Introduction
MicroRNAs (miRs) are endogenous, small non-coding
RNAs that have been identified as post-transcriptional regulators
of gene expression. miRs exert their function by binding to the 3′
untranslated regions (UTRs) of target mRNAs, which prevent mRNA
translation or cause target degradation, leading to a
downregulation in the expression of their downstream target genes
(1). Accumulating evidence reveals
that miRs are crucial in a variety of cellular and biological
processes. In addition, deregulation of miRs has been demonstrated
in numerous types of human diseases, including cancer (2). During tumor development and progression,
miRs function as oncogenes or tumour suppressor genes (3). Gene expression profiling studies have
revealed that miR expression signatures are associated with
specific tumour subtypes and clinical outcomes (4). In certain clinical circumstances, miR
profiling may be superior to mRNA profiling to classify tumor
subtypes and provide a prognosis for patients (5,6).
Additional studies have indicated that numerous cancer-associated
miRs are frequently identified at genomic breakpoint regions
(3).
Acute promyelocytic leukemia (APL) is a subtype of
acute myeloid leukemia (AML), and is characterized by a reciprocal
translocation that involves chromosomes 15 and 17. The specific
chromosome translocation t(15;17)(q24;q11-12) results in the fusion
of retinoic acid receptor-α (RARα) gene on chromosome 17 with
promyelocytic leukemia (PML) gene on chromosome 15, subsequently
contributing to a maturation arrest at the promyelocytic stage of
development and leukemogenesis (7).
The remission rate and disease-free survival time are usually
higher in patients with APL compared with patients with other
subtypes of AML, and relapse remains an important factor affecting
the long term disease-free survival of patients with APL (8). Consequently, the molecular pathogenesis
of APL is complicated and requires elucidation. Previous studies
have investigated the genes and proteins involved in the
development and progression of APL, and studies have revealed that
miRs are important in leukemia with APL progression, which have led
to novel diagnostic and therapeutic methodologies (9–11). miR-299
locating at 14q32 has been reported to be upregulated in APL
(12). However, little is known
concerning the potential functions and exact mechanistic roles of
miR-299 in APL. In a previous study conducted by the present
authors, 28 miRs, including miR-299-5p, were demonstrated to
modulate p21Cip1/Waf1 expression by directly targeting its 3′UTR
(13). Therefore, the present study
hypothesizes that an overexpression of miR-299 contributes to APL
progression by targeting p21Cip1/Waf1 post-transcriptionally.
The present study demonstrated that miR-299 was
overexpressed in APL cells, and its expression significantly
promoted APL cell growth and cell cycle progression at G1/S
transition, primarily involving miR-299-5p, but not miR-299-3p.
Additional experiments revealed that miR-299 downregulates protein,
but not mRNA, levels of p21Cip1/Waf1. Overall, the present results
indicate that p21Cip1/Waf1 is a direct functional target of miR-299
in APL.
Materials and methods
Patients samples
A total of 45 patients with newly diagnosed APL (25
males and 20 females; median age, 35 years; age range, 14–65 years)
were recruited from the Department of Hematology, Affiliated Union
Hospital of Fujian Medical University (Fuzhou, China). The
diagnosis of APL was based on the 2008 World Health Organization
criteria (14). In total, 18 healthy
donors (10 males and 8 females; median age, 28 years; age range,
16–62 years) were used as controls for normalization. Written
informed consent was obtained from all patients.
RNA extraction and quantitative
polymerase chain reaction (qPCR)
Mononuclear cells (MNCs) were separated from bone
marrow or whole peripheral blood from the patients and healthy
donors using Ficoll-Hypaque density gradient centrifugation
(Inno-Train Diagnostik GmbH, Kronberg im Taunus, Germany). Total
RNA was extracted from MNCs and human promyelocytic leukemia NB4
and HL-60 cells (American Type Culture Collection, Manassas, VA,
USA) infected with miR-299-5p/3p and anti-miR-299-5p/3p, using
TRIzol reagent (Invitrogen™; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). To determine miR expression, total RNA (1
µl/sample) was reverse-transcribed using miR-specific stem-loop
reverse transcription (RT) primers, reverse transcriptase, RT
buffer, dNTPs and an RNase inhibitor, according to the
manufacturer's protocol (TaqMan® MicroRNA Reverse
Transcription kit; Applied Biosystems™; Thermo Fisher Scientific,
Inc.). qPCR was performed using an Applied Biosystems™ StepOnePlus™
Real-Time PCR System (Thermo Fisher Scientific, Inc.). The 20-µl
reaction system contained the corresponding complementary DNA (2
µl), miRNA-specific TaqMan® primers (1 µl),
TaqMan® Universal PCR Master Mix (10 µl) and
ddH2O (7 µl) (Applied Biosystems™; Thermo Fisher
Scientific, Inc.). The PCR conditions were 95°C for 10 min,
followed by 50 cycles at 95°C for 15 sec and 60°C for 1 min. RNU6B
was used as an endogenous housekeeping control for data
normalization of miR levels. The endogenous mRNA levels of p21 were
detected using the SYBR Green PCR Master Mix kit, according to the
manufacturer's protocol (Takara Bio, Inc., Otsu Japan). The RT
reactions contained 500 ng total RNA extracted from the samples, 2
µl 5X PrimeScript™ Buffer (Takara Bio, Inc.), 0.5 µl 1X
PrimeScript™ RT Enzyme Mix I (Takara Bio, Inc.) and 0.5 µl
oligo(dT) primer. The 10-µl reactions were incubated for 42 min at
37°C, followed by 30 sec of incubation at 85°C and subsequent
exposure to 4°C. The PCR primers (Invitrogen; Thermo Fisher
Scientific, Inc.) were as follows: p21, forward
5′-TGATTAGCAGCGGAACAAG-3′ and reverse 5′-AAACAGTCCAGGCCAGTATG-3′;
β-actin, forward 5′-TTGTTACAGGAAGTCCCTTGCC-3′ and reverse
5′-ATGCTATCACCTCCCCTGTGTG-3′. The comparative threshold cycle (Cq)
method was used to measure the relative changes in expression;
2−∆∆Ct represents the fold-change in expression
(15).
Cell culture
Human embryonic kidney (HEK)-293T cells were
obtained from the American Type Culture Collection and cultured in
Dulbecco's modified Eagle's medium (Invitrogen; Thermo Fisher
Scientific, Inc.) with 10% fetal bovine serum (FBS; Invitrogen;
Thermo Fisher Scientific, Inc.). Human promyelocytic leukemia HL-60
and NB4 cells were cultured in RPMI-1640 (Invitrogen; Thermo Fisher
Scientific, Inc.) with 10% FBS, 50 U/ml penicillin and 50 µg/ml
streptomycin (Invitrogen; Thermo Fisher Scientific).
Lentivirus production and
infection
Pri-miR-299 sequences were amplified and cloned into
the pWPXL lentiviral vector (a gift from Professor Didier Trono,
School of Life Sciences, Ecole Polytechnique Fédérale de Lausanne,
Lausanne, Switzerland). Briefly, a ~700 bp fragment carrying
pri-miR-299 was amplified from human genomic DNA by the
Phusion® High-Fidelity DNA Polymerase enzyme (New
England Biolabs, Inc., Ipswich, MA, USA) using the following PCR
primers: Forward 5′-CGGGATCCATGACGTGGTTGACTACGC-3′ and reverse
5′-GCGTCGACTCTTCAATTACTCCAGAGG-3′ (Invitrogen; Thermo Fisher
Scientific, Inc.). The amplified fragment was first cloned into a
pMD-18T vector (Takara Bio Inc.) with fusion green fluorescent
protein expression and then subcloned (BamHI + EcoRV; Takara Bio
Inc.) into a pWPXL lentivirus vector. The pWPXL vectors were
transfected into HEK-293T cells with the packaging plasmid psPAX2
and the VSV-G envelope plasmid pMD2.G (obtained from Dr Didier
Trono) using Lipofectamine® 2000 reagent (Invitrogen™;
Thermo Fisher Scientific, Inc.). Cell supernatants were collected
at 48 h post-transfection and passed through a 0.22-mm filter. The
titer of purified virus was 2.0×108 IU/ml. In total,
1×105 NB4 and HL-60 cells were infected with
1×106 recombinant lentivirus-transducing units plus 6
µg/ml polybrene (Sigma-Aldrich, St. Louis, MO, USA).
Cell proliferation
Cell proliferation was measured using cell counting
kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.,
Kumamoto, Japan). In brief, 4,000 cells were plated into each well
of a 96-well plate and 10 µl CCK-8 solution was added to 90 µl
culture medium. The cells were subsequently incubated for 2 h at
37°C and optical density was measured at 450 and 650 nm using a
microplate reader (ELx808; Bio-Tek Instruments, Inc., Winooski, VT,
USA). Three independent experiments were performed.
Fluorescence-activated cell sorting
(FACS)
Cells were collected and fixed in ice-cold 70%
ethanol overnight. The fixed cells were washed with
phosphate-buffered saline (PBS) and stained with a freshly-prepared
solution containing 25 µg/ml propidium iodide (Sigma-Aldrich), 10
µg/ml RNaseA (Sigma-Aldrich), 0.05 mM ethylene diamine
(Sigma-Aldrich) and 0.2% Triton X-100 tetra-acetic acid
(Sigma-Aldrich) in PBS for 30 min in the dark. For each sample,
≥20,000 cells were analyzed using FACS cytometry (Epics Altra;
Beckman Coulter, Inc., Brea, CA, USA) and Multicycle AV for Windows
5.0 (Phoenix Flow Systems, Inc., San Diego, CA, USA).
Oligonucleotide transfection
miR-299-5p and miR-299-3p mimics and control were
synthesized by Genepharma Co., Ltd. (Shanghai, China). The
sequences were as follows: miR-229-5p mimic,
5′-UGGUUUACCGUCCCACAUACAU-3′; miR-229-3p mimic,
5′-UAUGUGGGAUGGUAAACCGCUU-3′; and negative control,
5′-UUCUCCGAACGUGUCACGUTT-3′. miR-299-3p and miR-299-5p inhibitors
(2′-O-methyl modification) were synthesized by Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). miR-299-5p/3p and anti-miR-299-5p/3p
were transfected into NB4 and HL-60 cells. Oligonucleotide
transfection was performed by using Lipofectamine® 2000
transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol.
Luciferase reporter constructs and
luciferase assay
The wild-type and mutant 3′UTR of p21Cip1/Waf1 were
cloned downstream of a cytomegalovirus (CMV) promoter-driven
firefly luciferase cassette in a pCDNA3.0 vector (Thermo Fisher
Scientific, Inc.). The primers were as follows: p21Cip1/Waf1
wild-type, forward 5′-AGTGGACGTTCCCCGAGTT-3′ and reverse
5′-TCCCAAAAGCCCATTTATT-3′; p21Cip1/Waf1 mutant, forward
5′-CTAGAAAGAAGATGTCCCCCATCATATACCCCTAAC-3′ and reverse
5′-GATGGGGGACATCTTCTTTCTAGGAGGGAGACACTG-3′ (Invitrogen; Thermo
Fisher Scientific, Inc.). For the luciferase assay, HL-60 cells
were cultured in 24-well plates and cotransfected with 20 pmol of
RNA (negative control, miR-299-5p or miR-299-3p mimics), 200 ng
luciferase reporter construct and 20 ng pRL-CMV Renilla luciferase
reporter construct (Promega Corporation, Madison, WI, USA) using
Lipofectamine 2000 transfection reagent according to the
manufacturer's protocol. After 48 h, luciferase activity was
measured using the Dual-Luciferase® Reporter Assay
System (Promega Corporation, Madison, WI, USA).
Western blot analysis
Proteins were separated on 12% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and then transferred to
a nitrocellulose membrane (Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The membrane was blocked with 5% non-fat milk and
incubated with mouse anti-p21Cip1/Waf1 monoclonal antibody
(dilution, 1:1,000; catalog no., DCS60; Cell Signaling Technology,
Inc., Danvers, MA, USA) and rabbit polyclonal anti-β-actin
(dilution, 1:1,000; catalog no., ab119716; Abcam, Cambridge, MA,
USA). After the membranes were washed, they were incubated for 1 h
at room temperature with mouse anti-rabbit (catalog no., bs-0295M;
dilution, 1:1,000; Bioss Inc., Woburn, MA, USA) and rabbit
anti-mouse (catalog no. bs-0296R; dilution, 1:1,000; Bioss Inc.)
immunoglobulin G secondary antibodies. The proteins were visualized
and quantified using Pierce™ ECL Western Blotting Substrate (Thermo
Fisher Scientific, Inc.).
Statistical analysis
Statistical analyses were performed using GraphPad
Prism version 5.0 software (GraphPad Software, Inc., La Jolla, CA,
USA). The results are presented as the mean ± standard error of the
mean. The data were subjected to two-tailed Student's t-test or
one-way analysis of variance when more than two groups were
compared. P<0.05 was considered to indicate a statistically
significant difference.
Results
Increased expression of miR-299
promotes APL cell growth and cell cycle G1/S transition
In order to investigate the function of miR-299 in
APL, a lentivirus vector cloned with miR-299 was constructed, and
two stable APL cell lines with lentivirus transduction were
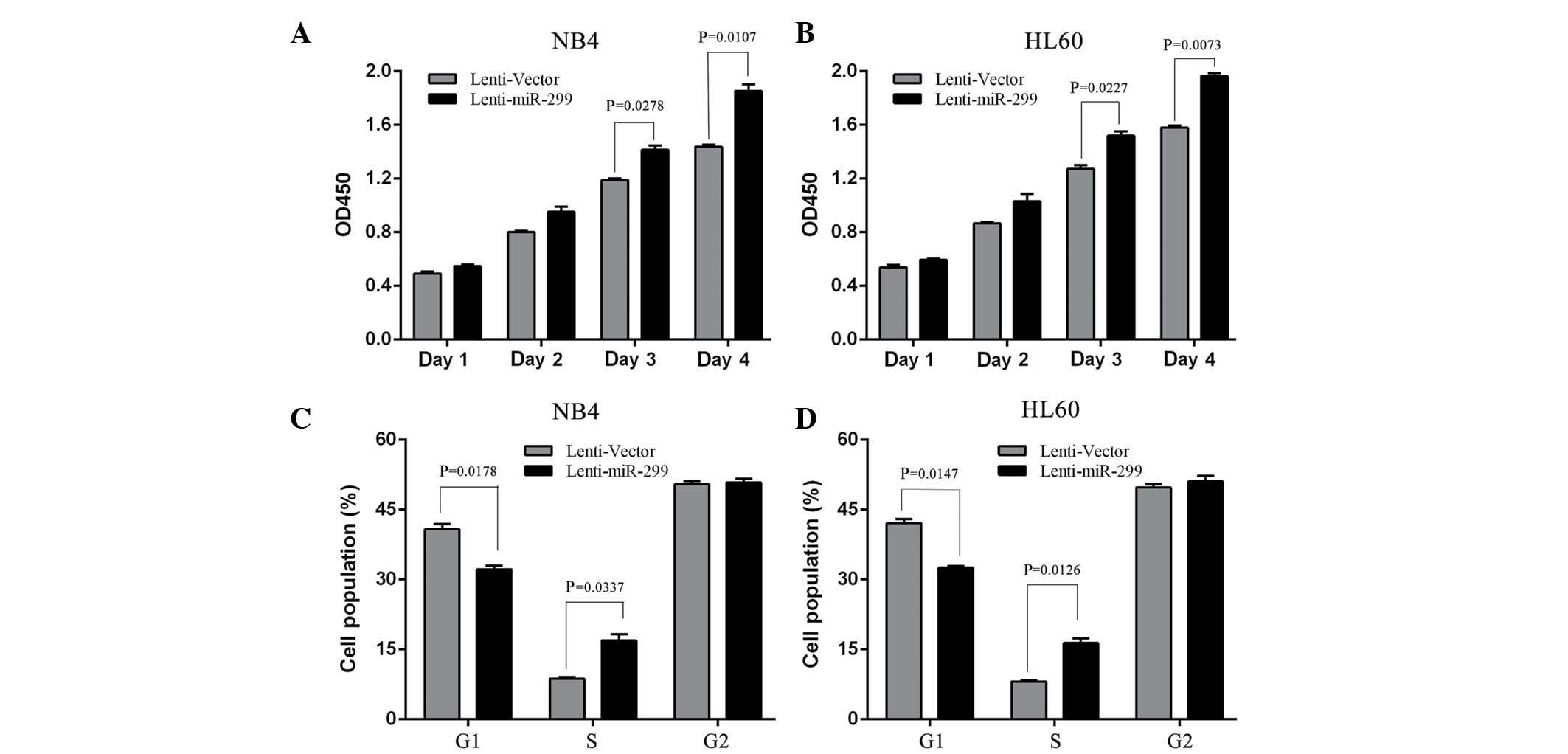
established: NB4–299 and HL-60-299. Cell proliferation assays
revealed that an increased expression of miR-299 lead to a
significant increase in cell growth of NB4–299 and HL-60–299 cells
compared with vector control cells subsequent to 3 and 4 days of
growth (Fig. 1A and B; P<0.05).
Since, miR-299 promotes APL cell proliferation, the effect that
miR-299 has on APL cell cycle progression was investigated. There
was a significant reduction in the number HL-60–299 and NB4–299
cells in the G1 phase and a clear increase in cells in the S phase
compared with the vector control cells (Fig. 1C and D; P<0.05). Therefore, miR-299
induced the growth of APL cells, possibly by enhancing cell cycle
progression at the G1/S transition.
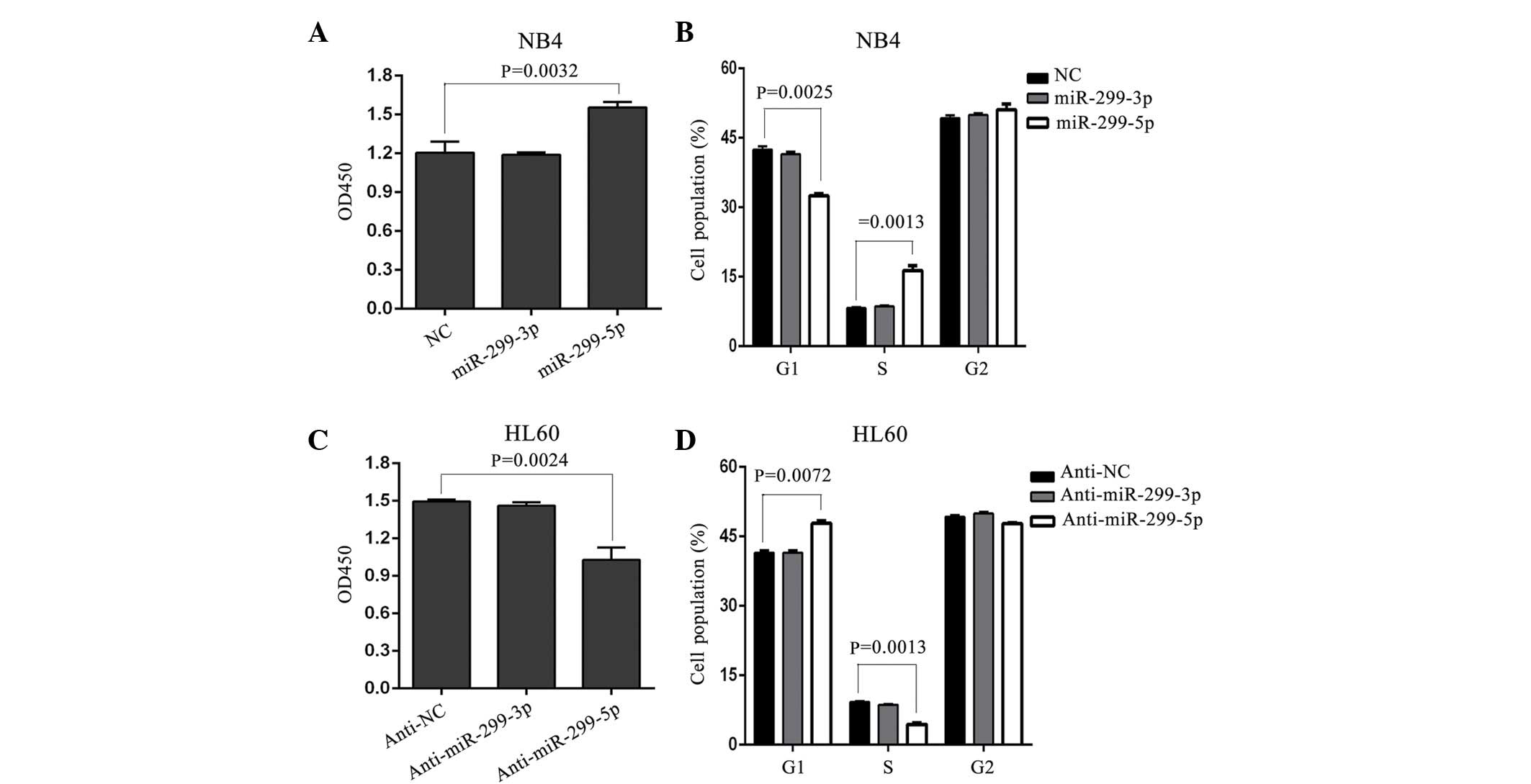
miR-299-5p, but not miR-299-3p,
promotes the growth and cell cycle progression of APL cells
According to miRBase sequences (www.mirbase.org/), miR-299 consists of two mature
sequences: miR-299-5p and miR-299-3p. Consequently, to additionally
investigate which mature sequence is involved in APL cell growth,
synthesized miR mimics of miR-299-5p and miR-299-3p were introduced
into NB4 cells, which did not express miR-299, but did express
PML/RARα. In addition, in a previous study, an anti-miR mimic of
miR-299-5p and miR-299-3p were introduced into HL-60 cells, which
did not express PML/RARα, but did have a relatively high expression
of miR-299 (12). The present results
demonstrated that miR-299-5p, but not miR-299-3p, promoted APL cell
growth and led to a significant increase in cell cycle arrest at G1
in NB4 cells (Fig. 2A and B).
Additionally, a significant decrease in cell growth and increased
cell cycle arrest at G1 was observed following silencing of
miR-299-5p in HL60 cells compared with control cells (P<0.05),
whereas miR-299-3p silencing had no significant effect (Fig. 2C and D). Overall, these results reveal
that APL cell proliferation and cell cycle progression is enhanced
by miR-299-5p, but not miR-299-3p.
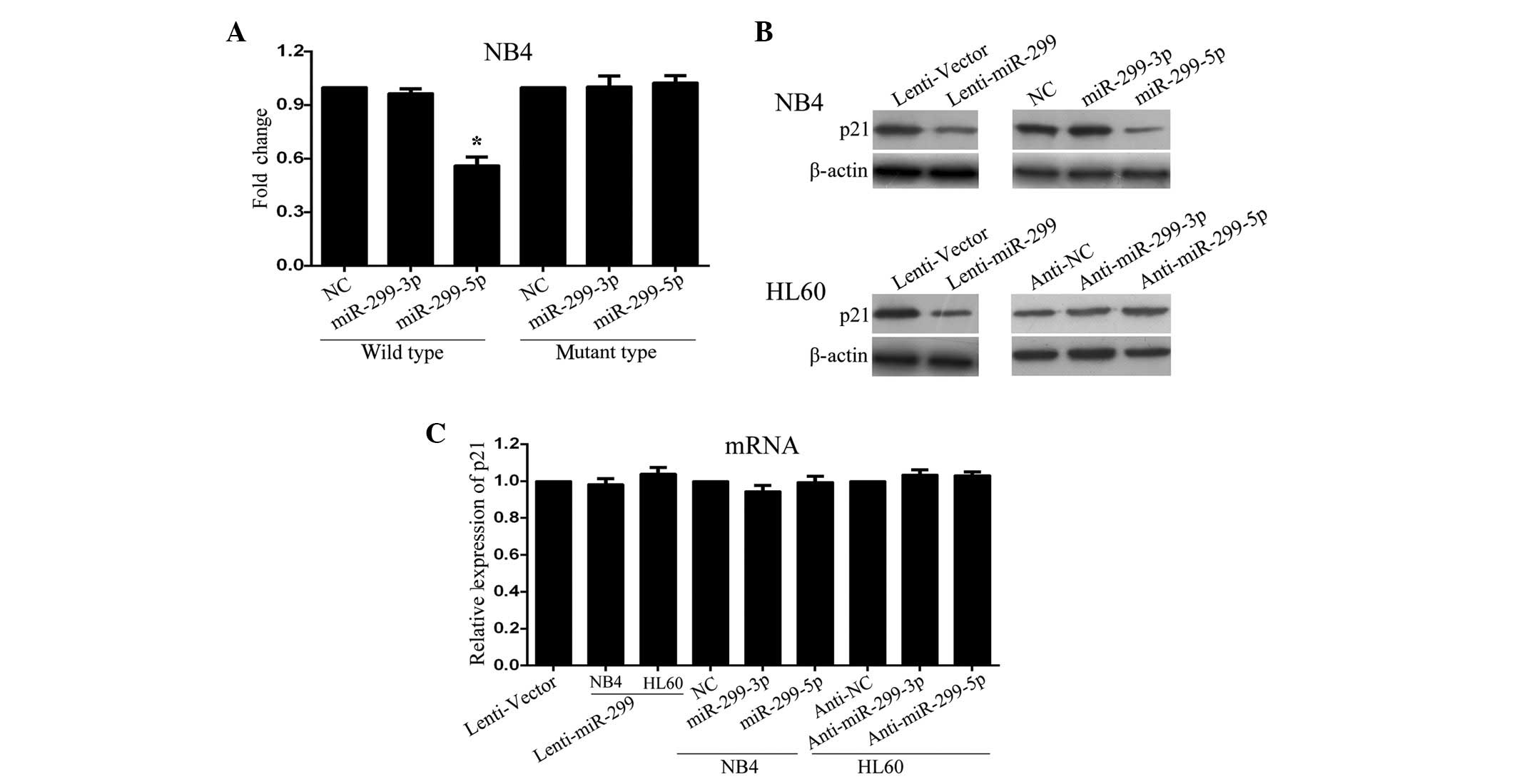
miR-299-5p downregulates p21Cip1/Waf1
expression by directly targeting the 3′UTR
It is well known that miRs is functions by causing a
downregulation in the expression of downstream target genes. The
present authors previously demonstrated that p21Cip1/Waf1 is
directly targeted by ~28 miRs in HEK-293 cells, using a
high-throughput luciferase reporter screen (13). Notably, miR-299-5p was one of the 28
miRs identified. Therefore, to determine whether miR-299-5p exerts
its function by downregulating the expression of p21Cip1/Waf1
through direct binding to its 3′UTR, full-length fragments of
p21Cip1/Waf1 mRNA 3′UTR (either wild-type or mutant-type) were
constructed and inserted downstream of the luciferase reporter
gene. For the luciferase assays, either the miR-299-5p or the
miR-299-3p mimic was cotransfected with various luciferase 3′ UTR
constructs in NB4 cells.
The present results revealed that the relative
luciferase activity with the wild-type 3′UTR of p21Cip1/Waf1 was
decreased by expression of miR-299-5p, but not miR-299-3p (Fig. 3A; P=0.034). Additional analysis
revealed that this regulation was sequence specific, since the
relative luciferase activity did not decrease as clearly in UTRs
with mutant-binding sites compared with wild-type-binding sites
(Fig. 3A). In concordance with these
results, a clear decrease in endogenous p21Cip1/Waf1 protein was
observed in NB4 and HL-60 cells following transfection with
miR-299-expressing lentivirus. Following transfection with the
miR-299-5p mimic, p21Cip1/Waf1 protein expression was significantly
decreased in NB4 cells, whereas no clear alterations were observed
following transfection with miR-299-3p mimic (Fig. 3B). Furthermore, inhibition of
miR-299-5p, but not miR-299-3p, significantly increased the
expression levels of p21Cip1/Waf1 protein in HL60 cells (Fig. 3B). Notably, overexpression of
miR-299-5p or miR-299-3p did not affect the expression level of
p21Cip1/Waf1 mRNA (Fig. 3C). Overall,
these results suggest that miR-299-5p downregulates p21Cip1/Waf1
expression post-transcriptionally by directly targeting its
3′UTR.
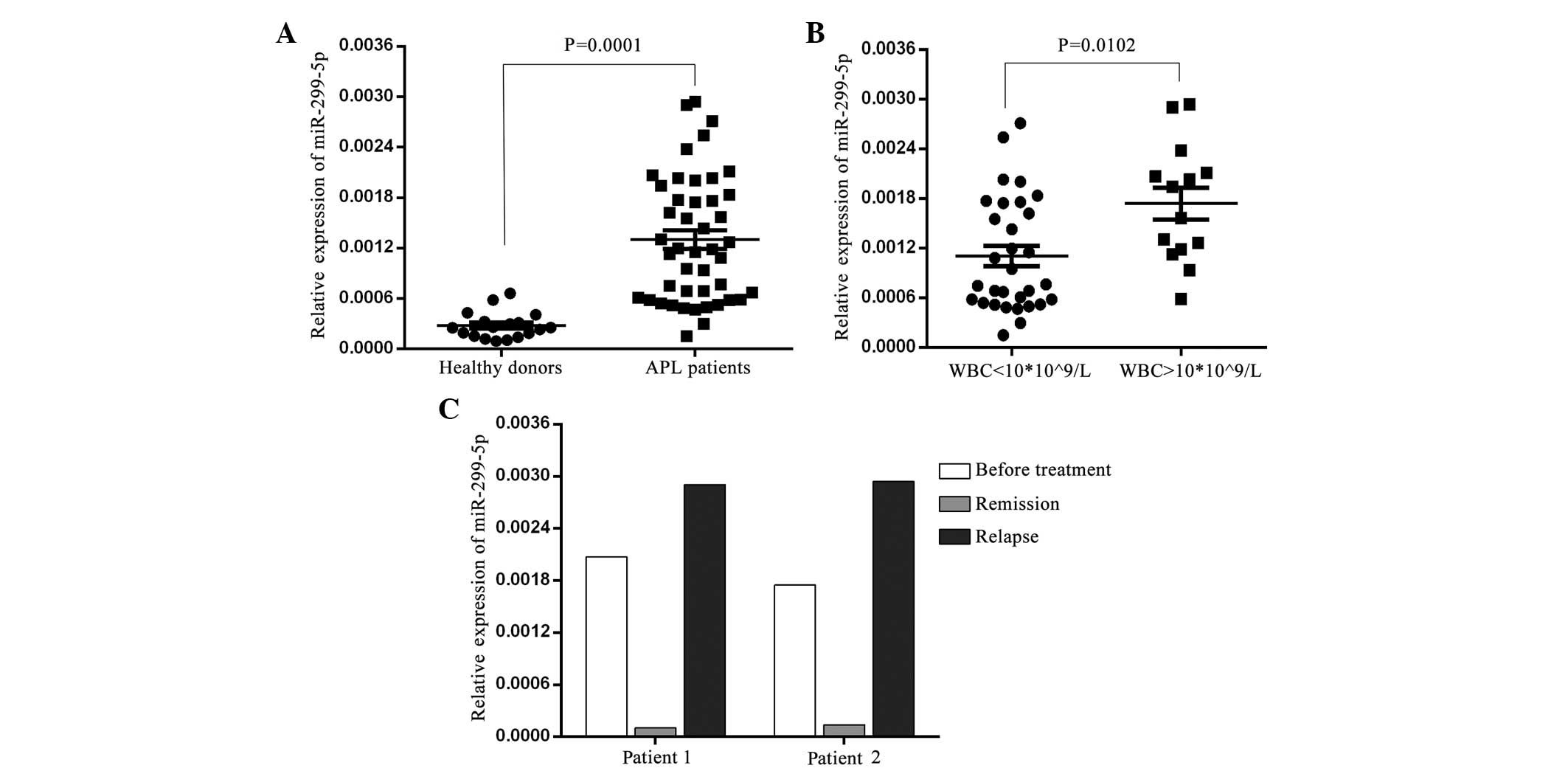
miR-299-5p is overexpressed in
patients with APL
Finally, the expression levels of miR-299-5p and
miR-299-3p were determined in bone marrow MNCs from newly diagnosed
APL patients and healthy donors using TaqMan qPCR. The results
revealed that miR-299-5p was significantly increased in APL
patients compared to healthy donors (Fig.
4A; P<0.01).miR-299-3p was not detected in APL patients
(data not shown). Prior to treatment, APL patients were divided
into high [white blood cell (WBC) count, >10×109/l]
and low-risk (WBC count, ≤10×109/l) groups.
Subsequently, miR-299-5p expression was analyzed in the high-risk
and low-risk groups, and the results revealed that miR-299-5p
expression was significantly higher in MNCs from high-risk APL
patients compared with low-risk patients (Fig. 4B; P<0.05). Furthermore, miR-299-5p
expression was determined in certain patients following treatment
remission and relapse. These patients were treated with all
trans-retinoic acid (ATRA; 45mg/m2 per dose) and
remained on ATRA for ~30 days following the attainment of complete
clinical remission. After discontinuing ATRA treatment, 3 cycles of
cytarabine and idarubicin (or daunorubicin) were administered as
consolidation therapy. As shown in Fig.
4C, miR-299-5p expression decreased following remission and
increased following relapse. Taken together, these results
demonstrate that the expression levels of miR-299-5p are increased
in APL patients; therefore, miR-299-5p may serve as a potential
prognostic indicator for APL.
Discussion
miR-299 consists of two mature sequences, miR-299-5p
and miR-299-3p, and is located at 14q32.31, which encodes 40 miRs
(16). Previously, it was reported
that miR-299-5p was downregulated in spheroid-derived breast cancer
cells, resulting in an increased expression of osteopontin
(17). Another study revealed that
miR-299-5p is involved in the regulation of hematopoietic
progenitor fate by modulating megakaryocytic-granulocytic from
erythroid-monocytic differentiation (18). In addition, a dysregulation of
miR-299-5p is observed in primary biliary cirrhosis (19) and prostate cancer (16). miR-299 expression is upregulated in
APL patient samples with a t(15;17) translocation, but is not
identified in NB4 cells with a t(15;17) translocation (12). However, there is little information
concerning the function of miR-299-5p.
The present study demonstrated that ectopic
expression of miR-299 promotes cell growth and cell cycle
progression at the G1/S transition in APL cells. As it is known,
miR-299 consists of two mature sequences, miR-299-3p and
miR-299-5p; therefore, the miR-299 sequence involved in APL cell
growth remains to be elucidated. Consequently, the present study
overexpressed miR-299-5p and miR-299-3p in HL-60 and NB4 cells by
introducing synthesized miR mimics. The present results revealed
that an overexpression of miR-299-5p promoted APL cell growth and
resulted in a significant increase in cell cycle arrest at G1,
whereas an overexpression of miR-299-3p did not. These results
indicate that miR-299 enhances APL cell proliferation and cell
cycle progression via miR-299-5p. In addition, the present study
identified that miR-299-5p was overexpressed in APL patients and,
therefore, may be used as a predictor of disease conditions, unlike
miR-299-3p.
Downstream target genes are essential when miRs
exert their function (20). The
present authors previously identified, using high-throughput
screening, 28 miRs, including miR-299-5p, which regulated
p21Cip1/Waf1 expression by directly targeting its 3′UTR (13). The present study also identified
p21Cip1/Waf1 as a direct functional downstream target of miR-299-5p
in APL cells. One potential target site of miR-299-5p was
identified in p21Cip1/Waf1 3′UTR, and cotransfection of miR-299-5p
with wild-type 3′UTR led to a decrease of the relative luciferase
activity. However, when the potential target site was mutated,
luciferase activity increased to a level similar to that observed
with the control vector. In addition, endogenous p21Cip1/Waf1
protein levels were downregulated by miR-299-5p in APL cells.
Notably, the expression level of p21Cip1/Waf1 mRNA did not alter
following an overexpression of miR-299-5p. Therefore, miR-299-5p
possibly regulates p21Cip1/Waf1 expression
post-transcriptionally.
In conclusion, the present study has demonstrated
for the first time, to the best of our knowledge, that miR-299,
primarily miR-299-5p, exerts growth-promoting effects in APL cells
by downregulating the expression of the tumor suppressor
p21Cip1/Waf1. The present results suggest an oncogenic function and
a potential therapeutic application for miR-299 in APL.
Acknowledgements
The authors are grateful to Professor Didier Trono
for providing the pWPXL, psPAX2 and pMD2.G lentivirus plasmids.
This study was supported by the National Natural Science Foundation
of China (grant no. 81201872), Natural Science Foundation of Fujian
Province (grant nos. 2010J01164 and 2013J01308) and Foundation of
Fujian Key Laboratory of Hematology (grant no. 2009J1004).
References
|
1
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhong X, Coukos G and Zhang L: miRNAs in
human cancer. Methods Mol Biol. 822:295–306. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
He L and Hannon GJ: MicroRNAs: Small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Avvisati G, ten Cate JW and Mandelli F:
Acute promyelocytic leukaemia. Br J Haematol. 81:315–320. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mathews V, George B, Lakshmi KM,
Viswabandya A, Bajel A, Balasubramanian P, Shaji RV, Srivastava VM,
Srivastava A and Chandy M: Single-agent arsenic trioxide in the
treatment of newly diagnosed acute promyelocytic leukemia: Durable
remissions with minimal toxicity. Blood. 107:2627–2632. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mi S, Lu J, Sun M, Li Z, Zhang H, Neilly
MB, Wang Y, Qian Z, Jin J, Zhang Y, et al: MicroRNA expression
signatures accurately discriminate acute lymphoblastic leukemia
from acute myeloid leukemia. Proc Natl Acad Sci USA.
104:19971–19976. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Georgantas RW III, Hildreth R, Morisot S,
Alder J, Liu CG, Heimfeld S, Calin GA, Croce CM and Civin CI: CD34+
hematopoietic stem-progenitor cell microRNA expression and
function: A circuit diagram of differentiation control. Proc Natl
Acad Sci USA. 104:2750–2755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jongen-Lavrencic M, Sun SM, Dijkstra MK,
Valk PJ and Löwenberg B: MicroRNA expression profiling in relation
to the genetic heterogeneity of acute myeloid leukemia. Blood.
111:5078–5085. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Dixon-McIver A, East P, Mein CA, Cazier
JB, Molloy G, Chaplin T, Andrew Lister T, Young BD and Debernardi
S: Distinctive patterns of microRNA expression associated with
karyotype in acute myeloid leukaemia. PLoS One. 3:e21412008.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wu S, Huang S, Ding J, Zhao Y, Liang L,
Liu T, Zhan R and He X: Multiple microRNAs modulate p21Cip1/Waf1
expression by directly targeting its 3′ untranslated region.
Oncogene. 29:2302–2308. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Vardiman JW, Thiele J, Arber DA, Brunning
RD, Borowitz MJ, Porwit A, Harris NL, Le Beau MM,
Hellström-Lindberg E, Tefferi A and Bloomfield CD: The 2008
revision of the World Health Organization (WHO) classification of
myeloid neoplasms and acute leukemia: Rationale and important
changes. Blood. 114:937–951. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Formosa A, Markert EK, Lena AM, Italiano
D, Finazzi-Agro' E, Levine AJ, Bernardini S, Garabadgiu AV, Melino
G and Candi E: MicroRNAs, miR-154, miR-299-5p, miR-376a, miR-376c,
miR-377, miR-381, miR-487b, miR-485-3p, miR-495 and miR-654-3p,
mapped to the 14q32.31 locus, regulate proliferation, apoptosis,
migration and invasion in metastatic prostate cancer cells.
Oncogene. 33:5173–5182. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lowery AJ, Miller N, Devaney A, McNeill
RE, Davoren PA, Lemetre C, Benes V, Schmidt S, Blake J, Ball G and
Kerin MJ: MicroRNA signatures predict oestrogen receptor,
progesterone receptor and HER2/neu receptor status in breast
cancer. Breast Cancer Res. 11:R272009. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tenedini E, Roncaglia E, Ferrari F,
Orlandi C, Bianchi E, Bicciato S, Tagliafico E and Ferrari S:
Integrated analysis of microRNA and mRNA expression profiles in
physiological myelopoiesis: Role of hsa-mir-299-5p in CD34+
progenitor cells commitment. Cell Death Dis. 1:e282010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Padgett KA, Lan RY, Leung PC, Lleo A,
Dawson K, Pfeiff J, Mao TK, Coppel RL, Ansari AA and Gershwin ME:
Primary biliary cirrhosis is associated with altered hepatic
microRNA expression. J Autoimmun. 32:246–253. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|