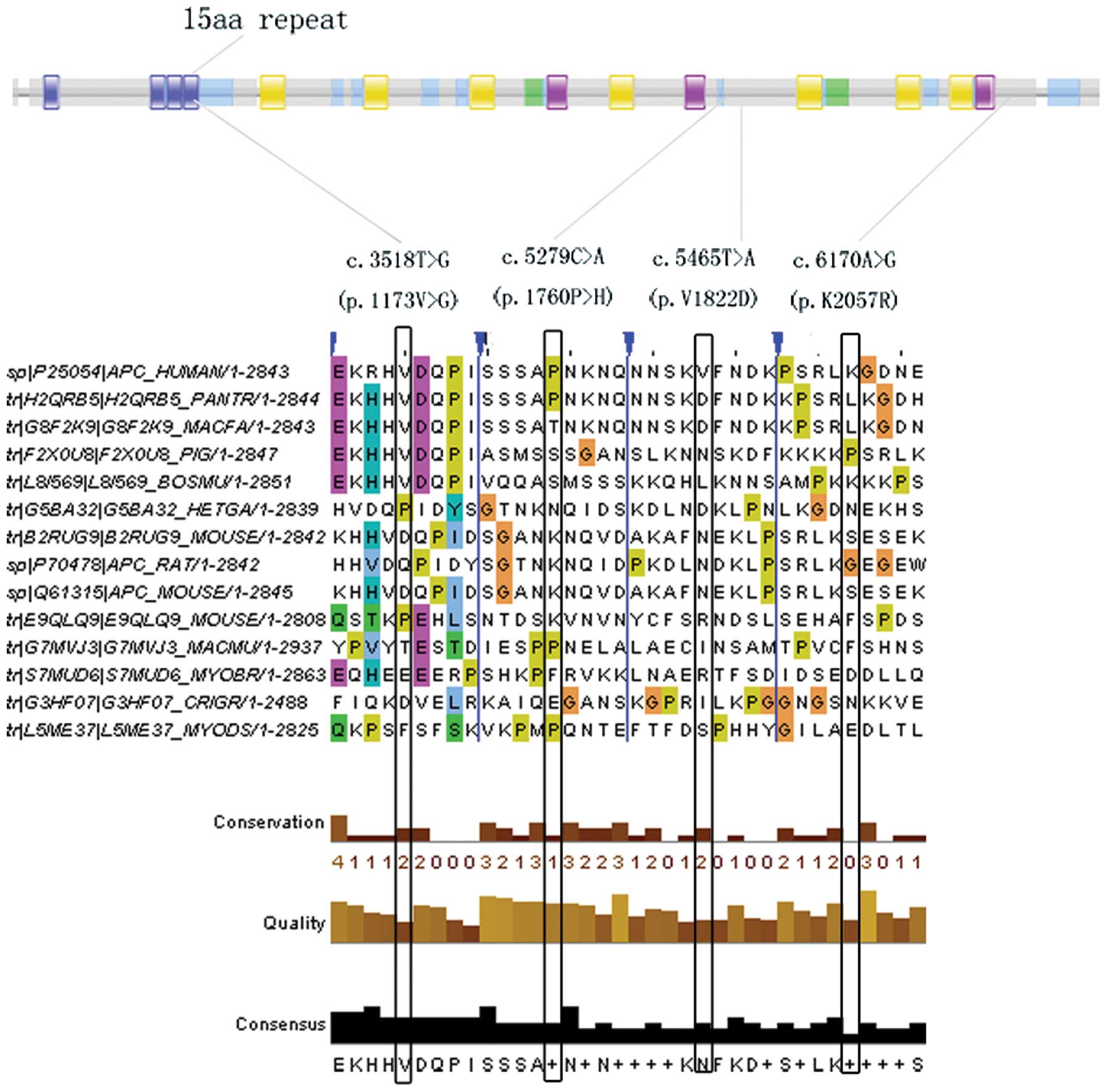
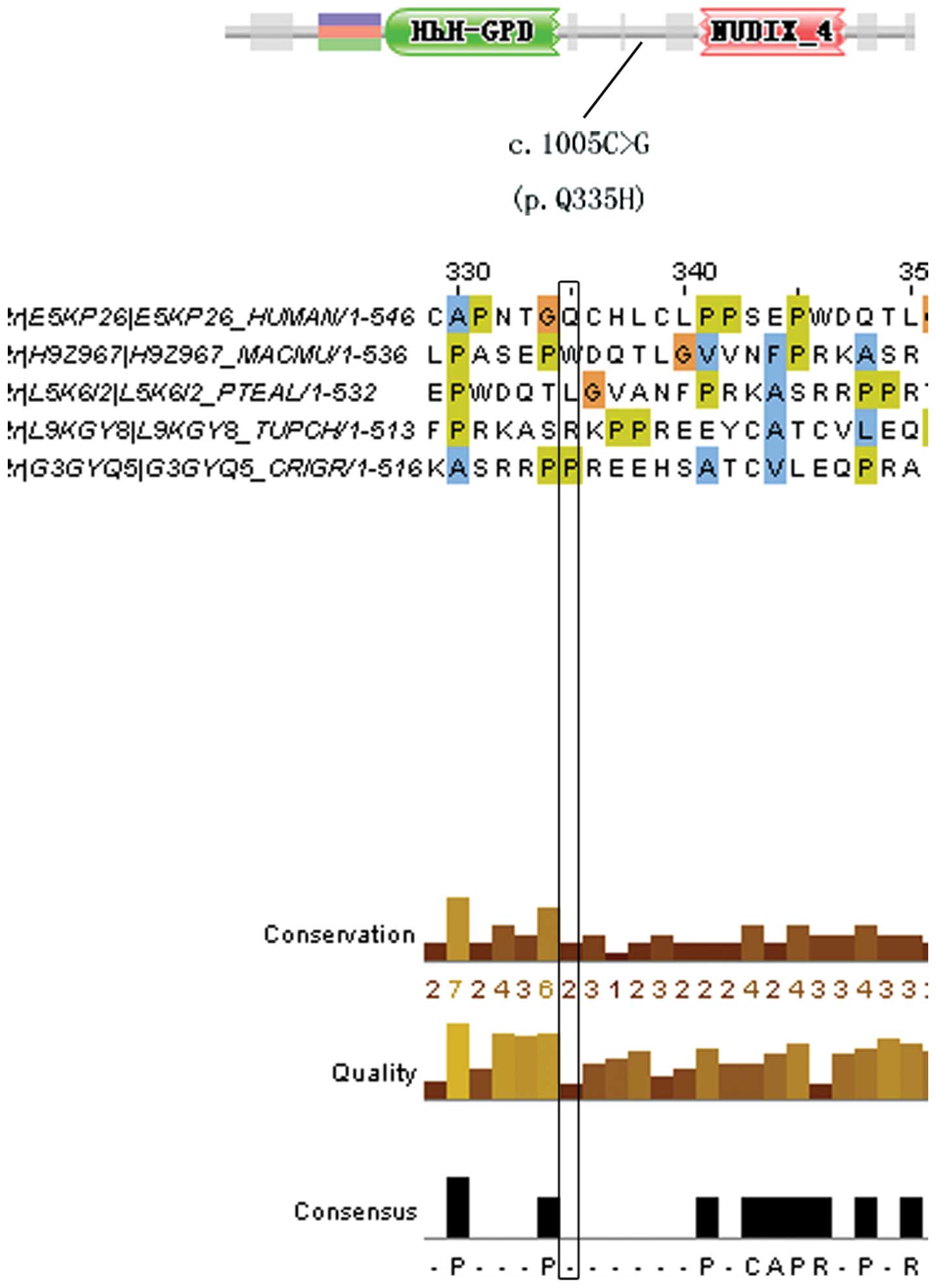
|
1
|
Gardner EJ, Burt RW and Freston JW:
Gastrointestinal polyposis: syndromes and genetic mechanisms. West
J Med. 132:488–499. 1980.PubMed/NCBI
|
|
2
|
Bianchi LK, Buerke CA, Bennett AE, Lopez
R, Hasson H and Church JM: Fundic gland polyp dysplasia is common
in familial adenomatous polyposis. Clin Gastroenterol Heptatol.
6:180–185. 2008. View Article : Google Scholar
|
|
3
|
Nugent KP and Philips RK: Rectal cancer
risk in older patients with adenomatous polyposis and ileorectal
anastomosis: a cause for concern. Br J Surg. 79:1204–1206. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kinzler KW, Nilbert MC, Vogelstein B,
Bryan TM, Levy DB, Smith KJ, Preisinger AC, Hamilton SR, Hedge P
and Markham A: Identification of a gene located at chromosome 5q21
that is mutated in colorectal cancers. Science. 251:1366–1370.
1991. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Groden J, Thliveris A, Samowitz W, Carlson
M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L and
Robertson M: Identification and characterization of the familial
adenomatous polyposis coli gene. Cell. 66:589–600. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu XR, Shan XN, Friedl W, Uhlhaas S, Li
JT, Propping P and Wang YP: Detection of germline mutations in the
APC gene with the protein truncation test. Yi Chuan Xue Bao.
32:903–908. 2005.(In Chinese). PubMed/NCBI
|
|
7
|
Gan Y, Zheng S and Cai X: Detection of a
gene mutation in familial adenomatous polyposis families by
PCR-RFLP method. Zhonghua Yi Xue Za Zhi. 74:352–354. 1994.(In
Chinese). PubMed/NCBI
|
|
8
|
Chen S, Zhou J, Zhang X, Zhou X, Zhu M,
Zhang Y, Ma G and Li J: Mutation analysis of the APC gene in a
Chinese FAP pedigree with unusual phenotype. ISRN Gastroenterol.
2011:9091212011.PubMed/NCBI
|
|
9
|
Sheng JQ, Cui WJ, Fu L, Jin P, Han Y, Li
SJ, Fan RY, Li AQ, Zhang MZ and Li SR: APC gene mutations in
Chinese familial adenomatous polyposis patients. World J
Gastroenterol. 16:1522–1526. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vandrovcová J, Štekrová J, Kebrdlová V and
Kohoutová M: Molecular analysis of the APC and MYH genes in Czech
families affected by FAP or multiple adenomas: 13 novel mutations.
Hum Mutat. 23:3972004. View Article : Google Scholar
|
|
11
|
Gómez-Fernández N, Castellví-Bel S,
Fernández-Rozadilla C, Balaguer F, Muñoz J, Madrigal I, Milà M,
Graña B, Vega A, Castells A, et al: Molecular analysis of the APC
and MUTYH genes in Galician and Catalonian FAP families: a
different spectrum of mutations? BMC Med Genet. 10:572009.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rivera B, González S, Sánchez-Tomé E,
Blanco I, Mercadillo F, Letón R, Benítez J, Robledo M, Capellá G
and Urioste M: Clinical and genetic characterization of classical
forms of familial adenomatous polyposis: a Spanish population
study. Ann Oncol. 22:903–909. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fostira F, Thodi G, Sandaltzopoulos R,
Fountzilas G and Yannoukakos D: Mutational spectrum of APC and
genotype-phenotype correlations in Greek FAP patients. BMC Cancer.
10:3892010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Torrezan GT, da Silva FC, Santos EM,
Krepischi AC, Achatz MI, Aguiar S Jr, Rossi BM and Carraro DM:
Mutational spectrum of the APC and MUTYH genes and
genotype-phenotype correlations in Brazilian FAP, AFAP and MAP
patients. Orphanet J Rare Dis. 8:542013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kanter-Smoler G, Fritzell K, Rohlin A,
Engwall Y, Hallberg B, Bergman A, Meuller J, Grönberg H, Karlsson
P, Björk J and Nordling M: Clinical characterization and the
mutation spectrum in Swedish adenomatous polyposis families. BMC
Med. 6:102008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Al-Tassan N, Chmiel NH, Maynard J, Fleming
N, Livingston AL, Williams GT, Hodges AK, Davies DR, David SS,
Sampson JR and Cheadle JP: Inherited variants of MYH associated
with somatic G:C->T:A mutations in colorectal tumors. Nat Genet.
30:227–232. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sieber OM, Lipton L, Crabtree M, Heinimann
K, Fidalgo P, Phillips RK, Bisgaard ML, Orntoft TF, Aaltonen LA,
Hodgson SV, et al: Multiple colorectal adenomas, classic
adenomatous polyposis and germ-line mutations in MYH. N Engl J Med.
348:791–799. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Renkonen ET, Nieminen P, Abdel-Rahman WM,
Moisio AL, Järvelä I, Arte S, Järvinen HJ and Peltomäki P:
Adenomatous polyposis families that screen APC mutation-negative by
conventional methods are genetically heterogeneous. J Clin Oncol.
23:5651–5659. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lammi L, Arte S, Somer M, Jarvinen H,
Lahermo P, Thesleff I, Pirinen S and Nieminen P: Mutations in AXIN2
cause familial tooth agenesis and predispose to colorectal cancer.
Am J Hum Genet. 74:1043–1050. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wei SC, Su YN, Tsai-Wu JJ, Wu CH, Huang
YL, Sheu JC, Wang CY and Wong JM: Genetic analysis of the APC gene
in Taiwanese familial adenomatous polyposis. J Biomed Sci.
11:260–265. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Prior TE and Bridgeman SJ: Identifying
mutations for MYH-associated polyposis. Current Protocols in Human
Genetics 64: 10.13.1–10.13.14. January 1–2010.DOI:
10.1002/0471142905.hg1013s64. View Article : Google Scholar
|
|
22
|
Pan M, Cong P, Wang Y, Lin C, Yuan Y, Dong
J, Banerjee S, Zhang T, Chen Y, Zhang T, et al: Novel LOVD
databases for hereditary breast cancer and colorectal cancer genes
in the Chinese population. Hum Mutat. 32:1335–1340. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang T, Moss A, Cong P, Pan M, Chang B,
Zheng L, Fang Q, Zareba W, Robinson J, Lin C, et al: Long QT
International Registry Investigators; HVP-China Investigators: LQTS
gene LOVD database. Hum Mutat. 31:E1801–E1810. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sherry ST, Ward MH, Kholodov M, Baker J,
Phan L, Smigielski EM and Sirotkin K: DbSNP: The NCBI database of
genetic variation. Nucleic Acids Res. 29:308–311. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Hubbard TJ, Aken BL, Beal K, Ballester B,
Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et
al: Ensembl 2007. Nucleic Acids Res. 35(Database issue): D610–D617.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ng PC and Henikoff S: Predicting
deleterious amino acid substitutions. Genome Res. 11:863–874. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Calabrese R, Capriotti E, Fariselli P,
Martelli PL and Casadio R: Functional annotations improve the
predictive score of human disease-related mutations in proteins.
Hum Mutat. 30:1237–1244. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Capriotti E and Altman RB: Improving the
prediction of disease-related variants using protein
three-dimensional structure. BMC Bioinformatics. 12(Suppl 4):
S32011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Capriotti E, Calabrese R, Fariselli P,
Martelli PL, Altman RB and Casadio R: WS-SNPs&GO: a web server
for predicting the deleterious effect of human protein variants
using functional annotation. BMC Genomics. 14(Suppl 3): S62013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cartegni L, Wang J, Zhu Z, Zhang MQ and
Krainer AR: ESEfinder: a web resource to identify exonic splicing
enhancers. Nucleic Acids Res. 31:3568–3571. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Punta M, Coggill PC, Eberhardt RY, Mistry
J, Tate J, Boursnell C, Pang N, Forslund K, Ceric G, Clements J, et
al: The Pfam protein families database. Nucleic Acids Res.
40(Database issue): D290–D301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Larkin MA, Blackshields G, Brown NP,
Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm
A, Lopez R, et al: Clustal W and Clustal X version 2.0.
Bioinformatics. 23:2947–2948. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Waterhouse AM, Procter JB, Martin DM,
Clamp M and Barton GJ: Jalview Version 2 - a multiple sequence
alignment editor and analysis workbench. Bioinformatics.
25:1189–1191. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yeo G, Burge CB, Hoon S and Venkatesh B:
Variation in sequence and organization of splicing regulatory
elements in vertebrate genes. Proc Natl Acad Sci USA.
101:15700–15705. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fairbrother WG, Yeh RF, Sharp PA and Burge
CB: Predictive identification of exonic splicing enhancers in human
genes. Science. 297:1007–1013. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang XH, Kangsamaksin T, Chao MS,
Banerjee JK and Chasin LA: Exon inclusion is dependent on
predictable exonic splicing enhancers. Mol Cell Biol. 25:7323–7332.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Adzhubei IA, Schmidt S, Peshkin L,
Ramensky VE, Gerasimova A, Bork P, Kondrashov AS and Sunyaev SR: A
method and server for predicting damaging missense mutations. Nat
Methods. 7:248–249. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Friedl W and Aretz S: Familial adenomatous
polyposis: experience from a study of 1164 unrelated german
polyposis patients. Hered Cancer Clin Pract. 3:95–114. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liang J, Lin C, Hu F, Wang F, Zhu L, Yao
X, Wang Y and Zhao Y: APC polymorphisms and the risk of colorectal
neoplasia: a HuGE review and meta-analysis. Am J Epidemiol.
177:1169–1179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Guerreiro CS, Cravo ML, Brito M, Vidal PM,
Fidalgo PO and Leitão CN: The V1822D APC polymorphism interacts
with fat, calcium and fiber intakes in modulating the risk of
colorectal cancer in Portuguese persons. Am J Clin Nutr.
85:1592–1597. 2007.PubMed/NCBI
|
|
41
|
Menéndez M, González S, Blanco I, Guinó E,
Peris M, Peinado MA, Capellá G and Moreno V: Colorectal cancer risk
and the APC V1822D variant. Int J Cancer. 112:161–163. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nakagawa H, Murata Y, Koyama K, Fujiyama
A, Miyoshi Y, Monden M, Akiyama T and Nakamura Y: Identification of
a brain-specific APC homologue, APCL and its interaction with
beta-catenin. Cancer Res. 58:5176–5181. 1998.PubMed/NCBI
|
|
43
|
Wachsmannova-Matelova L, Stevurkova V,
Adamcikova Z, Holec V and Zajac V: Polymorphisms in the adenomatous
polyposis coli gene in Slovak families suspected of FAP. Neuro
Endocrinol Lett. 30(Suppl 1): S25–S28. 2009.
|
|
44
|
Herrmann SM, Adler YD, Schmidt-Petersen K,
Nicaud V, Morrison C, Paul M and Zouboulis ChC: The concomitant
occurrence of multiple epidermal cysts, osteomas and thyroid gland
nodules is not diagnostic for Gardner syndrome in the absence of
intestinal polyposis: a clinical and genetic report. Br J Dermatol.
149:877–883. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shu Z, Yanqin H and Ying Y: Hereditary
colorectal cancer in china. Hered Cancer Clin Pract. 3:155–164.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Cartegni L, Chew SL and Krainer AR:
Listening to silence and understanding nonsense: exonic mutations
that affect splicing. Nat Rev Genet. 3:285–298. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Pagani F and Baralle FE: Genomic variants
in exons and introns: Identifying the splicing spoilers. Nat Rev
Genet. 5:389–396. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Grosso AR, Martins S and Carmo-Fonseca M:
The emerging role of splicing factors in cancer. EMBO Rep.
9:1087–1093. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bechtel JM, Rajesh P, Ilikchyan I, Deng Y,
Mishra PK, Wang Q, Wu X, Afonin KA, Grose WE, Wang Y, et al: The
Alternative Splicing Mutation Database: a hub for investigations of
alternative splicing using mutational evidence. BMC Res Notes.
1:32008. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Covaciu C, Grosso F, Pisaneschi E,
Zambruno G, Gregersen PA, Sommerlund M, Hertz JM and Castiglia D: A
founder synonymous COL7A1 mutation in three Danish families with
dominant dystrophic epidermolysis bullosa pruriginosa identifies
exonic regulatory sequences required for exon 87 splicing. Br J
Dermatol. 165:678–682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Tournier I, Vezain M, Martins A,
Charbonnier F, Baert-Desurmont S, Olschwang S, Wang Q, Buisine MP,
Soret J, Tazi J, et al: A large fraction of unclassified variants
of the mismatch repair genes MLH1 and MSH2 is associated with
splicing defects. Hum Mutat. 29:1412–1424. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Pagani F, Raponi M and Baralle FE:
Synonymous mutations in CFTR exon 12 affect splicing and are not
neutral in evolution. Proc Natl Acad Sci USA. 102:6368–6372. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Krawczak M, Reiss J and Cooper DN: The
mutational spectrum of single base-pair substitutions in mRNA
splice junctions of human genes: causes and consequences. Hum
Genet. 90:41–54. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Nouri N, Fazel-Najafabadi E, Behnam M,
Nouri N, Aryani O, Ghasemi M, Nasiri J and Sedghi M: Use of in
silico tools for classification of novel missense mutations
identified in dystrophin gene in developing countries. Gene.
535:250–254. 2014. View Article : Google Scholar : PubMed/NCBI
|