Introduction
Head and neck squamous cell carcinoma is the sixth
most commonly observed cancer worldwide, and its mortality rate is
~50% (1). Approximately 600,000 new
cases are reported worldwide each year (1). Laryngeal squamous cell carcinoma (LSCC)
accounts for ~25% of all head and neck squamous cell carcinomas
(2). Although the 5-year survival
rate of LSCC is ~60% (3), outcomes
have not improved over the previous three decades for the majority
of patients (1). One of the major
reasons for the poor outcome is the low rate of diagnosis (4). The progression of LSCC may undergo
several different phases: Dysplasia (including mild dysplasia,
moderate dysplasia and severe dysplasia), cancer in situ
(CIS) and LSCC (5). To identify novel
biomarkers that indicate the early stages of LSCC, or specific
biomarkers for various individuals, is urgently required for the
early detection of LSCC and the development of individualized
therapies. Therefore, the role of microRNAs (miRs) as possible
biomarkers and targets for therapy has been extensively
investigated in a number of types of cancer (6–8).
miR is a class of gene regulator that is able to
suppress the expression of proteins via base pairing with the
3′-untranslated region of target messenger RNA (9–11).
Accumulating evidence has indicated that miRs have significant
roles in diverse biological processes, and the dysfunction of miRs
may be implicated in a number of diseases, including cancer
(12–17). Altered miR expression patterns have
been reported in LSCC. For example, miR-21 and miR-106b are
upregulated in LSCC cancerous tissues compared with adjacent
non-tumor tissues (18). miR-34a/c,
miR-370 and miR-206 have been reported to be downregulated in human
LSCC tissues (19–21). miR-203 has additionally been reported
to be downregulated in laryngeal squamous cell carcinoma and is
able to suppress proliferation and induce apoptosis of tumors
(22). However, the aberrant
expression of miRs in LSCC patients and their expression during the
earlier stages of the disease are poorly understood.
According to microarray data of miR expression in
tumor and dysplasia tissues of 10 LSCC patients with dysplasia
(23), miR-148a and miR-375 were
differentially expressed in the dysplasia and tumor tissues of 1
patient. In the present study, TaqMan probe stem-loop quantitative
polymerase chain reaction (qPCR) was utilized to accurately measure
the amount of miR-148a and miR-375 in LSCC cancer tissues, CIS
tissues, mild dysplasia, moderate dysplasia and severe dysplasia
tissues, as well as normal controls. It was observed that miR-148a
and miR-375 were significantly upregulated in LSCC tissues, and the
increased level of miR-375 in LSCC was significantly associated
with a more aggressive tumor phenotype. Receiver-operating
characteristic (ROC) curve analysis suggested that expression
levels of miR-375 may be used as markers, with high sensitivity and
specificity for LSCC diagnosis. Furthermore, miR-148a and miR-375
levels increased gradually during laryngeal carcinogenesis, and an
increased expression level of miR-148a or miR-375 may predict LSCC
progression and could serve as an early biomarker of LSCC.
Materials and methods
Specimens
A total of 179 formalin-fixed, paraffin-embedded
tissue sections were prepared from resected tissues obtained from
Beijing Tongren Hospital (Beijing, China) between April 2011 and
August 2014. The specimens included 29 laryngeal squamous cell
carcinoma, 19 mild dysplasia, 29 moderate dysplasia, 34 severe
dysplasia, 36 CIS and 32 normal controls from vocal cord polyps.
The samples were obtained from 164 males and 15 females, with an
average age of 57.1±0.90 years (range, 27–86 years). No patients
had received chemoradiotherapy prior to surgery. The tumors were
staged according to the revised International Union for
International Cancer Control/Tumor-Node-Metastasis staging system
(24), with 20 patients classified as
I–II and 9 patients classified as III–IV. A total of 169 patients
had no previous medical history, whilst 4 patients had a history of
high blood pressure and 6 patients had a history of dyslipidemia.
The present study was approved by the ethical board of Beijing
Tongren Hospital.
RNA extraction
Total RNA was extracted from formalin-fixed,
paraffin-embedded tissue sections with the miRNeasy FFPE kit
(Qiagen China Co., Ltd., Shanghai, China) according to the
manufacturer's protocol. Tissue sections were sliced (5–20 µm
thick) and the first 2–3 sections were discarded. Deparaffinization
solution was added to deparaffinize the paraffin-embedded tissue at
56°C for 3 min, and subsequently buffer PKD was added and mixed by
vortexing. Following centrifugation for 1 min at 11,000 × g, 10 µl
proteinase K was added to the lower, clear phase and incubated at
56°C for 15 min, followed by incubation at 80°C for 15 min with
buffer PKD. The incubation at 80°C in buffer PKD partially reversed
formaldehyde modification of nucleic acids. The lower, clear phase
was transferred into a fresh microcentrifuge tube and incubated on
ice for 3 min. The supernatant was subsequently transferred to a
fresh microcentrifuge tube following centrifugation for 15 min at
20,000 × g, taking care not to disturb the pellet. DNase booster
buffer was added equivalent to a tenth of the total sample volume
and 10 µl DNase I stock solution was additionally added and
incubated at room temperature for 15 min. Buffer RBC was added to
adjust the binding conditions, and ethanol (100%) was added to the
sample. Precipitates were visible following the addition of
ethanol. The sample, including any precipitate, was transferred to
an RNeasy MinElute spin column and centrifuged for 15 sec at ~8000
× g, following by washing with buffer RPE. Finally, RNase-free
water was directly added to the spin column membrane to elute the
RNA.
Complementary DNA (cDNA)
synthesis
cDNA was synthesized using Moloney Murine Leukemia
Virus (M-MLV) reverse transcriptase (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). A stem-loop reverse
transcription primer was utilized for the reverse transcription of
miR. Primers were designed using DNAMAN software (Lynnon Biosoft,
San Ramon, CA, USA) and were synthesized by Life Technologies
(Thermo Fisher Scientific, Inc.). Primer sequences are presented in
Table I. Following RNase-free DNase
treatment (5 U/1µg RNA), 500 ng total RNA was mixed with reverse
transcription primer and dNTPs, incubated at 65°C for 5 min and
then placed immediately on ice. Subsequently, the mixture
containing reverse transcription buffer, DL-Dithiothreitol, M-MLV
reverse transcriptase and RNase inhibitor was added and incubated
at 37°C for 50 min, followed by a final reverse transcriptase
inactivation step at 75°C for 5 min. cDNA samples were stored at
−80°C until required for PCR analysis.
 | Table I.Sequence of primers used in the
reverse transcription-quantitative polymerase chain reaction. |
Table I.
Sequence of primers used in the
reverse transcription-quantitative polymerase chain reaction.
| Primer | Sequence (5′→3′) |
|---|
| miR-148a-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACACAAAGT |
| miR-148a-forward |
AGCTGTTCAGTGCACTACAGA |
| miR-148a-reverse | GTGCAGGGTCCGAGGT |
| miR-148a-probe |
FAM-CTGGATACGACACAAAG-MGB |
| miR-375-RT |
GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACTCACGCG |
| miR-375-forward |
CACAAAATTTGTTCGTTCGGCT |
| miR-375-reverse | GTGCAGGGTCCGAGGT |
| miR-375-probe |
FAM-CTGGATACGACTCACGC-MGB |
| U6-RT |
AAAATATGGAACGCTTCACGAATTTG |
| U6-forward |
CTCGCTTCGGCAGCACATATACT |
| U6-reverse |
ACGCTTCACGAATTTGCGTGTC |
| U6-probe |
FAM-CCATGCTAATCTTCTCTGTA-MGB |
qPCR assays
qPCR was performed using a CFX96 Real-Time PCR
Detection System (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
with TaqMan probes (Applied Biosystems; Thermo Fisher Scientific,
Inc.) according to the manufacturers protocol. No cDNA template
reactions were set up as negative controls and U6 snRNA was
amplified as an endogenous control. All reactions were performed in
triplicates. The PCR conditions were as follows: 95°C for 30 sec,
followed by 40 cycles of 95°C for 5 sec and 60°C for 34 sec. The
experiments were repeated three times and the data were normalized
using the endogenous U6 small nucleolar RNA. The 2−ΔΔCq
method was utilized for the normalization of PCR data (25). Primers were designed using DNAMAN
software (Lynnon Biosoft) and were synthesized by Life Technologies
(Thermo Fisher Scientific, Inc.). Primer sequences are presented in
Table I.
Statistical analysis
The comparison between miR-148a and miR-375
expression in laryngeal cancer, mild dysplasia, moderate dysplasia,
severe dysplasia, CIS and normal epithelial tissues was evaluated
using the independent samples t-test (two-tailed). Correlations of
miR-148a and miR-375 expression with patient tumor stages were
performed using the t-test followed by the Bonferroni
multiple-comparison correction. P≤0.05 was considered to indicate a
statistically significant difference. The ROC curve analysis and
all other statistical tests were performed using GraphPad Prism
version 5 (GraphPad Software, Inc., La Jolla, CA, USA).
Results
miR-148a and miR-375 are significantly
upregulated in LSCC
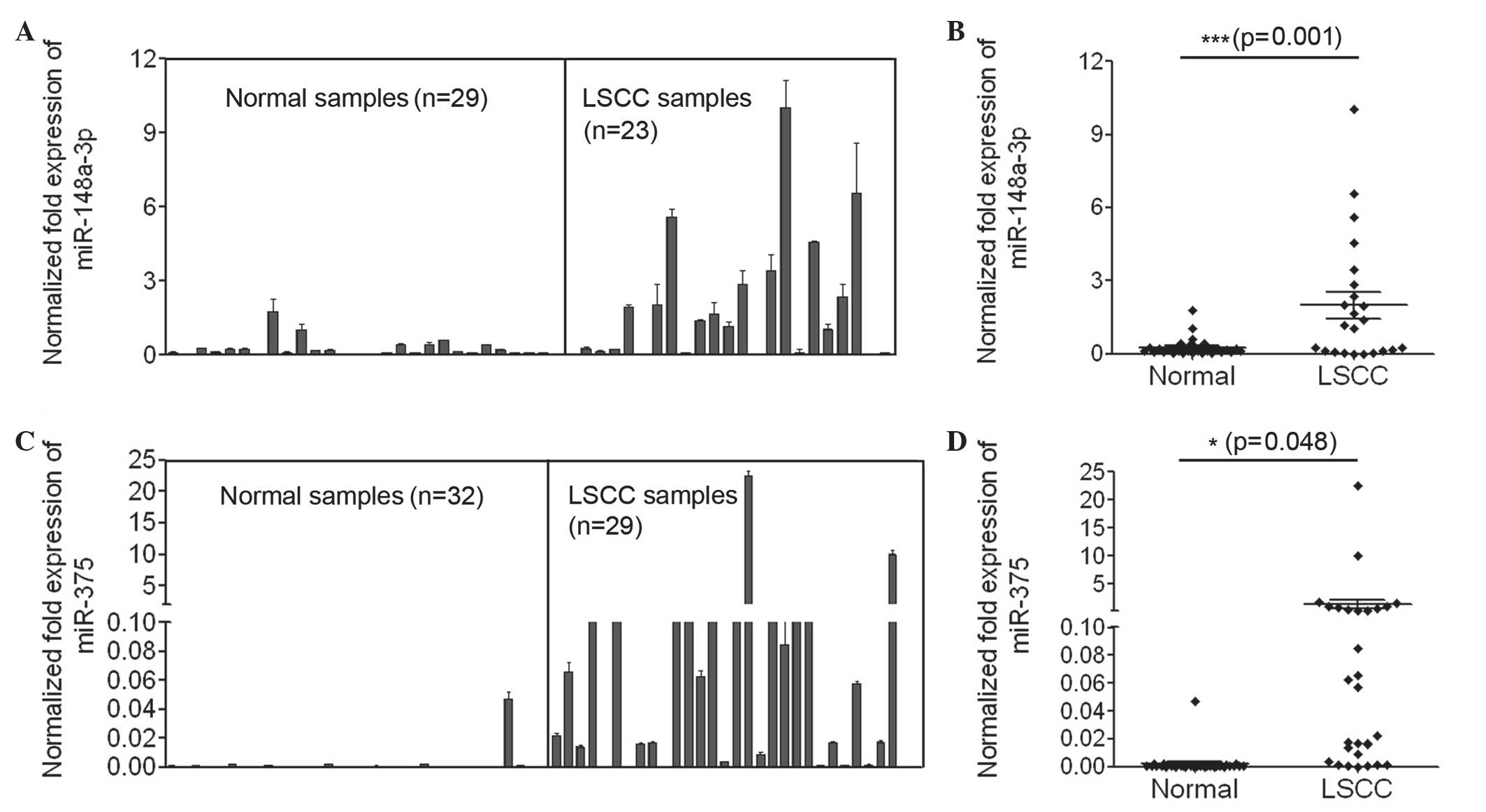
To accurately evaluate the expression of miR-148a
and miR-375 in LSCC, qPCR using TaqMan probes was performed to
measure the amount of miR-148a and miR-375. The present study
initially investigated the expression of miR-148a in the laryngeal
cancer tissues of 23 LSCC patients and 29 non-matched normal
epithelial tissues. The expression level of miR-148a in the
majority of the laryngeal cancer tissues was increased compared
with that in normal epithelial tissues, and the average expression
of miR-148a in laryngeal cancer samples was significantly increased
compared with that in the normal controls (P<0.001; Fig. 1A and B). In addition, the present
study examined the expression of miR-375 in the laryngeal cancer
tissues of 29 LSCC patients and 32 normal epithelial tissues. The
number of clinical samples used to measure the expression levels of
miR-148a and miR-375 differed, as clinical samples with poor
amplification results were not analyzed. The expression of miR-375
in normal epithelial tissues was extremely low, and its expression
in the majority of laryngeal cancer tissue samples was increased
compared with that in normal tissues (Fig. 1C). The average expression of miR-375
in laryngeal cancer samples was significantly increased compared
with that in the normal controls (P=0.048; Fig. 1D). In summary, the data indicated that
miR-148a and miR-375 were significantly upregulated in laryngeal
tumor tissues and may have significant roles in LSCC.
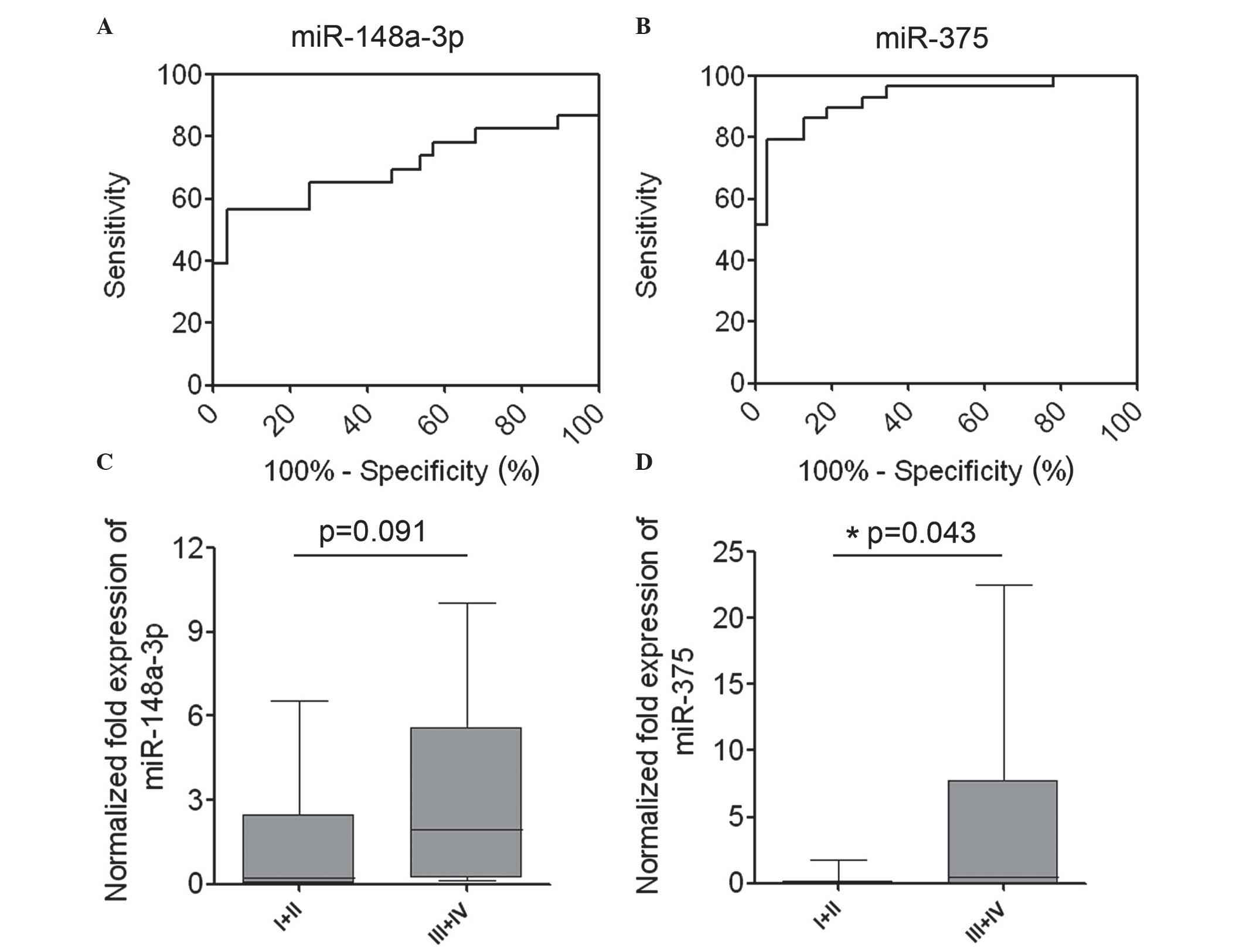
miR-375 may serve as a biomarker for
LSCC diagnosis
As miR-148a and miR-375 were significantly
upregulated in laryngeal cancer tissues, ROC curves were used to
analyze the sensitivity and specificity of miR-148a and miR-375 as
candidate biomarkers for LSCC diagnosis. The results from the test
samples gave area under the curve values of 0.7050 for miR-148a
(P=0.012) and 0.927 for miR-375 (P<0.0001). These results
suggested that the expression levels of miR-375 may be useful as a
marker, with high sensitivity and specificity for LSCC diagnosis
(Fig. 2A and B).
High levels of expression of miR-148a
and miR-375 are associated with aggressive phenotypes of LSCC
To additionally investigate the association between
the expression of miR-148a and miR-375 and patient
clinicopathological characteristics, the relative expression of
miR-148a and miR-375 in LSCC tissues was statistically analyzed.
Association analysis revealed that the mean expression level of
miR-148a in LSCC tissues of stage III/IV was increased compared
with that in tissues of stage I/II; however, this difference was
not significant (P=0.091; Fig. 2C).
High levels of miR-375 in LSCC were significantly associated with a
more aggressive tumor phenotype (P=0.043; stage I/II vs. III/IV;
Fig. 2D). The association between
increased levels of miR-375 and a more aggressive phenotype of LSCC
indicates that miR-375 may have a significant role in LSCC
carcinogenesis.
miR-148a and miR-375 levels increase
during various stages of LSCC carcinogenesis
The progression of LSCC undergoes several different
phases: Dysplasia (including mild dysplasia, moderate dysplasia and
severe dysplasia), CIS and LSCC (Fig.
3A). To additionally investigate the potential role of miR-148a
and miR-375 in LSCC carcinogenesis, the present study measured the
levels of miR-148a and miR-375 in these various phases. The present
study initially examined the expression of miR-148a in 19 mild
dysplasia, 26 moderate dysplasia, 32 severe dysplasia and 35 CIS
tissues, and additionally compared the expression level with that
in the normal controls and LSCC tissues described previously. The
results revealed that the expression level of miR-148a in mild
dysplasia tissues was significantly increased compared with that in
normal epithelial tissues (P<0.0001). The mean expression of
miR-148a increased gradually during LSCC progression from the
moderate dysplasia stage. The level of miR-148a in severe dysplasia
tissues was increased compared with that in moderate dysplasia
tissues (P=0.047); the level in LSCC was additionally increased
compared with that in moderate dysplasia tissues (P=0.049; Fig. 3B). The expression level of miR-375 was
additionally examined in 19 mild dysplasia, 29 moderate dysplasia,
34 severe dysplasia and 36 CIS tissues, and additionally compared
the expression level with that in the normal controls and LSCC
tissues described previously. The results revealed that the mean
expression of miR-375 increased gradually during LSCC progression
from dysplasia. The expression level of miR-375 in mild dysplasia
tissues was significantly increased compared with that in normal
epithelial tissues (P=0.027); the level in LSCC was additionally
higher compared with that in mild dysplasia tissues (P=0.048;
Fig. 3C). These results suggested
that miR-148a and miR-375 may have significant roles in dysplasia
and LSCC progression.
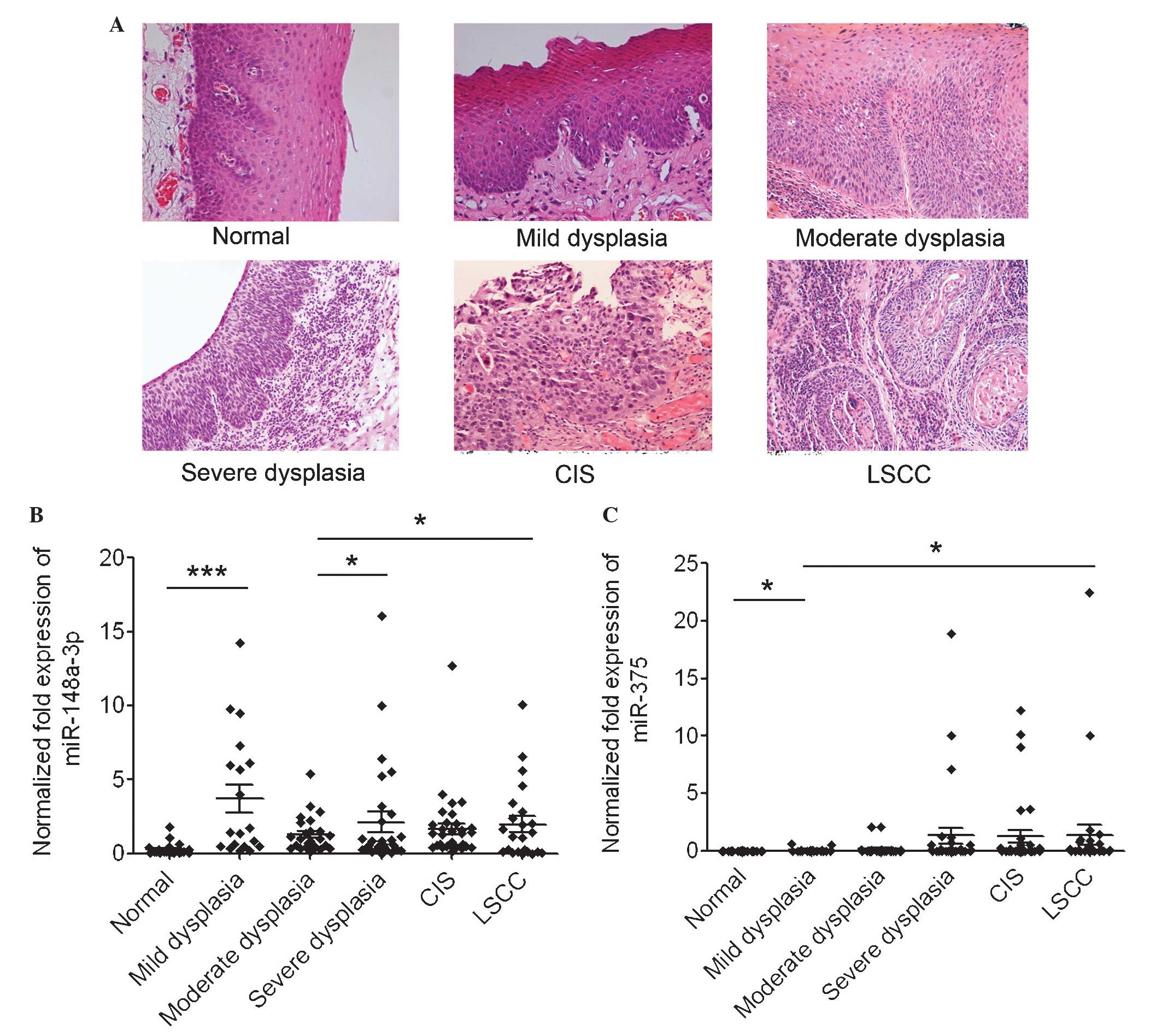 | Figure 3.miR-148a and miR-375 increase during
various stages of LSCC carcinogenesis. (A) Representative
pathological biopsies of normal epithelial, mild dysplasia,
moderate dysplasia and severe dysplasia, CIS and LSCC tissues.
Hematoxylin and eosin staining; magnification, ×200. (B) Normalized
expression of miR-148a in normal epithelial tissues and abnormal
tissues of various stages of LSCC carcinogenesis. *P<0.05,
severe dysplasia group and LSCC group vs. moderate dysplasia group;
*** P<0.001, mild dysplasia group vs. normal control group. (C)
Normalized expression of miR-375 in normal epithelial tissues and
abnormal tissues of various stages of LSCC carcinogenesis.
*P<0.05, mild dysplasia group vs. normal control and LSCC group.
miR, microRNA; LSCC, laryngeal squamous cell carcinoma; CIS, cancer
in situ. |
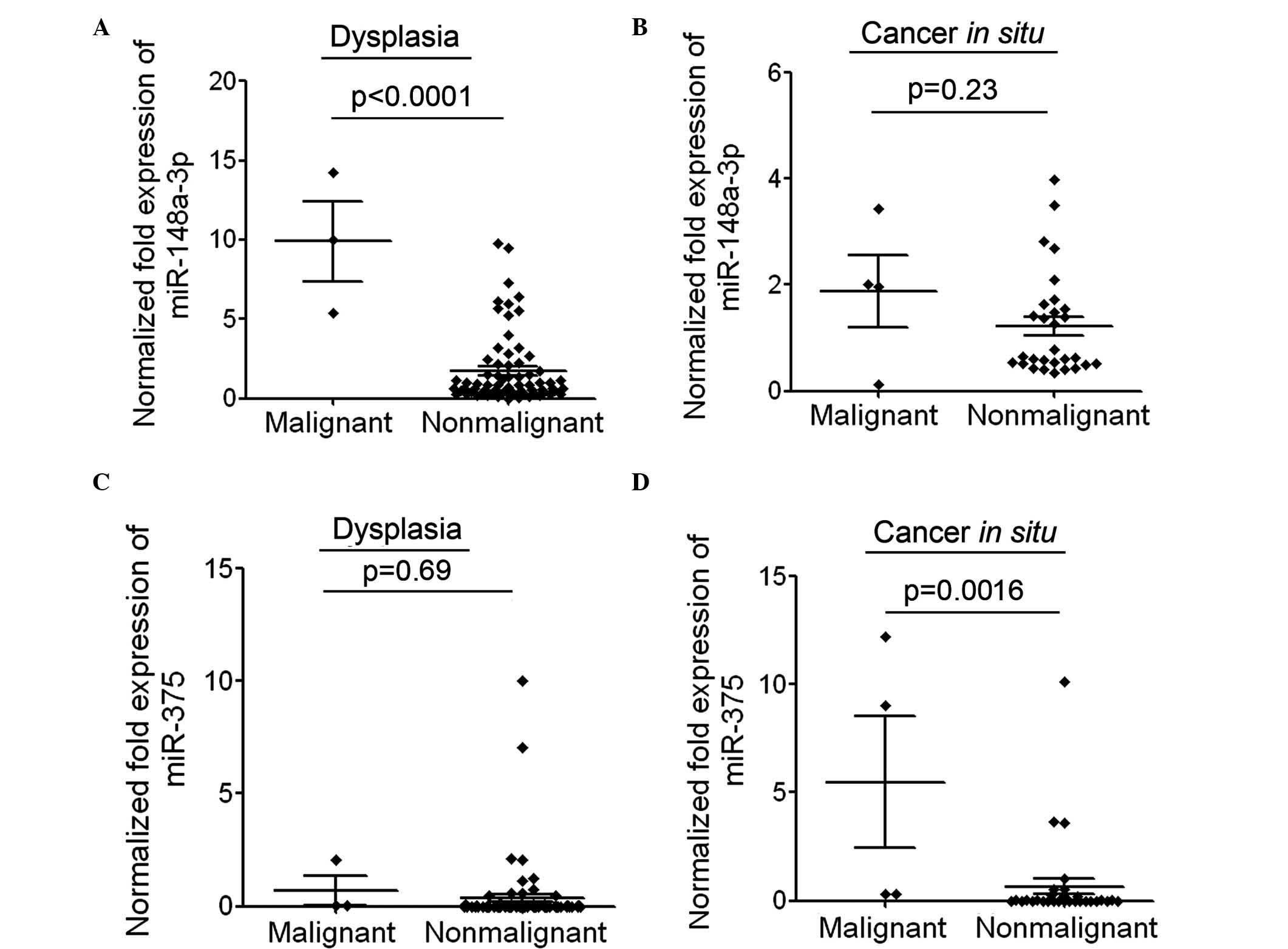
Positive association between the
expression level of miR-148a of miR-375 and malignant
transformation
To additionally investigate whether increased
expression of miR-148a or miR-375 in patients with dysplasia is
able to predict disease progression, the present study analyzed the
expression of miR-148a and miR-375 in the dysplasia tissues of a
number of malignantly transformed patients. Among the patients with
dysplasia or CIS, only a small number of patients progressed to
LSCC and thus, the number of malignantly transformed patients and
non-malignantly transformed patients was different. The mean
expression level of miR-148a in the dysplasia tissues of 3
malignantly transformed patients was significantly increased
compared with that in currently nonmalignantly transformed patients
(P<0.0001; Fig. 4A). The
expression level of miR-148a in CIS tissues of 4 malignantly
transformed patients was also slightly increased compared with that
in currently nonmalignantly transformed patients (Fig. 4B). The mean expression level of
miR-375 in the dysplasia tissues of 3 malignantly transformed
patients was slightly higher compared with that in currently
nonmalignantly transformed patients (Fig.
4C). The expression level of miR-375 in CIS tissues of 4
malignantly transformed patients was significantly increased
compared with that in currently nonmalignantly transformed patients
(P=0.0016; Fig. 4D). These results
suggested that patients with increased expression of miR-148a or
miR-375 may be more prone to malignant transformation. The
association between the expression level of miR-148a or miR-375 and
malignant transformation requires additional investigation in an
increased number of clinical samples.
Discussion
In the present study, the expression of miR-148a and
miR-375 was examined in a number of patients with laryngeal
dysplasia or LSCC, and the potential application of the expression
of these miRs in LSCC diagnosis was investigated. The expression
profiles of miR-148a and miR-375 in LSCC indicated that they are
significantly upregulated in LSCC, and their increased levels were
additionally associated with more aggressive tumor phenotypes.
Additional analysis suggested that miR-375 may serve as a potential
biomarker for LSCC diagnosis. Several innate properties of miRs
mean that they are attractive as potential biomarkers. miRs are
small and stable against degradation and can be detected easily by
specific and sensitive qPCR in small quantities of sample. miRs are
additionally detectable in bodily fluids, including serum, plasma,
saliva, urine and tears (26,27). Furthermore, expression profiles of
miRs in the plasma and/or serum of cancer patients may reflect the
change in expression of miRs in tumor cells (28). Circulating miRs may represent a novel
class of non-invasive biomarkers for cancer diagnostic and
prognostic information. Therefore, the present study examined the
expression levels of miR-148a and miR-375 in the serum of LSCC
patients and investigated the potential application of circulating
miRs in LSCC diagnosis.
The present study investigated the expression of
miR-148a and miR-375 during various phases of LSCC progression and
observed that miR-148a and miR-375 increased during different
stages of LSCC carcinogenesis. Notably, the association analysis of
miR expression with disease progression indicated that increased
expression of miR-148a or miR-375 in patients with laryngeal
dysplasia may predict subsequent malignant transformation and may
therefore serve as an early biomarker of LSCC. Early diagnosis of
cancer is crucial for improving cancer therapy. To intervene or
administer treatment at early or precancerous stages may prevent
disease progression or carcinogenesis, and is important for
increasing recovery rates and improving patient quality of life
(29,30). Therefore, association between the
expression levels of miR-148a or miR-375 and malignant
transformation require additional investigation in a larger number
of clinical samples and the potential application of miR-148a or
miR-375 as early biomarkers of LSCC requires further
investigation.
The aberrant expression of miR-148a has been
reported in various forms of cancer, including hepatocellular
carcinoma, bladder, breast and gastric cancer (31–34).
However, no previous studies have examined the expression level of
miR-148a in laryngeal carcinoma. To the best of our knowledge, the
present study is the first to report the aberrant expression of
miR-148a in LSCC. Recently, miR-148a was identified as a target of
H19 and involved in regulating LSCC progression (35). The detailed function and mechanism of
miR-148a in regulating LSCC progression required further
investigation. The expression level of miR-375 in paired-LSCC
tissues has been previously investigated (18). Yu et al (18) reported that miR-375 was downregulated
in LSCC tissues, which was inconsistent with the results of the
present study. This previous study compared the expression of
miR-375 in LSCC tissue with adjacent normal tissues (18). Increasing evidence has suggested that
normal tissues adjacent to cancer are not necessarily normal. For
example, gene expression data from pancreatic cancer and adjacent
normal tissue specimens demonstrated that the adjacent normal
tissues had already acquired a number of transcriptional
alterations and was not an appropriate baseline for comparison with
cancer (36). A similar phenomenon
has been observed in colorectal cancer patients (37). Therefore, the change in expression of
miR-375 between LSCC tissues and adjacent normal tissues was not in
accordance with that between LSCC tissues and normal laryngeal
tissues. The authors of the present study propose that the
expression profile of miR-375 in a number of tissues in the various
phases of LSCC progression presented in the current paper may
reflect the expression changes of miR-375 during LSCC progression
in vivo more appropriately. The detailed function and
underlying mechanism of miR-375 regulation of LSCC carcinogenesis
requires additional investigation.
In conclusion, the present study determined that
miR-148a and miR-375 levels are increased during LSCC progression,
and high levels of expression of these two miRs was associated with
aggressive phenotypes of LSCC. Notably, an increased expression
level of miR-148a or miR-375 in patients with laryngeal dysplasia
may predict subsequent malignant transformation, and these results
suggest that miR-148a and miR-375 may serve as potential predictive
biomarkers for early diagnosis of LSCC.
Acknowledgements
The present study was supported by grants from the
Research Fund for the Doctoral Program of Higher Education of
China, Beijing, China (grant no. 20131107110004) and the Scientific
and Technological Innovation Base for Training and Development of
Engineering Projects, Beijing, China (grant no.
Z141107004414028).
Glossary
Abbreviations
Abbreviations:
|
miRs
|
microRNAs
|
|
LSCC
|
laryngeal squamous cell carcinoma
|
|
CIS
|
cancer in situ
|
|
ROC
|
receiver-operating characteristic
|
References
|
1
|
Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I,
Wang C, Liu X and Zhou X: Down-regulation of the microRNA-99 family
members in head and neck squamous cell carcinoma. Oral Oncol.
48:686–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cai K, Wang Y and Bao X: MiR-106b promotes
cell proliferation via targeting RB in laryngeal carcinoma. J Exp
Clin Cancer Res. 30:732011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: GLOBOCAN 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Fleskens SA, Bergshoeff VE, Voogd AC, van
Velthuysen ML, Bot FJ, Speel EJ, Kremer B, Takes R and Slootweg P:
Interobserver variability of laryngeal mucosal premalignant
lesions: A histopathological evaluation. Mod Pathol. 24:892–898.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Furusaka T, Susaki Y, Saito T, Katsura Y
and Ikeda M: Long-term follow-up and salvage surgery in patients
with T2N0M0 squamous cell carcinoma of the glottic larynx following
concurrent chemoradiation therapy with cisplatin and 5-fluorouracil
for laryngeal preservation. Acta Otolaryngol. 133:91–98. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kimura S, Naganuma S, Susuki D, Hirono Y,
Yamaguchi A, Fujieda S, Sano K and Itoh H: Expression of microRNAs
in squamous cell carcinoma of human head and neck and theesophagus:
miR-205 and miR-21 are specific markers for HNSCC and ESCC. Oncol
Rep. 23:1625–1633. 2010.PubMed/NCBI
|
|
7
|
Xie L, Qian X and Liu B: MicroRNAs: Novel
biomarkers for gastrointestinal carcinomas. Mol Cell Biochem.
341:291–299. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Alencar AJ, Malumbres R, Kozloski GA,
Advani R, Talreja N, Chinichian S, Briones J, Natkunam Y, Sehn LH,
Gascoyne RD, et al: MicroRNAs are independent predictors of outcome
in diffuse large B-cell lymphoma patients treated with R-CHOP. Clin
Cancer Res. 17:4125–4135. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Borel C, Deutsch S, Letourneau A,
Migliavacca E, Montgomery SB, Dimas AS, Vejnar CE, Attar H,
Gagnebin M, Gehrig C, et al: Identification of cis- and
trans-regulatory variation modulating microRNA expression levels in
human fibroblasts. Genome Res. 21:68–73. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Macfarlane LA and Murphy PR: MicroRNA:
Biogenesis, function and role in cancer. Curr Genomics. 11:537–561.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Davidson B, Tropé CG and Reich R: The
clinical and diagnostic role of microRNAs in ovarian carcinoma.
Gynecol Oncol. 133:640–646. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nugent M: MicroRNA function and
dysregulation in bone tumors: The evidence to date. Cancer Manag
Res. 6:15–25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu C, Yu J, Yu S, Lavker RM, Cai L, Liu
W, Yang K, He X and Chen S: MicroRNA-21 acts as an oncomir through
multiple targets in human hepatocellular carcinoma. J Hepatol.
53:98–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Y, Kim S and Kim IM: Regulation of
metastasis by microRNAs in ovarian cancer. Front Oncol. 4:1432014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Han C, Yu Z, Duan Z and Kan Q: Role of
microRNA-1 in human cancer and its therapeutic potentials. Biomed
Res Int. 2014:4283712014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu X, Wu Y, Liu Y, Deng H, Shen Z, Xiao B
and Guo J: miR-21, miR-106b and miR-375 as novel potential
biomarkers for laryngeal squamous cell carcinoma. Curr Pharm
Biotechnol. 15:503–508. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li W, Ma H and Sun J: MicroRNA-34a/c
function as tumor suppressors in Hep-2 laryngeal carcinoma cells
and may reduce GALNT7 expression. Mol Med Rep. 9:1293–1298.
2014.PubMed/NCBI
|
|
20
|
Yungang W, Xiaoyu L, Pang T, Wenming L and
Pan X: miR-370 targeted FoxM1 functions as a tumor suppressor in
laryngeal squamous cell carcinoma (LSCC). Biomed Pharmacother.
68:149–154. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang T, Liu M, Wang C, Lin C, Sun Y and
Jin D: Down-regulation of MiR-206 promotes proliferation and
invasion of laryngeal cancer by regulating VEGF expression.
Anticancer Res. 31:3859–3863. 2011.PubMed/NCBI
|
|
22
|
Tian L, Li M, Ge J, Guo Y, Sun Y, Liu M
and Xiao H: MiR-203 is downregulated in laryngeal squamous cell
carcinoma and can suppress proliferation and induce apoptosis of
tumours. Tumour Biol. 35:5953–5963. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang HK and Liu HG: Is severe dysplasia
the same lesion as carcinoma in situ? 10-year follow-up of
laryngeal precancerous lesions. Acta Otolaryngol. 2:325–328. 2012.
View Article : Google Scholar
|
|
24
|
Barnes L, Eveson JW, Reichart P and
Sidransky D: World Health Organization Classification of Tumours.
Pathology and Genetics of Head and Neck Tumours. IARC Press.
(Lyon). 10–80. 2001.
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kosaka N, Iguchi H and Ochiya T:
Circulating microRNA in body fluid: A new potential biomarker for
cancer diagnosis and prognosis. Cancer Sci. 101:2087–2092. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body fluids -
the mix of hormones and biomarkers. Nat Rev Clin Oncol. 8:467–477.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Taylor DD and Gercel-Taylor C: MicroRNA
signatures of tumor-derived exosomes as diagnostic biomarkers of
ovarian cancer. Gynecol Oncol. 110:13–21. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhu Y, Yang SR, Wang PP, Savas S, Wish T,
Zhao J, Green R, Woods M, Sun Z, Roebothan B, et al: Influence of
pre-diagnostic cigarette smoking on colorectal cancer survival:
Overall and by tumour molecular phenotype. Br J Cancer.
110:1359–1366. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Nan H, Giovannucci EL, Wu K, Selhub J,
Paul L, Rosner B, Fuchs CS and Cho E: Pre-diagnostic leukocyte
genomic DNA methylation and the risk of colorectal cancer in women.
PLoS One. 8:e594552013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Liu L, Ye JX, Qin YZ, Chen QH and Ge LY:
Evaluation of miR-29c, miR-124, miR-135a and miR-148a in predicting
lymph node metastasis and tumor stage of gastric cancer. Int J Clin
Exp Med. 8:22227–22236. 2015.PubMed/NCBI
|
|
32
|
Jiang Q, He M, Ma MT, Wu HZ, Yu ZJ, Guan
S, Jiang LY, Wang Y, Zheng DD, Jin F and Wei MJ: MicroRNA-148a
inhibits breast cancer migration and invasion by directly targeting
WNT-1. Oncol Rep. 35:1425–1432. 2016.PubMed/NCBI
|
|
33
|
Ma L, Xu Z, Xu C and Jiang X:
MicroRNA-148a represents an independent prognostic marker in
bladder cancer. Tumour Biol. Dec 23–2015.[Epub ahead of print].
|
|
34
|
Jung KH, Zhang J, Zhou C, Shen H, Gagea M,
Rodriguez-Aguayo C, Lopez-Berestein G, Sood AK and Beretta L:
Differentiation therapy for hepatocellular carcinoma: Multifaceted
effects of miR-148a on tumor growth and phenotype and liver
fibrosis. Hepatology. 63:864–879. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wu T, Qu L, He G, Tian L, Li L, Zhou H,
Jin Q, Ren J, Wang Y, Wang J, et al: Regulation of laryngeal
squamous cell cancer progression by the lncRNA
H19/miR-148a-3p/DNMT1 axis. Oncotarget. 7:11553–11566.
2016.PubMed/NCBI
|
|
36
|
Wu Y, Wang X, Wu F, Huang R, Xue F, Liang
G, Tao M, Cai P and Huang Y: Transcriptome profiling of the cancer,
adjacent non-tumor and distant normal tissues from a colorectal
cancer patient by deep sequencing. PLoS One. 7:e410012012.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Clare SE, Pardo I, Mathieson T, Lillemoe
HA, Blosser RJ, Choi M, Sauder CAM, Doxey DK, Badve S, Storniolo
AMV, et al: Abstract P1-03-02: ‘Normal’ tissue adjacent to breast
cancer is not normal. Cancer Res. 72:P103–02. 2012. View Article : Google Scholar
|