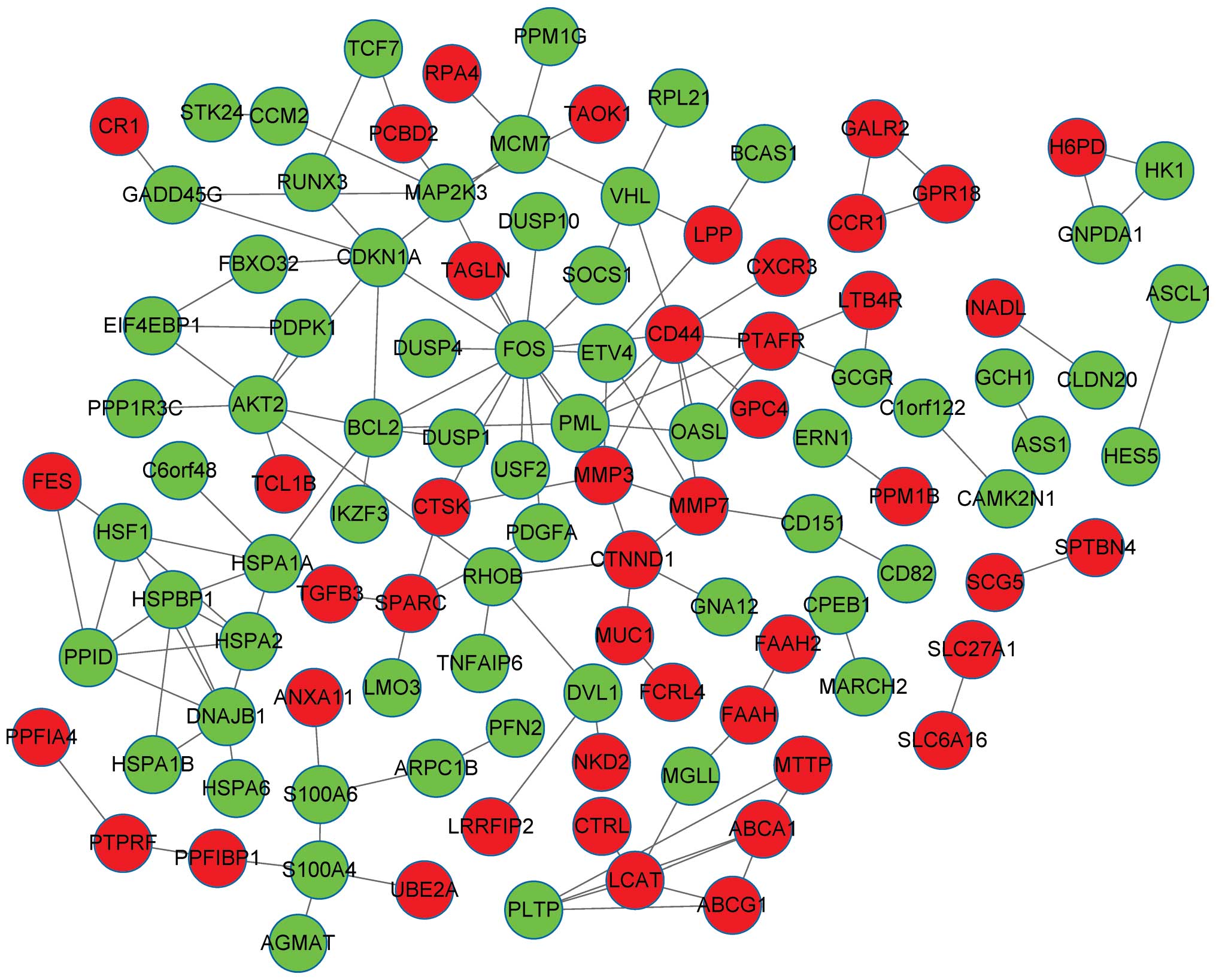
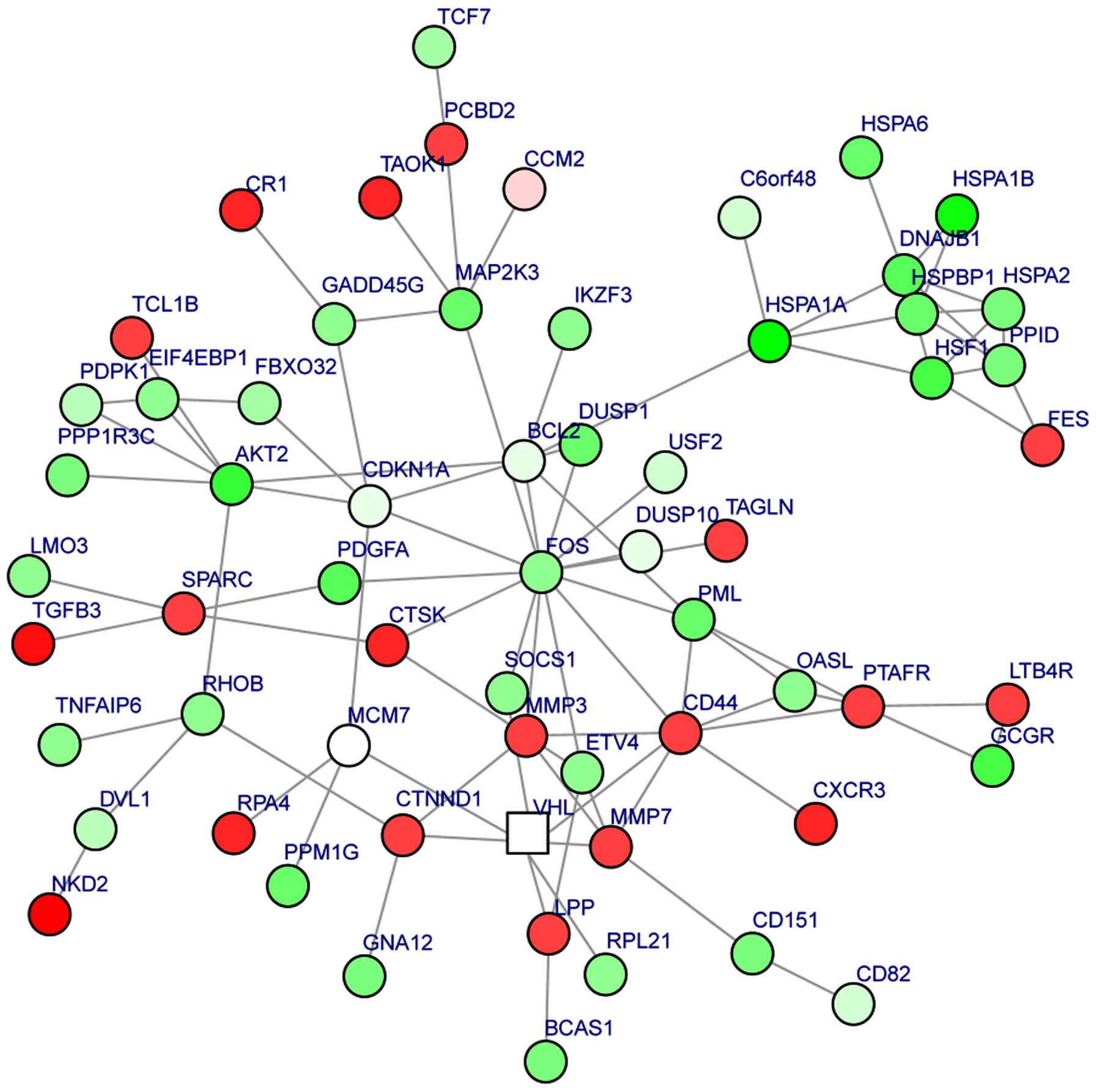
|
1
|
Scully RE: Pathology of ovarian cancer
precursors. J Cell Biochem Suppl. 23:208–218. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iorio MV and Croce CM: MicroRNA profiling
in ovarian cancer. Methods Mol Biol. 1049:187–197. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yuan ZQ, Sun M, Feldman RI, Wang G, Ma X,
Jiang C, Coppola D, Nicosia SV and Cheng JQ: Frequent activation of
AKT2 and induction of apoptosis by inhibition of
phosphoinositide-3-OH kinase/Akt pathway in human ovarian cancer.
Oncogene. 19:2324–2330. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yin BW and Lloyd KO: Molecular cloning of
the CA125 ovarian cancer antigen: Identification as a new mucin,
Muc16. J Biol Chem. 276:27371–27375. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Scotton CJ, Wilson JL, Milliken D, Stamp G
and Balkwill FR: Epithelial cancer cell migration: A role for
chemokine receptors? Cancer Res. 61:4961–4965. 2001.PubMed/NCBI
|
|
6
|
Strieter RM: Chemokines: Not just
leukocyte chemoattractants in the promotion of cancer. Nat Immunol.
2:285–286. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Scotton CJ, Wilson JL, Scott K, Stamp G,
Wilbanks GD, Fricker S, Bridger G and Balkwill FR: Multiple actions
of the chemokine CXCL12 on epithelial tumor cells in human ovarian
cancer. Cancer Res. 62:5930–5938. 2002.PubMed/NCBI
|
|
8
|
Campbell IG, Russell SE, Choong DY,
Montgomery KG, Ciavarella ML, Hooi CS, Cristiano BE, Pearson RB and
Phillips WA: Mutation of the PIK3CA gene in ovarian and breast
cancer. Cancer Res. 64:7678–7681. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Karakas B, Bachman K and Park B: Mutation
of the PIK3CA oncogene in human cancers. Br J Cancer. 94:455–459.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao H, Shen J, Wang D, Guo Y, Gregory S,
Medico L, Hu Q, Yan L, Odunsi K, Lele S and Liu S: Associations
between gene expression variations and ovarian cancer risk alleles
identified from genome wide association studies. PloS One.
7:e479622012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Smyth GK: Limma: Linear models for
microarray data. Bioinformatics and Computational Biology Solutions
using R and Bioconductor. Springer. (New York). 397–420. 2005.
View Article : Google Scholar
|
|
13
|
Zhao M, Sun J and Zhao Z: TSGene: A web
resource for tumor suppressor genes. Nucleic Acids Res.
41:D970–D976. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen JS, Hung WS, Chan HH, Tsai SJ and Sun
HS: In silico identification of oncogenic potential of fyn-related
kinase in hepatocellular carcinoma. Bioinformatics. 29:420–427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Palla G, Derényi I, Farkas I and Vicsek T:
Uncovering the overlapping community structure of complex networks
in nature and society. Nature. 435:814–818. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu M, Zhang Q, Deng M, Miao J, Guo Y, Gao
W and Cui Q: An analysis of human microRNA and disease
associations. PloS One. 3:e34202008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Smoot ME, Ono K, Ruscheinski J, Wang PL
and Ideker T: Cytoscape 2.8: New features for data integration and
network visualization. Bioinformatics. 27:431–432. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
He X and Zhang J: Why do hubs tend to be
essential in protein networks? PLoS Genet. 2:e882006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Beisser D, Klau GW, Dandekar T, Müller T
and Dittrich MT: BioNet: An R-Package for the functional analysis
of biological networks. Bioinformatics. 26:1129–1130. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Baekelandt M, Kristensen GB, Nesland JM,
Tropé CG and Holm R: Clinical significance of apoptosis-related
factors p53, Mdm2 and Bcl-2 in advanced ovarian cancer. J Clin
Oncol. 17:20611999.PubMed/NCBI
|
|
22
|
White E: Life, death and the pursuit of
apoptosis. Genes Dev. 10:1–15. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Eliopoulos AG, Kerr DJ, Herod J, Hodgkins
L, Krajewski S, Reed JC and Young LS: The control of apoptosis and
drug resistance in ovarian cancer: Influence of p53 and Bcl-2.
Oncogene. 11:1217–1228. 1995.PubMed/NCBI
|
|
24
|
Miyashita T and Reed JC: Bcl-2 gene
transfer increases relative resistance of S49. 1 and WEHI7. 2
lymphoid cells to cell death and DNA fragmentation induced by
glucocorticoids and multiple chemotherapeutic drugs. Cancer Res.
52:5407–5411. 1992.PubMed/NCBI
|
|
25
|
Marx D, Binder C, Meden H, Lenthe T,
Ziemek T, Hiddemann T, Kuhn W and Schauer A: Differential
expression of apoptosis associated genes bax and bcl-2 in ovarian
cancer. Anti Cancer Res. 17:2233–2240. 1997.
|
|
26
|
Witham J, Valenti MR, De-Haven-Brandon AK,
Vidot S, Eccles SA, Kaye SB and Richardson A: The Bcl-2/Bcl-XL
family inhibitor ABT-737 sensitizes ovarian cancer cells to
carboplatin. Clin Cancer Res. 13:7191–7198. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Teng CS: Protooncogenes as mediators of
apoptosis. Int Rev Cytol. 197:137–202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mahner S, Baasch C, Schwarz J, Hein S,
Wölber L, Jänicke F and Milde-Langosch K: C-Fos expression is a
molecular predictor of progression and survival in epithelial
ovarian carcinoma. Br J Cancer. 99:1269–1275. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Le XF, Claret FX, Lammayot A, Tian L,
Deshpande D, LaPushin R, Tari AM and Bast RC Jr: The role of
cyclin-dependent kinase inhibitor p27Kip1 in anti-HER2
antibody-induced G1 cell cycle arrest and tumor growth inhibition.
J Biol Chem. 278:23441–23450. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kamb A, Gruis NA, Weaver-Feldhaus J, Liu
Q, Harshman K, Tavtigian SV, Stockert E, Day RS III, Johnson BE and
Skolnick MH: A cell cycle regulator potentially involved in genesis
of many tumor types. Science. 264:436–440. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dong Y, Walsh MD, McGuckin MA, Gabrielli
BG, Cummings MC, Wright RG, Hurst T, Khoo SK and Parsons PG:
Increased expression of cyclin-dependent kinase inhibitor 2
(CDKN2A) gene product P16INK4A in ovarian cancer is associated with
progression and unfavourable prognosis. Int J Cancer. 74:57–63.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gartel AL, Serfas MS and Tyner AL:
p21–negative regulator of the cell cycle. Proc Soc Exp Biol Med.
213:138–149. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Wu H, Wade M, Krall L, Grisham J, Xiong Y
and Van Dyke T: Targeted in vivo expression of the cyclin-dependent
kinase inhibitor p21 halts hepatocyte cell-cycle progression,
postnatal liver development and regeneration. Genes Dev.
10:245–260. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ostrovsky O and Bengal E: The
mitogen-activated protein kinase cascade promotes myoblast cell
survival by stabilizing the cyclin-dependent kinase inhibitor,
p21WAF1 protein. J Biol Chem. 278:21221–21231. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Strait KA, Dabbas B, Hammond EH, Warnick
CT, Ilstrup SJ and Ford CD: Cell cycle blockade and differentiation
of ovarian cancer cells by the histone deacetylase inhibitor
trichostatin A are associated with changes in p21, Rb and Id
proteins. Mol Cancer Ther. 1:1181–1190. 2002.PubMed/NCBI
|
|
36
|
Aruffo A, Stamenkovic I, Melnick M,
Underhill CB and Seed B: CD44 is the principal cell surface
receptor for hyaluronate. Cell. 61:1303–1313. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fraser JR, Laurent TC and Laurent UB:
Hyaluronan: Its nature, distribution, functions and turnover. J
Intern Med. 242:27–33. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cannistra SA, Kansas GS, Niloff J,
DeFranzo B, Kim Y and Ottensmeier C: Binding of ovarian cancer
cells to peritoneal mesothelium in vitro is partly mediated by
CD44H. Cancer Res. 53:3830–3838. 1993.PubMed/NCBI
|
|
39
|
Cannistra SA, Abu-Jawdeh G, Niloff J,
Strobel T, Swanson L, Andersen J and Ottensmeier C: CD44 variant
expression is a common feature of epithelial ovarian cancer: Lack
of association with standard prognostic factors. J Clin Oncol.
13:1912–1921. 1995.PubMed/NCBI
|
|
40
|
Bourguignon LY, Zhu H, Chu A, Iida N,
Zhang L and Hung MC: Interaction between the adhesion receptor,
CD44 and the oncogene product, p185 HER2, promotes human ovarian
tumor cell activation. J Biol Chem. 272:27913–27918. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang S, Balch C, Chan MW, Lai HC, Matei
D, Schilder JM, Yan PS, Huang TH and Nephew KP: Identification and
characterization of ovarian cancer-initiating cells from primary
human tumors. Cancer Res. 68:4311–4320. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang YC, Yo YT, Lee HY, Liao YP, Chao TK,
Su PH and Lai HC: ALDH1-bright epithelial ovarian cancer cells are
associated with CD44 expression, drug resistance and poor clinical
outcome. Am J Pathol. 180:1159–1169. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Overall CM and López-Otín C: Strategies
for MMP inhibition in cancer: Innovations for the post-trial era.
Nat Rev Cancer. 2:657–672. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
44
|
Vihinen P and Kähäri VM: Matrix
metalloproteinases in cancer: Prognostic markers and therapeutic
targets. Int J Cancer. 99:157–166. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Schmalfeldt B, Prechtel D, Härting K,
Späthe K, Rutke S, Konik E, Fridman R, Berger U, Schmitt M, Kuhn W
and Lengyel E: Increased expression of matrix metalloproteinases
(MMP)-2, MMP-9 and the urokinase-type plasminogen activator is
associated with progression from benign to advanced ovarian cancer.
Clin Cancer Res. 7:2396–2404. 2001.PubMed/NCBI
|
|
46
|
Wang FQ, So J, Reierstad S and Fishman DA:
Matrilysin (MMP-7) promotes invasion of ovarian cancer cells by
activation of progelatinase. Int J Cancer. 114:19–31. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tanimoto H, Underwood LJ, Shigemasa K,
Parmley TH, Wang Y, Yan Y, Clarke J and O'Brien TJ: The matrix
metalloprotease pump-1 (MMP-7, Matrilysin): A candidate
marker/target for ovarian cancer detection and treatment. Tumor
Biol. 20:88–98. 1999. View Article : Google Scholar
|