Introduction
Flavonoids have recently drawn extensive attention
due to their interesting biological activities (1). This family of compounds comprises a
large class of low-molecular weight, natural products of plant
origin that are ubiquitously distributed in edible fruits and
vegetables. Quercetin is one of the most important flavonoids and
exhibits a wide range of biological activities. The significant
antitumor, anti-allergy and anti-inflammatory effects of quercetin
have been extensively reviewed (2,3). Evidence
indicates that quercetin is able to target various types of
malignant tumor cells, including leukemia, breast, esophagus,
colon, prostate, nasopharyngeal, endometrial and lung cancers
(1,4).
The proliferation of these malignant cells may be inhibited by
quercetin; however, the exact molecular mechanisms underlying the
effects of quercetin are unknown.
Recently, a new quantitative method has demonstrated
great promise for the simultaneous, accurate identification and
quantitation of complex protein mixtures. This method is termed
stable isotope labeling by amino acids in cell culture (SILAC)-mass
spectrometry (MS), and identifies proteins by MS based on stable
isotopes (5). Using this method, we
have previously studied total proteomic expression changes of HepG2
cells and identified certain significant differences in HepG2 cells
following their cultivation with quercetin. These proteins may
serve important roles in multiple cellular processes, including
protein synthesis, signal transduction, cytoskeleton formation and
metabolism (6).
One of the most important goals of anticancer
research is to inhibit proliferation of malignant cells. Normal
cell proliferation requires successful transition through cell
cycle checkpoints (7). Access to the
mitotic stages of the cell cycle is strictly controlled by
growth-promoting and growth-inhibitory signals; these signalling
pathways ensure that cells do not unnecessarily commit to DNA
replication and cell division (8). In
eukaryotes, cyclins and cyclin-dependent kinases (CDKs) are
principally responsible for controlling cell cycle regulation
(7). Cyclins are the regulatory
subunits, while CDKs are the catalytic subunits, and each cyclin
contains a special region that allows binding to form a cyclin-CDK
complex (8). When this complex is in
an active state, it permits cells to move past the check points of
particular cell cycle phases through the phosphorylation of unique
protein substrates. The G1/S transition is the most important check
point of the cell cycle that is regulated by cyclin-CDK complexes
(8).
In mammalian cells, G1 cyclins serve a major a role
in cell cycle regulation. The G1 cyclins are composed of two
families of phase regulatory proteins: D-type cyclins and cyclin E
(9). Cyclin D consists of three
subunits that are central to the transition of cells from one phase
to the next: Cyclin D1 (CCND1), D2 (CCND2), and D3 (CCND3). These
proteins share a cyclin box and PEST sequence, but bind to
different chromosomes (10).
Recently, the majority of research on the structure and function of
cyclin proteins has focused on CCND1. Overexpression and
rearrangement of CCND1 has been found to be associated with tumor
progression in numerous different tumor types, including carcinomas
of the esophagus, prostate and breast (11,12). In
the present study, total protein identification by SILAC-MS
revealed that the peak of CCND1 in cells treated with quercetin was
significantly different compared with the CCND1 expression in
control HepG2 cells. Due to the essential function of CCND1, the
influence of quercetin on cell cycle distribution of HepG2 cells
was investigated. Combined with the SILAC-MS results, the current
study reports the possible mechanism by which quercetin may inhibit
the proliferation of HepG2 cells.
Materials and methods
Materials
Quercetin (PHR1488), all normal L-amino acids,
trypan blue, dimethyl sulfoxide, Tris-HCl, Triton X-100 and
propidium iodide were obtained from Sigma-Aldrich (St. Louis, MO,
USA). The human hepatoma cell line HepG2 was obtained from the
American Type Culture Collection (Manassas, VA, USA). Deuterated
leucine (Leu-d3,5,5,5-D3, 98%, lot: l1.8473) was obtained from
Cambridge Isotope Laboratories, Inc. (Tewksbury, MA, USA).
Potassium phosphate and Tris base were purchased from Merck
Millipore (Darmstadt, Germany). Rabbit polyclonal anti-CCND1 IgG,
(sc-753) and mouse monoclonal anti-β-actin IgG (sc-130300)
antibodies were purchased from Santa Cruz Biotechnology Inc. (Santa
Cruz, CA, USA).
Cell culture and SILAC
HepG2 cells were cultured and labeled as described
previously (6). Briefly, Eagle's
minimum essential medium (Yubo Biotech Ltd., Shanghai, China),
including appropriate amounts of amino acids with the exception of
leucine, was mixed with 50 mg/l each of the antibiotics gentamicin,
penicillin and streptomycin, reconstituted according to the
manufacturer's specifications. The leucine-free medium was divided
into two equal portions. L-leucine (Leu-d0) and L-leucine-d3
(Leu-d3) were added to each media to reach a final leucine
concentration of ~52 mg/l. The mixture with a full complement of
amino acids was filtered through a sterile 0.22 µm filter (EMD
Millipore, Bedford, MA, USA). Control HepG2 cells were cultured in
normal Dulbecco's modified Eagle's medium (DMEM; Yubo Biotech Ltd.)
with Leu-d0, and cells treated with quercetin were put in
d3-labeling media with Leu-d3. The morphological changes of the
HepG2 cells were observed following the addition of quercetin.
Briefly, the growth rate and cell morphology of HepG2 cells was
evaluated using an inverted microscope (IX50; Olympus Corporation,
Tokyo, Japan) after 24, 48 and 72 h. The results of our previous
research (6) indicated that the
proliferation and apoptotic properties of HepG2 cells were
significantly different when the cells were treated with 50 µmol/l
quercetin for 48 h compared with untreated cells. Thus, this
condition was selected to perform the Leu-d3 labeling treatment.
Cells were grown in DMEM containing 10% fetal bovine serum in
humidified air with 5% CO2 at 37°C. The d3-labeling
medium was replaced after four days, and cells were passaged for
>10 generations in order to produce adequate proportions of
Leu-d3-labeled cells.
Protein preparation and gel
digestion
As described in our previous study (6), HepG2 cells were treated with 50 µM
quercetin for 48 h and collected to analyze protein changes. HepG2
cells were cultured with and without quercetin and lysed using
radioimmunoprecipitation assay buffer [10 mM Tris-Cl (pH8.0); 1 mM
ethylenediaminetetraacetic acid; 0.5 mM ethylene glycol tetraacetic
acid; 1% Triton X-100; 0.1% sodium deoxycholate; 0.1% sodium
dodecyl sulfate (SDS); 140 mM NaCl] to extract the protein. Equal
amounts of the protein extraction were then separated with 12%
Tris-glycine SDS-polyacrylamide gel electrophoresis (PAGE), stained
with Coomassie Blue. The gel lanes were cut to 1 mm3
pieces and destained with 1:1 acetonitrile (ACN) and 50 mM ammonium
bicarbonate solution, followed by dehydration in 100% ACN for ~15
min. The gel pieces were completely dried under vacuum
centrifugation. Subsequently, samples were incubated overnight in a
10 ng/µl MS-grade trypsin solution (Promega Corporation, Madison,
WI, USA). Finally, peptides were extracted with a formic acid
solution (5% formic acid, 50% ACN) and resuspended in 50% ACN and
0.1% trifluroacetic acid for the MS analysis.
Liquid chromatography (LC)-tandem MS
(MS/MS) analysis and protein quantification
LC-MS/MS analysis was designed as described in our
previous paper (6). Briefly, LC-MS/MS
analysis was performed with nano-ultra-performance liquid
chromatography (nanoUPLC) and a 4x tandem mass spectrometer
(UPLC-Q-TOF; Waters Corporation, Milford, MA, USA) coupled with an
electrospray ionization (ESI) source. NanoUPLC separations were
performed on a nanoAcquity™ C18 column 3.5 µm, 0.75 µm × 100 mm
(Waters Corporation). The chromatographic separation was performed
at a flow rate of 0.3 µl/min with the mobile phase, consisting of
solvents A [water:formic acid, 100:0.1 (v/v)] and B [ACN:formic
acid, 100:0.1 (v/v)], using a linear gradient elution from 95% A:5%
B to 60% A:40% B in a 45-min initial step; then decreased to 10%
A:90% B at 55 min, and finally increased to 95% A:5% B at 60 min.
Subsequently, 2 µl of sample was injected for the nanoUPLC-MS/MS
analysis. The ESI source was performed in positive ion mode at a
spray voltage and cone voltage of 2.8 kV and 40 kV. The source
temperature was set at 90°C, and 0.45 l/h argon was used as the
collision gas. The speed of nano gas flow was controlled at 0.36
l/h. Spectra were accumulated until a satisfactory signal/noise
ratio had been obtained from a range of 400–1,600 mz. Three MS/MS
ions with charged states of 2+ and 3+ were selected for each
replicate; trypsin autolysis products and keratin-derived precursor
ions were automatically excluded.
The MS/MS data (pkl files), including mass values,
intensity and charge of precursor ions, were acquired by the
software ProteinLynx v2.25 (Waters Corporation). Mascot Program 2.0
(Matrix Science Ltd., London, UK) was used to analyze the pkl
files. A strict standard was set to ensure a high confidence level
for identification. Peptides with Mascot scores below the threshold
value were excluded from the data (P<0.01). Protein abundance
was calculated as the peak intensity of the fragment ions from the
labeled vs. unlabeled peptides. A protein was considered
significantly different after treatment with quercetin if its
average change ratio was 5x higher or lower than the average
standard variation of all quantified proteins.
Reverse transcription-polymerase chain
reaction (RT-PCR) and western blot analysis of CCND1 expression
following treatment with quercetin
Semi-quantitative RT-PCR and western blot analyses
were conducted to further explore the change in CCND1 expression
after the results of the LC-MS/MS analysis were obtained. Briefly,
Trizol reagent was used to isolate RNA from control HepG2 cells and
HepG2 cells treated with 50 µM quercetin for 48 h. Trizol (1 ml)
was added to each tube of 107 cell pellets and incubated
for 5 min at 4°C. The lysate was then extracted with 0.2 ml
phenol/chloroform for 15 sec and centrifuged at 12,000 × g and 4°C
for 15 min. The concentration of RNA was determined by the
OD260/280 absorption value of the solution. Total RNA quality was
evaluated by 1% agarose gel electrophoresis.
RT-PCR was performed using a One-Step RNA PCR kit
(Promega Corporation) with 500 ng total RNA by using the following
primers in the following conditions: 45 min at 45°C; 2 min at 95°C;
followed by 25 cycles of 30 sec at 94°C, 30 sec at 54°C and 30 sec
at 72°C; and a final elongation step of 10 min at 72°C. The CCND1
(forward, 5′-AGGAACAGAAGTGCGAGGAG-3′; reverse,
5′-CACAGAGGGCAACGAAGGT-3′) β-actin (forward,
5′-ACCCCCACTGAAAAAGATGA-3′; reverse, 5′-ATCTTCAAACCTCCATGATG-3′)
primers were synthesized by Invitrogen (Thermo Fisher Scientific,
Inc., Carlsbad, CA, USA). Approximately 5 µl of amplified product
was separated on a 1% agarose gel, stained with ethidium bromide
and observed under ultraviolet light. The DNA ladder was obtained
from Nanjing Jinsirui Biotechnology Co., Ltd. (Nanjing, China).
β-actin was used as a positive control for RT-PCR. The Quantiscan
3.0 software used for densitometry was obtained from Azure
Biosystems, Inc. (Dublin, CA, USA).
Western blot analysis was performed as described
previously (6). Protein extractions
from control HepG2 cells and HepG2 cells treated with quercetin for
48 h were electrophoresed on 12% SDS-PAGE gels and transferred onto
nitrocellulose membranes (GE Healthcare Life Sciences, Piscataway,
NJ, USA). The membranes were washed in phosphate-buffered saline
(PBS) containing Tween 20 and incubated with anti-CCND1 and
anti-β-actin primary antibodies (dilution, 1:300) after blocking
with 5% skimmed milk. Membranes were incubated with polyclonal
horseradish peroxidase-conjugated rabbit anti-goat IgG antibody
(cat. no. ab6741; Abcam, Cambridge, MA, USA) for 1 h. Finally,
membranes were developed with a Western Lightning Chemiluminescence
Substrate Kit (PerkinElmer, Inc., Shanghai, China) according to the
manufacturer's specifications.
HepG2 cell cycle distribution
following treatment with quercetin
Flow cytometric analysis was performed to explore
changes in the HepG2 cell cycle following treatment with quercetin.
HepG2 cells (~106/ml) were exposed to 50 µM quercetin
for either 12, 24, 48 or 72 h. After washing in PBS, the cells were
fixed in ice-cold 70% ethanol and stored at −20°C. Then they were
stained with propidium in a hypotonic buffer and suspended in 1 ml
of hypotonic fluorochrome solution containing 50 µg propidium
iodide per ml in 0.1% sodium citrate and 0.1% Triton X-100. All
samples were analyzed with a flow cytometer (Elite ESP;
Beckman-Coulter, Miami, FL, USA). The cell cycle distribution was
estimated with Listmode software (version 3.0;
Beckman-Coulter).
Results
Influence of quercetin on HepG2
morphology and proliferation
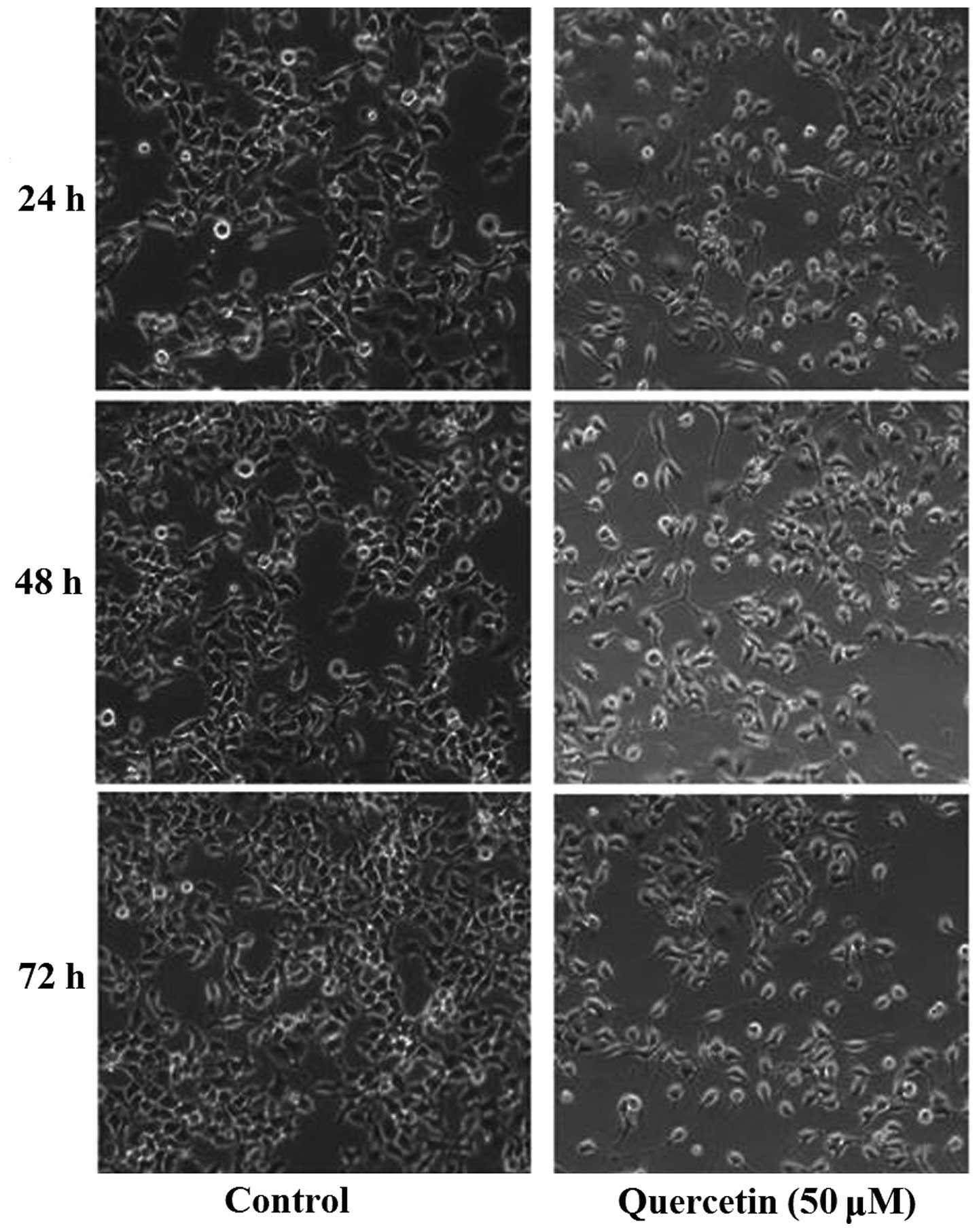
Our previous studies demonstrated that quercetin can
inhibit proliferation of HepG2 cells. Using an inverted microscope
(IX50; Olympus Corporation), substantial morphological alteration
of the HepG2 cells treated with quercetin was observed. The
quercetin-treated cells became circular, detached from their
substrate and proliferated slowly (Fig.
1). According to the results of a 3-(4,
5-dimethyl-2-thiazolyl)-2, 5-diphenyl-2H-tetrazolium bromide assay,
quercetin inhibited the proliferation of HepG2 cells in a time- and
dosage-dependent manner (6).
The Leu-d3 labeling ratio detection was performed as
previously described. After 10 passages of HepG2 cells, the
Leu-d3-labeling ratio was nearly 100% (6).
To ensure the accuracy of the results, the two HepG2
samples (the control HepG2 cells and those treated with 50 µM
quercetin) were mixed at a 1:1 ratio. The sequence SYELPDGQVITIGNER
of β-actin was used for quantification purposes. The details of the
Leu-d3 labeling and ratio identification are explained in Zhou
et al (6).
Quantification of CCND1 by
SILAC-MS
In a previous paper, we laid out a classification
system to group proteins with significantly different expression
levels (6). During the development of
tumors, D-type cyclins serve a central role in cell cycle
transitions. Recent studies have focussed on the correlations
between subunit CCND1 and the poor prognosis of malignant diseases,
such as gastric cancer and breast cancer (13). In the current study, changes in CCND1
expression were observed in hepatoma cells following treatment with
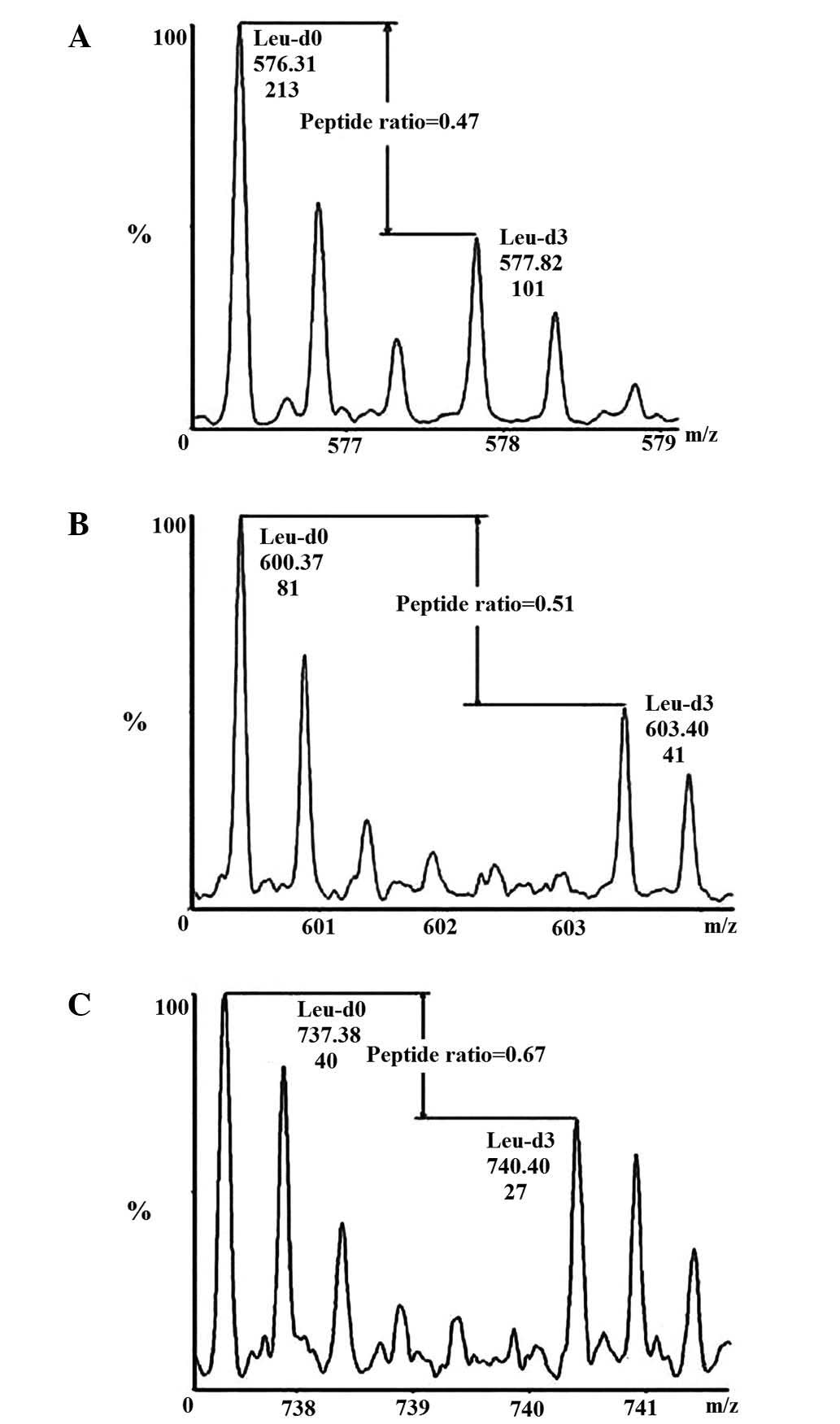
quercetin using a quantitative proteomics method. There were three
Leu-containing peptides of CCND1 to be detected and the MS score
was ~158. The average protein expression level of CCND1 was reduced
to 0.55 following quercetin treatment; a representative pair of
isotope labeling peaks of Leu-containing peptides are shown in
Fig. 2.
CCND1 expression following treatment
with quercetin measured by RT-PCR and western blot analyses
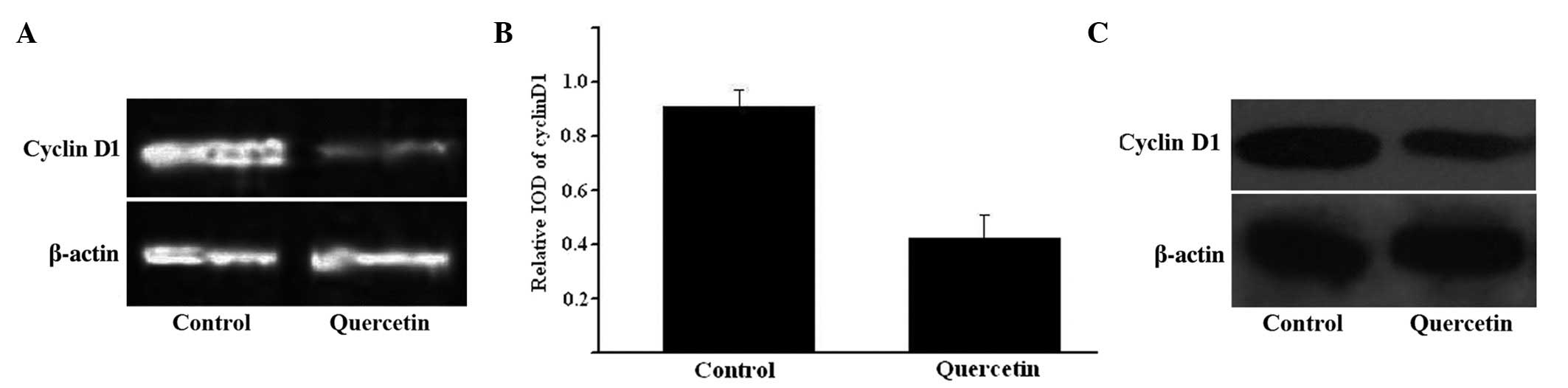
CCND1 gene expression was assessed using RT-PCR and
western blotting. The RNA and protein expression levels of CCND1
were consistently decreased in HepG2 cells when exposed to 50 µM
quercetin for 48 h, while no change was observed in β-actin
expression (Fig. 3). The relative
integrated optical density values of RT-PCR with β-actin are
indicated in Fig. 3C.
Cell cycle distribution of HepG2 cells
when treated with quercetin
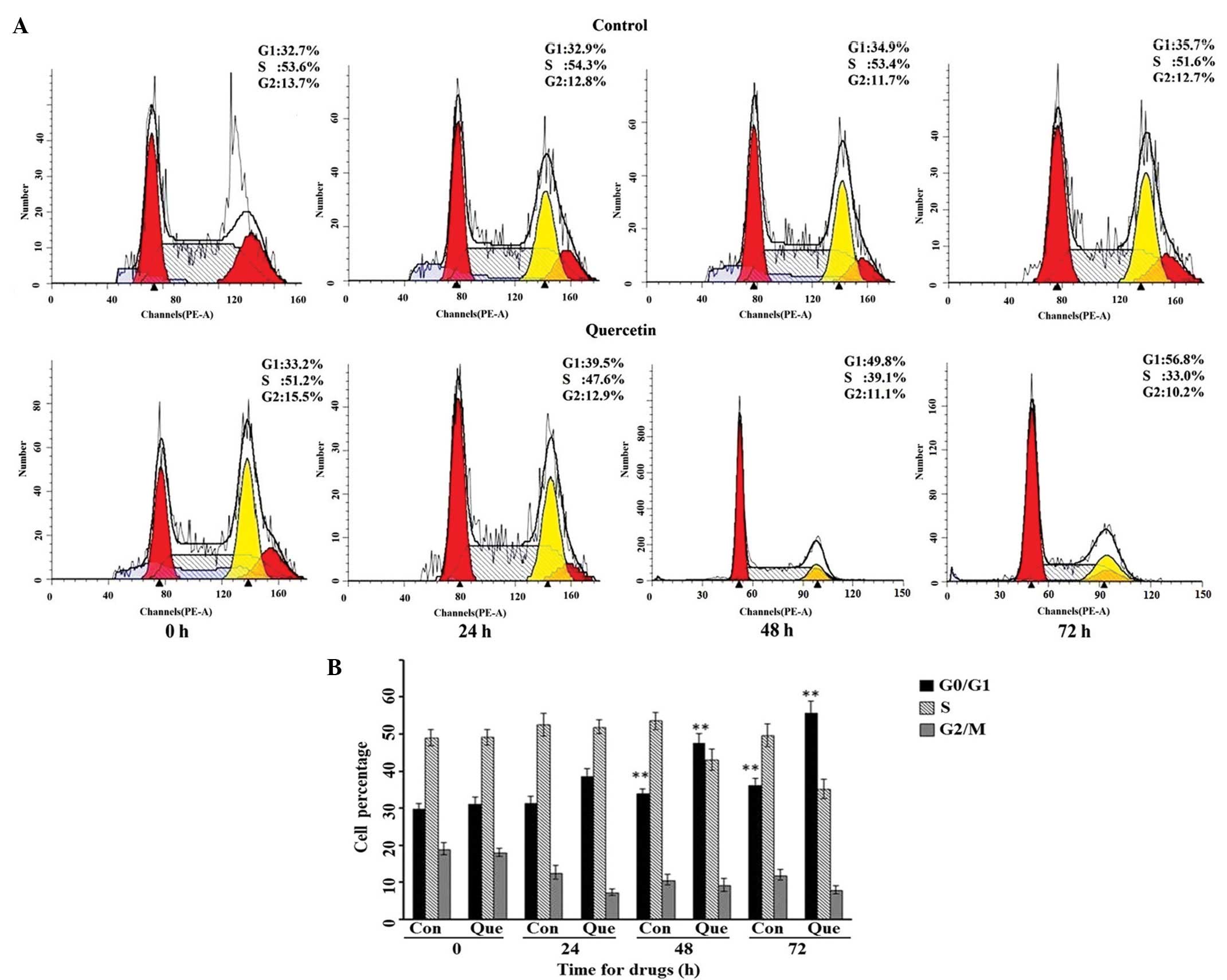
Based on the results of the MS quantification, we
suspected that quercetin may have effects on the cell cycle
distribution of HepG2 cells by influencing CCND1 activity, as the
CCND1 is important in cell cycle transitions. HepG2 cells were
treated with 50 µM quercetin for 24, 48 or 72 h, respectively, and
the cell cycle distribution was detected by flow cytometry. As
shown in Fig. 4A, the cell cycle
distribution of HepG2 significantly altered following treatment
with an effective concentration of quercetin; the percentage of
cells in the G0/G1 phase increased, and those in S phase gradually
decreased in comparison with the control HepG2 cells. The raw data
and statistical comparison can be seen in Fig. 4B.
Discussion
In recent years, the polyphenolic flavonoid compound
quercetin has attracted a great deal of attention due to its
wide-ranging biological activity and low toxicity (14). Quercetin is naturally found in plants
and is consumed in vegetables, beverages, fruits and numerous
herbal tonics. It serves important roles in various biological
processes, including antioxidant processes, multidrug resistance,
liver fibrosis and antitumorigenesis (15,16). Among
these activities, the effect of quercetin on carcinogenesis is a
highly promising research area. It has been demonstrated that
quercetin affects the development of several types of tumors,
including leukemia, breast, esophagus, colon, prostate,
nasopharyngeal, endometrial and lung cancers, and therefore has
potential value in clinical applications (17,18).
Quercetin may function in several ways to produce anticancer
effects; for example, it may influence apoptosis induction,
suppression of heat shock proteins, inhibition of oncogene
expression and antiangiogenesis, among others (19,20). Most
previous reports have focused on a certain gene or pathway, while
the exact mechanism of the effect of quercetin on these biological
processes remains unknown. In our studies, some of these mechanisms
were explored by identifying the differentially expressed proteins
in HepG2 cells following exposure to quercetin.
One of the conventional methods used to detect
protein expression levels in cells, tissues or organisms is
two-dimensional coupling with matrix-assisted laser desorption
ion-time of flight (MALDI-TOF) MS. However, this method provides
relatively low sensitivity and low accuracy results on protein
expression levels (5). A recently
developed protein identification method named SILAC-MS has the
benefit of significantly reducing the variation between replicates
and results in better quantitative statistics due to the large set
of uniformly labeled peptides. SILAC-MS is beginning to be used
extensively in the life sciences due to its simple, inexpensive and
accurate properties (21). We
utilized this method to tag leucine in HepG2 cells with d3, and
these cells were treated with quercetin (6). The isotope peaks of the
quercetin-treated HepG2 cells were then compared with control HepG2
cells in the same MS image. After three replicate experiments, 70
proteins with significantly different expression were identified,
following strictly defined standards. According to annotations in
the protein database ExPASy, these proteins can be classified into
the following categories: Cytoskeleton, signaling transduction,
protein synthesis, chaperone, metabolism and DNA/RNA binding. Some
proteins that were identified, such as members of the heat shock
protein (HSP) family, were consistent with other studies. Zanini
et al (22) reported that
quercetin is able to inhibit HSP expression in neuroblastoma cells.
This may contribute to causing higher sensitization to doxorubicin
and invert multidrug resistance. The expression of the Ras GTPase
activating-like protein IQGAP1 and β-tubulin, which takes part in
the construction of the cell skeleton, were observed to
significantly decrease following treatment with 50 µM quercetin for
48 h (6). Combined with a
scratch-induced migration assay, we suspected that quercetin may
regulate the migration ability of HepG2 cells through its
interaction with IQGAP1 and β-tubulin. Other proteins with
significant changes to their expression, such as the proto-oncogene
tyrosine-protein kinase ABL1 and α-enolase, deserve attention in
future research.
The goal of antitumor research is to inhibit the
proliferation of malignant cells, and cell proliferation requires
the successful transition through multiple cell cycle checkpoints
(7). In mammalian cells, D-type
cyclins play a central role in integrating extracellular
proliferation signals with cell cycle machinery (8). The D-type cyclin family comprises three
types of proteins: Cyclin D1, cyclin D2 and cyclin D3. The CCND1
gene is located at 11q13 and codes for the CCND1 protein. CCND1 can
be overexpressed in a number of tumor types, particularly in poorly
differentiated tumors. It has been reported that CCND1 is likely to
be a proto-oncogene (23);
deleterious changes to CCND1 may contribute to uncontrolled cell
growth characteristic of tumors, and overexpression or mutation of
CCND1 are considered to be associated with progression or poor
prognosis in numerous different malignant tumors, such as
carcinomas of the esophagus, breast, lung and pancreas, as well as
mantle cell lymphomas (24,13). In nasopharyngeal carcinoma patients,
the abnormal regulation of CCND1 may contribute to carcinogenesis
through enabling persistent infection with epstein-barr virus
(25). During the development of
lymphomas, an overexpression of CCND1 may shorten the time that a
cell spends in the resting G phase and accelerate the transition
process into the S phase, leading to malignant proliferation
(26). A mechanism study revealed
that the most common genetic abnormality affecting CCND1 has a
large impact on DNA transcription (26). CCND1 can act as a regulatory subunit
of CDK4 and CDK6, which are key enzymes in the cell cycle
transition process; it is able to phosphorylate CDKs and inhibit
the activity of the retinoblastoma protein pRb, leading to the
release of E2F-type transcription factors. These E2Fs can regulate
the transcription process of CCND1 so as to promote the cell cycle
progression. It is notable that CCND1 expression can also be
stimulated by underphosphorylated pRb. There may be an
autoregulatory loop within the cell cycle transition (26,27).
As is demonstrated by the present results, HepG2
cells treated with quercetin exhibit decreased expression of CCND1,
as well as significant changes in the cell cycle distribution in
comparison with control HepG2 cells. There have also been studies
on the influence of quercetin on the cell cycle distribution in
malignant cells, including HeLa human cervical cancer cells and
MDA-MB-453 breast cancer cells (28,29). In
certain cell types, quercetin may block the cells from
transitioning to the G1 or G2 phase. However, Jeong et al
(30) reported that a low dose of
quercetin (10 µM) may induce cell arrest in the G1 phase. In the
current study, the concentration of quercetin use was ~50 µM.
Although we have observed the same cell cycle arrest phenomenon,
the proportion of each cell cycle phase differed among the
quercetin concentration treatments. Whether there is a correlation
between the concentration of quercetin and malignant cell cycle
distribution is the subject of our next study.
In contrast to CCND1, there are relatively few
reports on CCND2 and CCND3 and their roles in carcinogenesis. CCND2
was once thought to reside in a chromosomal region that does not
readily undergo amplification. However, there has been new evidence
revising the location of CCND2. Yamak et al (31) reported that CCND2 may enhance the
activity of transcription factor GATA4, a key regulator of
cardiomyocyte growth and differentiation. Since human mutations in
this domain are linked to congenital heart disease, it has been
suggested that CCND2 may play an important role in cardiogenesis
(31). In chronic myeloid leukemia,
Jena et al (32) found that
BCR/ABL promotes the cell cycle progression by altering expression
of CCND2 (32). The overexpression of
CCND2 may also be detected in gastric cancer cases and correlates
positively with a poor prognosis (33). There have also been limited
investigations on CCND3. It is reported that CCND3 is expressed
differentially among lymphoma subtypes. In indolent lymphomas,
CCND3 overexpression was a marker of poor overall survival and poor
relapse-free survival (34). CCND3 is
also downregulated in human epidermal growth factor receptor 2
(HER2)-overexpressing breast cancer and is induced by rapamycin to
cause G1 arrest (35). Notably, a
lack of CCND1 is associated with a compensatory upregulation of
CCND3 in mice overexpressing wild-type HER2 (ErbB2) (36). There may be an association between the
regulation of these two cyclin D family members. As for the cyclin
E gene, its overexpression has been found to be an important
predictor of a poor prognosis for patients with tumors of various
organs and tissue sources. Over 10% of transgenic mice
overexpressing human cyclin E spontaneously developed mammary
carcinoma (37).
Given the critical role of D-type cyclins in cell
cycle regulation, their abnormal or untimely expression could
potentially disrupt the cell cycle and promote the proliferation of
malignant cells. In the current study of tumor cell proteomes using
the SILAC-MS method, it was found that the expression of CCND1
differed significantly following treatment with quercetin, but that
quercetin did not affect the other cyclin D family members. A
western blot assay on HepG2 cells with and without quercetin
confirmed the SILAC-MS results. We suspect that the effect of
quercetin on HepG2 cell proliferation was partly due to changes in
the expression of the CCND1 gene and the subsequent block of the G1
phase. In the future, we plan to study the effect of quercetin on
CCND1 and inoculated tumors in an animal model in order to further
test the impact of quercetin treatments on tumor proliferation.
Acknowledgements
This work was financially supported by grants from
Sichuan Province Program (nos. 2010100568 and 2011JYZ034).
Glossary
Abbreviations
Abbreviations:
|
SILAC
|
stable isotope labeling by amino acids
in cell culture
|
|
CCND1
|
cyclin D1
|
|
CCND2
|
cyclin D2
|
|
CCND3
|
cyclin D3
|
|
Leu-d3
|
deuterated leucine
|
|
Leu-d0
|
normal leucine
|
|
SDS-PAGE
|
sodium dodecyl sulfate-polyacrylamide
gel electrophoresis
|
|
ACN
|
acetonitrile
|
|
MS
|
mass spectrometry
|
|
ESI
|
electrospray ionization
|
|
LC
|
liquid chromatography
|
|
MS/MS
|
tandem mass spectrometry
|
|
MALDI-TOF
|
matrix-assisted laser desorption
ion-time of flight
|
References
|
1
|
Linsalata M, Orlando A, Messa C, Refolo MG
and Russo F: Quercetin inhibits human DLD-1 colon cancer cell
growth and polyamine biosynthesis. Anticancer Res. 30:3501–3507.
2010.PubMed/NCBI
|
|
2
|
Guazelli CF, Fattori V, Colombo BB,
Georgetti SR, Vicentini FT, Casagrande R, Baracat MM and Verri WA
Jr: Quercetin-loaded microcapsules ameliorate experimental colitis
in mice by anti-inflammatory and antioxidant mechanisms. J Nat
Prod. 76:200–208. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu H, Xue JX, Li X, Ao R and Lu Y:
Quercetin liposomes protect against radiation-induced pulmonary
injury in a murine model. Oncol Lett. 6:453–459. 2013.PubMed/NCBI
|
|
4
|
Chuang-Xin L, Wen-Yu W, Yao C, Xiao-Yan L
and Yun Z: Quercetin enhances the effects of
5-fluorouracil-mediated growth inhibition and apoptosis of
esophageal cancer cells by inhibiting NF-κB. Oncol Lett. 4:775–778.
2012.PubMed/NCBI
|
|
5
|
Ong SE, Blagoev B, Kratchmarova I,
Kristensen DB, Steen H, Pandey A and Mann M: Stable isotope
labeling by amino acids in cell culture, SILAC, as a simple and
accurate approach to expression proteomics. Mol Cell Proteomics.
1:376–386. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou J, Liang S, Fang L, et al:
Quantitative proteomic analysis of HepG2 cells treated with
quercetin suggests IQGAP1 involved in quercetin-induced regulation
of cell proliferation and migration. OMICS. 13:93–103. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Malumbres M and Barbacid M: Cell cycle,
CDKs and cancer: A changing paradigm. Nat Rev Cancer. 9:153–166.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hengstschläger M, Braun K, Soucek T,
Miloloza A and Hengstschläger-Ottnad E: Cyclin-dependent kinases at
the G1-S transition of the mammalian cell cycle. Mutat Res.
436:1–9. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee MH and Yang HY: Regulators of G1
cyclin-dependent kinases and cancers. Cancer Metastasis Rev.
22:435–449. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Musgrove EA, Caldon CE, Barraclough J,
Stone A and Sutherland RL: Cyclin D as a therapeutic target in
cancer. Nat Rev Cancer. 11:558–572. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
He Y, Franco OE, Jiang M, Williams K, Love
HD, Coleman IM, Nelson PS and Hayward SW: Tissue-specific
consequences of cyclin D1 overexpression in prostate cancer
progression. Cancer Res. 67:8188–8197. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mylona E, Tzelepis K, Theohari I,
Giannopoulou I, Papadimitriou C and Nakopoulou L: Cyclin D1 in
invasive breast carcinoma: Favourable prognostic significance in
unselected patients and within subgroups with an aggressive
phenotype. Histopathology. 62:472–480. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
El-Hafez AA, El Aaty Shawky A and Hasan B:
Cyclin D1 overexpression associates with favourable prognostic
factors in invasive breast carcinoma. Cancer Biomark. 12:149–154.
2012.PubMed/NCBI
|
|
14
|
Weng CJ and Yen GC: Flavonoids, a
ubiquitous dietary phenolic subclass, exert extensive in vitro
anti-invasive and in vivo. Cancer Metastasis Rev. 31:323–351. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yuan J, Wong IL, Jiang T, Wang SW, Liu T,
Wen BJ, Chow LM and Wan Sheng B: Synthesis of methylated quercetin
derivatives and their reversal activities on P-gp- and
BCRP-mediated multidrug resistance tumour cells. Eur J Med Chem.
54:413–422. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hernández-Ortega LD, Alcántar-Díaz BE,
Ruiz-Corro LA, Sandoval-Rodriguez A, Bueno-Topete M,
Armendariz-Borunda J and Salazar-Montes AM: Quercetin improves
hepatic fibrosis reducing hepatic stellate cells and regulating
pro-fibrogenic/anti-fibrogenic molecules balance. J Gastroenterol
Hepatol. 27:1865–1872. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao P, Mao JM, Zhang SY, Zhou ZQ, Tan Y
and Zhang Y: Quercetin induces HepG2 cell apoptosis by inhibiting
fatty acid biosynthesis. Oncol Lett. 8:765–769. 2014.PubMed/NCBI
|
|
18
|
Youn H, Jeong JC, Jeong YS, Kim EJ and Um
SJ: Quercetin potentiates apoptosis by inhibiting nuclear
factor-kappaB signaling in H460 lung cancer cells. Biol Pharm Bull.
36:944–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Psahoulia FH, Moumtzi S, Roberts ML,
Sasazuki T, Shirasawa S and Pintzas A: Quercetin mediates
preferential degradation of oncogenic Ras and causes autophagy in
Ha-RAS-transformed human colon cells. Carcinogenesis. 28:1021–1031.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pratheeshkumar P, Budhraja A, Son YO, Wang
X, Zhang Z, Ding S, Wang L, Hitron A, Lee JC, Xu M, et al:
Quercetin inhibits angiogenesis mediated human prostate tumor
growth by targeting VEGFR-2 regulated AKT/mTOR/P70S6K signaling
pathways. PLoS One. 7:e475162012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen CY, Chi LM, Chi HC, Tsai MM, Tsai CY,
Tseng YH, Lin YH, Chen WJ, Huang YH and Lin KH: Stable isotope
labeling with amino acids in cell culture (SILAC)-based
quantitative proteomics study of a thyroid hormone-regulated
secretome in human hepatoma cells. Mol Cell Proteomics.
11:M1112012. View Article : Google Scholar
|
|
22
|
Zanini C, Giribaldi G, Mandili G, Carta F,
Crescenzio N, Bisaro B, Doria A, Foglia L, di Montezemolo LC,
Timeus F and Turrini F: Inhibition of heat shock proteins (HSP)
expression by quercetin and differential doxorubicin sensitization
in neuroblastoma and Ewing's sarcoma cell lines. J Neurochem.
103:1344–1354. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tashiro E, Tsuchiya A and Imoto M:
Functions of cyclin D1 as an oncogene and regulation of cyclin D1
expression. Cancer Sci. 98:629–635. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Saini SS and Klein MA: Targeting cyclin D1
in non-small cell lung cancer and mesothelioma cells by antisense
oligonucleotides. Anticancer Res. 31:3683–3690. 2011.PubMed/NCBI
|
|
25
|
Shih LC, Tsai CW, Tsai MH, Tsou YA, Chang
WS, Li FJ, Lee MH and Bau DT: Association of cyclin D1 genotypes
with nasopharyngeal carcinoma risk. Anticancer Res. 32:1093–1098.
2012.PubMed/NCBI
|
|
26
|
Jares P, Campo E, Pinyol M, Bosch F,
Miquel R, Fernandez PL, Sanchez-Beato M, Soler F, Perez-Losada A,
Nayach I, et al: Expression of retinoblastoma gene product (pRb) in
mantle cell lymphomas. Correlation with cyclin D1 (PRAD1/CCND1)
mRNA levels and proliferative activity. Am J Pathol. 148:1591–1600.
1996.PubMed/NCBI
|
|
27
|
Coupland SE, Bechrakis N, Schüler A,
Anagnostopoulos I, Hummel M, Bornfeld N and Stein H: Expression
patterns of cyclin D1 and related proteins regulating G1-S phase
transition in uveal melanoma and retinoblastoma. Br J Ophthalmol.
82:961–970. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi EJ, Bae SM and Ahn WS:
Antiproliferative effects of quercetin through cell cycle arrest
and apoptosis in human breast cancer MDA-MB-453 cells. Arch Pharm
Res. 31:1281–1285. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Priyadarsini Vidya R, Senthil Murugan R,
Maitreyi S, Ramalingam K, Karunagaran D and Nagini S: The flavonoid
quercetin induces cell cycle arrest and mitochondria-mediated
apoptosis in human cervical cancer (HeLa) cells through p53
induction and NF-κB inhibition. Eur J Pharmacol. 649:84–91. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jeong JH, An JY, Kwon YT, Rhee JG and Lee
YJ: Effects of low dose quercetin: Cancer cell-specific inhibition
of cell cycle progression. J Cell Biochem. 106:73–82. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yamak A, Temsah R, Maharsy W, Caron S,
Paradis P, Aries A and Nemer M: Cyclin D2 rescues size and function
of GATA4 haplo-insufficient hearts. Am J Physiol Heart Circ
Physiol. 303:H1057–H1066. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jena N, Deng M, Sicinska E, Sicinski P and
Daley GQ: Critical role for cyclin D2 in BCR/ABL-induced
proliferation of hematopoietic cells. Cancer Res. 62:535–541.
2002.PubMed/NCBI
|
|
33
|
Takano Y, Kato Y, Masuda M, Ohshima Y and
Okayasu I: Cyclin D2, but not cyclin D1, overexpression closely
correlates with gastric cancer progression and prognosis. J Pathol.
189:194–200. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Filipits M, Jaeger U, Pohl G, Stranzl T,
Simonitsch I, Kaider A, Skrabs C and Pirker R: Cyclin D3 is a
predictive and prognostic factor in diffuse large B-cell lymphoma.
Clin Cancer Res. 8:729–733. 2002.PubMed/NCBI
|
|
35
|
García-Morales P, Hernando E,
Carrasco-García E, Menéndez-Gutierrez MP, Saceda M and
Martínez-Lacaci I: Cyclin D3 is down-regulated by rapamycin in
HER-2-overexpressing breast cancer cells. Mol Cancer Ther.
5:2172–2181. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang Q, Sakamoto K, Liu C, Triplett AA,
Lin WC, Rui H and Wagner KU: Cyclin D3 compensates for the loss of
cyclin D1 during ErbB2-induced mammary tumor initiation and
progression. Cancer Res. 71:7513–7524. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Akli S, Van Pelt CS, Bui T, Multani AS,
Chang S, Johnson D, Tucker S and Keyomarsi K: Overexpression of the
low molecular weight cyclin E in transgenic mice induces metastatic
mammary carcinomas through the disruption of the ARF-p53 pathway.
Cancer Res. 67:7212–7222. 2007. View Article : Google Scholar : PubMed/NCBI
|