovarian cancer. J Cancer Res Clin Oncol. 140:1457–1463.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
de Ibanez Caceres I, Battagli C, Esteller
M, Herman JG, Dulaimi E, Edelson MI, Bergman C, Ehya H, Eisenberg
BL and Cairns P: Tumor cell-specific BRCA1 and RASSF1A
hypermethylation in serum, plasma and peritoneal fluid from ovarian
cancer patients. Cancer Res. 64:6476–6481. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang H, Zhang S, Cui J, Zhang A, Shen L
and Yu H: Expression and promoter methylation status of mismatch
repair gene hMLH1 and hMSH2 in epithelial ovarian cancer. Aust N Z
J Obstet Gynaecol. 48:505–509. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Fang F, Balch C, Schilder J, Breen T,
Zhang S, Shen C, Li L, Kulesavage C, Snyder AJ, Nephew KP and Matei
DE: A phase 1 and pharmacodynamic study of decitabine in
combination with carboplatin in patients with recurrent,
platinum-resistant, epithelial ovarian cancer. Cancer.
116:4043–4053. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Bhagat R, Chadaga S, Premalata CS, Ramesh
G, Ramesh C, Pallavi VR and Krishnamoorthy L: Aberrant promoter
methylation of the RASSF1A and APC genes in epithelial ovarian
carcinoma development. Cell Oncol (Dordr). 35:473–479. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Bondurant AE, Huang Z, Whitaker RS, Simel
LR, Berchuck A and Murphy SK: Quantitative detection of RASSF1A DNA
promoter methylation in tumors and serum of patients with serous
epithelial ovarian cancer. Gynecol Oncol. 123:581–587. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Ma L, Zhang JH, Liu FR and Zhang X:
Hypermethylation of promoter region of RASSF1A gene in ovarian
malignant epithelial tumors. Zhonghua Zhong Liu Za Zhi. 27:657–659.
2005.(In Chinese). PubMed/NCBI
|
|
71
|
Liggett TE, Melnikov A, Yi Q, Replogle C,
Hu W, Rotmensch J, Kamat A, Sood AK and Levenson V: Distinctive DNA
methylation patterns of cell-free plasma DNA in women with
malignant ovarian tumors. Gynecol Oncol. 120:113–120. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Shen Y, Shen R, Ge L, Zhu Q and Li F:
Fibrillar type I collagen matrices enhance metastasis/invasion of
ovarian epithelial cancer via β1 integrin and PTEN signals. Int J
Gynecol Cancer. 22:1316–1324. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Qiao YH, Cheng J and Guo RX: Expression of
phosphorylated protein kinase B and PTEN protein in ovarian
epithelial cancer. Zhonghua Fu Chan Ke Za Zhi. 42:325–329. 2007.(In
Chinese). PubMed/NCBI
|
|
74
|
Che Y, Yao Q, Dai S, Luo B and Wang Y:
Study of the mutation and expression of PTEN gene in endometrial
carcinoma and epithelial ovarian cancer. Zhonghua Fu Chan Ke Za
Zhi. 37:608–611. 2002.(In Chinese). PubMed/NCBI
|
|
75
|
Dong HP, Kleinberg L, Silins I, Flørenes
VA, Tropé CG, Risberg B, Nesland JM and Davidson B: Death receptor
expression is associated with poor response to chemotherapy and
shorter survival in metastatic ovarian carcinoma. Cancer.
112:84–93. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fu S, Hu W, Iyer R, Kavanagh JJ, Coleman
RL, Levenback CF, Sood AK, Wolf JK, Gershenson DM, Markman M, et
al: Phase 1b-2a study to reverse platinum resistance through use of
a hypomethylating agent, azacitidine, in patients with
platinum-resistant or platinum-refractory epithelial ovarian
cancer. Cancer. 117:1661–1669. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Brait M, Maldonado L, Noordhuis MG, Begum
S, Loyo M, Poeta ML, Barbosa A, Fazio VM, Angioli R, Rabitti C, et
al: Association of promoter methylation of VGF and PGP9.5 with
ovarian cancer progression. PLoS One. 8:e708782013. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Al-Shabanah OA, Hafez MM, Hassan ZK,
Sayed-Ahmed MM, Abozeed WN, Alsheikh A and Al-Rejaie SS:
Methylation of SFRPs and APC genes in ovarian cancer infected with
high risk human papillomavirus. Asian Pac J Cancer Prev.
15:2719–2725. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Saran U, Arfuso F, Zeps N and Dharmarajan
A: Secreted frizzled-related protein 4 expression is positively
associated with responsiveness to cisplatin of ovarian cancer cell
lines in vitro and with lower tumour grade in mucinous ovarian
cancers. BMC Cell Biol. 13:252012. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Berchuck A, Iversen ES, Luo J, Clarke JP,
Horne H, Levine DA, Boyd J, Alonso MA, Secord AA, Bernardini MQ, et
al: Microarray analysis of early stage serous ovarian cancers shows
profiles predictive of favorable outcome. Clin Cancer Res.
15:2448–2455. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Raspaglio G, Petrillo M, Martinelli E, Li
Puma DD, Mariani M, De Donato M, Filippetti F, Mozzetti S, Prislei
S, Zannoni GF, et al: Sox9 and Hif-2α regulate TUBB3 gene
expression and affect ovarian cancer aggressiveness. Gene.
542:173–181. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Tanwar PS, Kaneko-Tarui T, Lee HJ, Zhang L
and Teixeira JM: PTEN loss and HOXA10 expression are associated
with ovarian endometrioid adenocarcinoma differentiation and
progression. Carcinogenesis. 34:893–901. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Dhillon VS, Shahid M and Husain SA: CpG
methylation of the FHIT, FANCF, cyclin-D2, BRCA2 and RUNX3 genes in
Granulosa cell tumors (GCTs) of ovarian origin. Mol Cancer.
3:332004. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Materna V, Surowiak P, Kaplenko I,
Spaczyński M, Duan Z, Zabel M, Dietel M and Lage H:
Taxol-resistance-associated gene-3 (TRAG-3/CSAG2) expression is
predictive for clinical outcome in ovarian carcinoma patients.
Virchows Arch. 450:187–194. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Su HY, Lai HC, Lin YW, Chou YC, Liu CY and
Yu MH: An epigenetic marker panel for screening and prognostic
prediction of ovarian cancer. Int J Cancer. 124:387–393. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Lum E, Vigliotti M, Banerjee N, Cutter N,
Wrzeszczynski KO, Khan S, Kamalakaran S, Levine DA, Dimitrova N and
Lucito R: Loss of DOK2 induces carboplatin resistance in ovarian
cancer via suppression of apoptosis. Gynecol Oncol. 130:369–376.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Karaca B, Atmaca H, Bozkurt E, Kisim A,
Uzunoglu S, Karabulut B, Sezgin C, Sanli UA and Uslu R: Combination
of AT-101/cisplatin overcomes chemoresistance by inducing apoptosis
and modulating epigenetics in human ovarian cancer cells. Mol Biol
Rep. 40:3925–3933. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Lawen A: Apoptosis - an introduction.
Bioessays. 25:888–896. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Singh M, Chaudhry P, Fabi F and Asselin E:
Cisplatin-induced caspase activation mediates PTEN cleavage in
ovarian cancer cells: a potential mechanism of chemoresistance. BMC
Cancer. 13:2332013. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Strathdee G, MacKean MJ, Illand M and
Brown R: A role for methylation of the hMLH1 promoter in loss of
hMLH1 expression and drug resistance in ovarian cancer. Oncogene.
18:2335–2341. 1999. View Article : Google Scholar : PubMed/NCBI
|