Introduction
Lipomas are common fat cell tumors that most often
occur in middle-aged or elderly patients. Lipomas may arise
subcutaneously (superficial lipomas) or in deep soft tissues
(deep-seated intra- or intermuscular lipomas). Superficial lipomas
are usually smaller (<5 cm) than their deep-seated counterparts
(>5 cm) (1). The genetic features
of lipomas have been characterized in considerable detail. The
karyotype of lipomas (2–4) is usually near- or pseudodiploid with
structural chromosomal rearrangements, the most common of which
involve the chromosomal segment 12q13~15 (5). According to the Mitelman Database of
Chromosome Aberrations and Gene Fusions in Cancer (http://cgap.nci.nih.gov/Chromosomes/Mitelman; database
updated on May 7, 2015), 476 lipomas have been reported with
chromosomes aberrations, with 12q13~15 being targeted in >300 of
them. The recombination with 12q13~15 occurs with multiple other
chromosome bands, the most common of which is 3q27~28,
corresponding to the translocation t(3;12)(q27~28;q14~15). Other
chromosome segments that are frequently recombined with 12q13~15
are 1p36, 1p32~34, 2p22~24, 2q35~37, 5q33, 9p21~22, 12p11~13 and
13q12~14. The rearrangements often target the high mobility
group AT-hook 2 (HMGA2) gene in 12q14.3, regardless of
whether the breakpoints occur within or outside the gene locus; the
essential outcome is the deregulation of HMGA2. The most
common gene fusing with HMGA2 in lipoma is
lipoma-preferred partner (LPP) (3q27) (6), but also other genes located in the
common breakpoint regions are known to recombine with HMGA2
recurrently. In addition, the chromosomal rearrangements may
occasionally lead to truncated HMGA2 transcripts (7).
According to the Mitelman Database of Chromosome
Aberrations and Gene Fusions in Cancer, the translocation
t(4;12)(q27;q15) and variants thereof have been reported in 6
lipomas: In 2 of them, as the sole chromosomal aberration; in other
2, as a three-way translocation; and in the last 2, as part of more
complex karyotypes (http://cgap.nci.nih.gov/Chromosomes/Mitelman; database
updated on May 7, 2015). The present study describes another 5
cases of lipoma carrying the t(4;12)(q27~28;q14~15) as a sole
anomaly and 1 case with a three-way translocation. Molecular
analyses were performed on 4 of the tumors in order to discover the
putative gene partner from 4q27~28 recombining with HMGA2,
but none was identified. Instead, the analysis revealed 4 truncated
forms of HMGA2 in the examined samples. The HMGA2
truncated transcripts included sequences located in the chromosomal
sub-band 4q28.1 in 3 of these cases.
Materials and methods
Ethics statement
The present study was approved by the Regional
Committee for Medical and Health Research of the University of Oslo
(Oslo, Norway; http://helseforskning.etikkom.no), and written
informed consent was obtained from the patients.
Patients
Patients were admitted to the Norwegian Radium
Hospital (Oslo, Norway) between January 1, 1998 and November 30,
2014. Table I shows the patients'
gender, age, diagnosis and tumor location. All tumors were
surgically removed.
 | Table I.Clinical, cytogenetics and molecular
data on the 6 lipomas. |
Table I.
Clinical, cytogenetics and molecular
data on the 6 lipomas.
| Case no. | Gender/age,
years | Diagnosis | Location | (cm) | Karyotype [no. cells
carrying karyotype] | Genome coordinates of
the fusiona |
|---|
| 1 | M/47 | Lipoma | Subcutaneous,
posterior axillary fold | 8.0 | 46,XY,t(4;12) | Chr12:65951415 |
|
|
|
|
|
|
(q27~28;q14~15)[cp15] | Chr4:1234122125 |
| 2 | F/73 | Lipoma | Intramuscular,
throat | 7.0 | 46,XX,t(1;4;12) | Chr12:65838592 |
|
|
|
|
|
|
(q21;q27~28;q14~15) | Chr4:126766222 |
|
|
|
|
|
|
[12]/45,idem,-20[3] |
|
| 3 | F/69 | Lipoma | Intramuscular,
flank | 10.0 |
46,XX,t(4;12)(q27~28; | Chr12:65838592 |
|
|
|
|
|
|
q14~15)[15]/46,XX[2] | Chr12:65915527 |
| 4 | F/33 | Lipoma (relapse) | Supraclavicular
fossa | 5.5 |
46,XX,t(4;12)(q27~28; | Chr12:65951415 |
|
|
|
|
|
| q14~15)[cp6] | Chr4:123450402 |
| 5 | M/61 | Lipoma | Intramuscular, right
thigh | 20.0 |
46,XY,t(4;12)(q27~28; | Chr12: Not
available |
|
|
|
|
|
|
q14~15)[27]/46,XY[3] | Chr4: Not
available |
| 6 | M/14 | Lipoma | Intramuscular, left
forearm | 11.0 |
46,XY,t(4;12)(q27~28; | Chr12: Not
available |
|
|
|
|
|
|
q14~15)[15]/46,XY[5] | Chr4: Not
available |
Chromosome banding analysis
Samples from the operation specimens were
mechanically and enzymatically disaggregated and short-term
cultured as described by Mandahl (8).
The cultures were harvested, and the chromosomes were G-banded
using Wright's stain (Sigma-Aldrich, St. Louis, MO, USA). The
subsequent cytogenetic analysis and karyotype description followed
the recommendations of the International System for Human
Cytogenetic Nomenclature (9).
RNA extraction
Tumor tissue adjacent to that used for cytogenetic
analysis and histologic examination had been frozen and stored at
−80°C from 4 tumors (cases 1–4). Total RNA was extracted using
miRNeasy kit, TissueLyser II and QIAcube according to the
manufacturer's protocol (Qiagen GmbH, Hilden, Germany). The
concentration and purity of the RNA was measured with the NanoVue
Spectrophotometer (GE Healthcare Bio-Sciences, Pittsburgh, PA,
USA).
3′-Rapid amplification of
complementary DNA (cDNA) ends (3′-RACE)
3′-RACE was performed using a protocol described
previously (10). Total RNA (100 ng)
was reverse-transcribed in a 20-µl reaction volume using A3RNV-RACE
(5′-ATCGTTGAGACTCGTACCAGCAGAGTCACGAGAGAGACTACACGGTACTGGTTTTTTTTTTTTTT-3′)
as a primer and iScript Select cDNA Synthesis kit (Bio-Rad
Laboratories AB, Oslo, Norway) according to the manufacturer's
protocol. cDNA (1 µl) was used as template and amplified in a
polymerase chain reaction (PCR) using the outer primer combination
HMGA2-846F1 (5′-CCACTTCAGCCCAGGGACAACCT-3′) and A3R-1New
(5′-TCGTTGAGACTCGTACCAGCAGAGTCAC-3′). PCR cycling conditions
consisted of an initial step of denaturation at 94°C for 30 sec,
followed by 35 cycles of 7 sec at 98°C, 2 min at 68°C and a final
extension for 5 min at 72°C. In total, 1 µl of the amplified
products was used as template in nested PCR with the primers
HMGA2-982F1 (5′-CAAGAGTCCCTCTAAAGCAGCTCA-3′) and A3R3
(5′-CGAGAGAGACTACACGGTACTGGT-3′). The nested PCR was performed
using the Touchdown-PCR conditions described by Korbie and Mattick
(11) in order to increase the
specificity of the PCR and improve the quality of the products. For
both PCRs, the 25-µl reaction volume contained 12.5 µl Premix Ex
Taq (Takara Bio Europe SAS, Saint-Germain-en-Laye, France),
template, and 0.4 µM of each of the forward and reverse primers.
PCR products (3 µl) were stained with GelRed™ (Biotium, Inc.,
Hayward, CA, USA) and analyzed by electrophoresis using 1.0%
agarose gels. The gel was scanned with G:BOX (Syngene, Frederick,
MD, USA), and the images were acquired using GeneSnap (Syngene).
The remaining 22 µl of the amplified fragments were purified using
the QIAquick PCR purification kit (Qiagen AB). Direct sequencing
was performed using the LIGHTRUN™ Sequencing Service of GATC
Biotech (Konstanz, Germany; www.gatc-biotech.com/en/products/sanger-services/lightrun-sequencing.html).
The Basic Local Alignment Search Tool (BLAST; http://blast.ncbi.nlm.nih.gov/Blast.cgi)
and BLAST-like alignment tool (http://genome.ucsc.edu/cgi-bin/hgBlat) programs were
used for computer analysis of the sequencing data.
Results
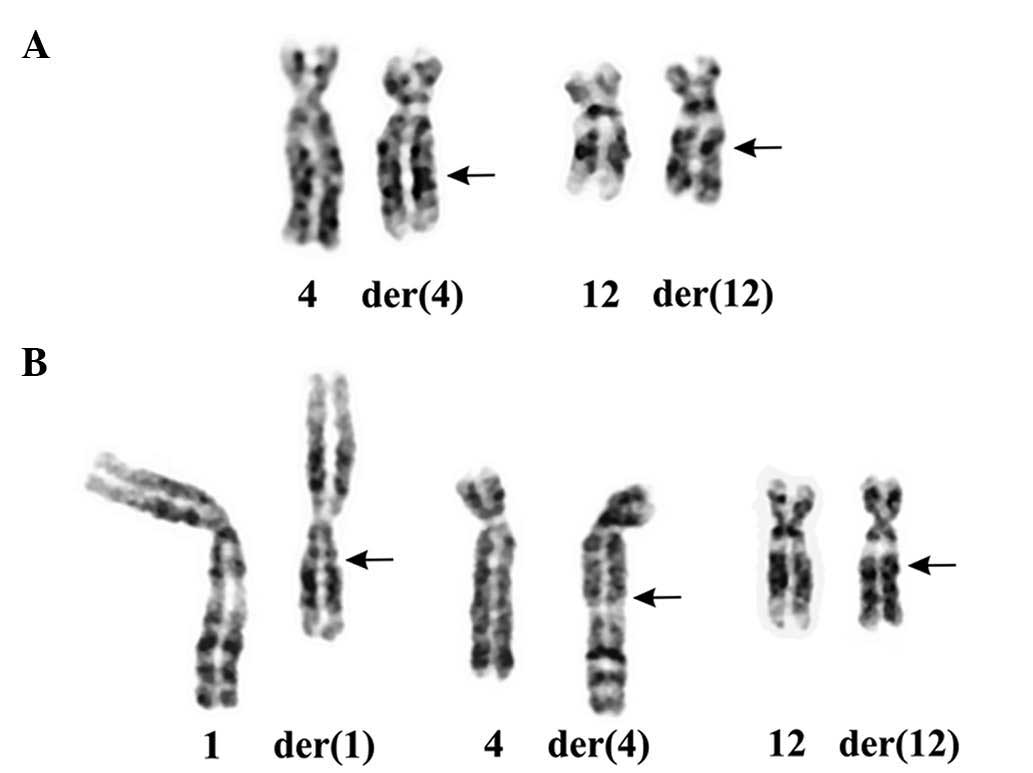
Pathology and cytogenetics
Table I contains the
patients' gender, age and diagnosis, and the location, karyotype,
HMGA2 expression and HMGA2 fusions of the examined
lipomatous tumors. In all 6 cases (3 males and 3 females), there
was recombination between the chromosome bands 12q14~15 and
4q27~28. In total, 5 cases carried t(4;12)(q14~15;q27~28) as the
sole karyotypic aberration, whereas 1 lipoma (case 2) had a
three-way translocation t(1;4;12)(q21;q27~28;q14~15) (Fig. 1).
Molecular genetics
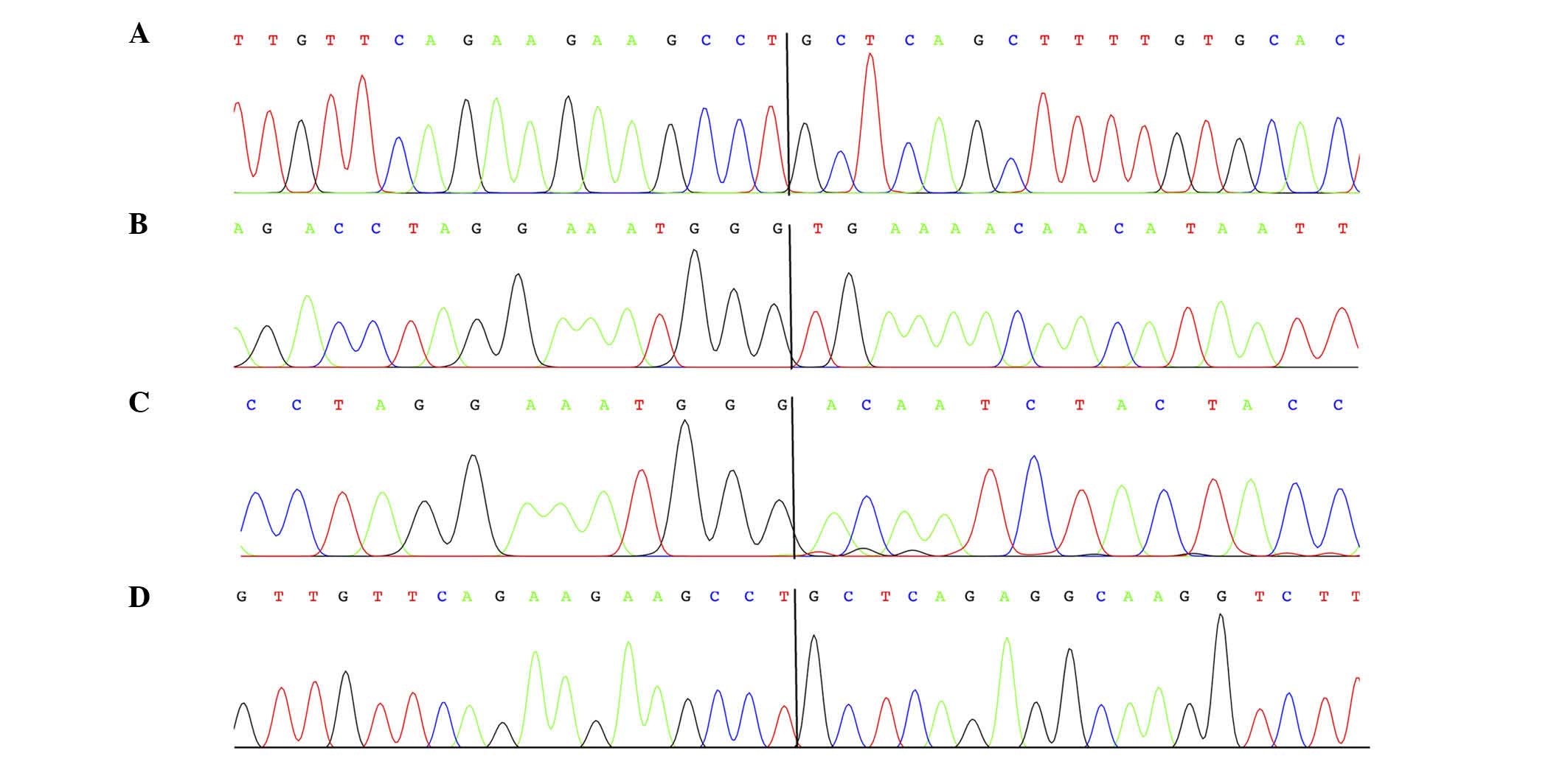
3′-RACE on the lipomas of cases 1–4 (Table I) amplified fragments which were
revealed to be chimeric HMGA2-cDNA fragments by Sanger
sequencing analysis (Fig. 2). In
lipomas 1, 2 and 4, HMGA2 was fused with sequences from
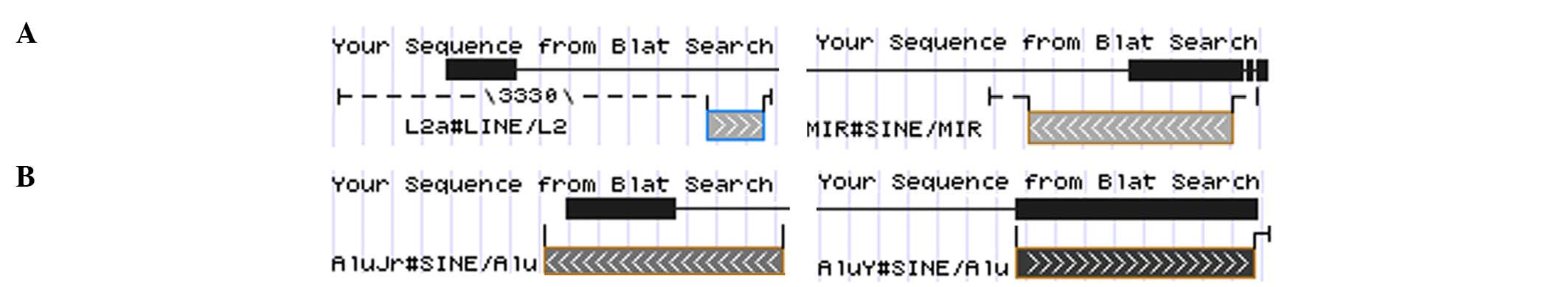
intergenic regions of 4q28.1. In lipoma 1, the exon 4 of
HMGA2 was fused with the transposable elements long
interspersed element L2a and mammalian interspersed repetitive
located in 4q28.1 (Fig. 3), whereas
lipoma 2 exhibited HMGA2 exon 3 fused with a circa 750-bp
fragment from band 4q28.1. In lipoma 4, HMGA2 exon 4 was
fused with two Alu sequences from chromosome band 4q28.1 (Fig. 3). In lipoma 3, the third exon of
HMGA2 was fused with two HMGA2 intron 3 sequences of
469 and 305 bp, respectively, with a distance between them of ~75
kbp.
Discussion
HMGA1 and HMGA2 are non-histone proteins involved in
a wide variety of nuclear processes, from chromatin dynamics to
gene regulation (12). HMGA
family genes are expressed during embryonic development (13), but are largely unexpressed in adult
normal tissues (14). High expression
levels of HMGA2 in tissues are usually associated with
neoplastic transformation (15).
Indeed, recent studies highlight a pivotal role of HMGA2 in
cancer pathogenesis and progression (16). The HMGA2 gene has been
previously observed to be disrupted, due to rearrangement of
chromosomal bands 12q13~15, in different connective tissue tumors,
including lipomas (7), pleiomorphic
adenomas of the salivary gland (17),
uterine leiomyomas (18) and lung
hamartomas (19). These alterations
usually involve exon 3 and cause deletion of downstream regions,
leading to a truncated transcript that can evade microRNA-dependent
gene silencing (20). Alternatively,
chromosomal rearrangements of 12q13~15 may lead to the formation of
a fusion gene (15). In lipoma
tumorigenesis, the most common fusion partner for HMGA2 is
LPP (3q27). Other frequent fusion partners of HMGA2
are C-X-C chemokine receptor type 7 (2q37), early B-cell
factor 1 (5q33) and neurofibromin 1B (9p23) (5).
The present analyses revealed that the translocation
t(4;12)(q27~28;q14~15) is recurrent in lipomas and leads to the
truncation of HMGA2. Notably, 4 of the 6 lipomas analyzed
were intramuscular and large; whether this represents a general
feature of t(4;12)-positive tumors remains to be investigated. In 3
lipomas (cases 1, 2 and 4), sequences located in 4q28.1 were fused
with HMGA2. The ensuing HMGA2-fusion transcripts
coded for putative proteins which contain amino acid residues 1–83
of HMGA2 protein (accession number NP_003474.1), corresponding to
exons 1–3 of the HMGA2 gene, and amino acid residues from
the fused sequences. This pattern is similar to that observed as a
result of other rearrangements of HMGA2 in lipomas, where
disruption of the HMGA2 locus leaves intact exons 1–3 of the
gene (which encode the AT-hooks domains) and separates them from
the 3′-terminal part of the gene (12). Notably, in cases 1 and 4, the
HMGA2 exon 4 was fused with transposable elements. The Human
Genome Project findings predicted that approximately half of the
genome consists of such elements, which are capable of integrating
at new genomic sites within the cell of origin (21), thus leading to transposable
element-driven transcript diversification mediated by alternative
splicing (22).
In conclusion, the present study characterized the
translocation t(4;12)(q27~28;q14~15) in four lipomas and showed
that it disrupts the HMGA2 locus separating exons 1–3 of the
gene from the 3′-untranslated region. This finding contributes to
the understanding of HMGA2 genetics in lipomas.
Acknowledgements
The present study was supported by The Norwegian
Radium Hospital Foundation (Oslo, Norway).
References
|
1
|
Hameed M: Pathology and genetics of
adipocytic tumors. Cytogenet Genome Res. 118:138–147. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mandahl N, Hoglund M, Mertens F, Rydholm
A, Willén H, Brosjö O and Mitelman F: Cytogenetic aberrations in
188 benign and borderline adipose tissue tumors. Genes Chromosomes
Cancer. 9:207–215. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sreekantaiah C, Leong SP, Karakousis CP,
McGee DL, Rappaport WD, Villar HV, Neal D, Fleming S, Wankel A,
Herrington PN, et al: Cytogenetic profile of 109 lipomas. Cancer
Res. 51:422–433. 1991.PubMed/NCBI
|
|
4
|
Bartuma H, Hallor KH, Panagopoulos I,
Collin A, Rydholm A, Gustafson P, Bauer HC, Brosjö O, Domanski HA,
Mandahl N and Mertens F: Assessment of the clinical and molecular
impact of different cytogenetic subgroups in a series of 272
lipomas with abnormal karyotype. Genes Chromosomes Cancer.
46:594–606. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mandahl N and Mertens F: Soft tissue
tumors. Cancer Cytogenetics (3rd). John Wiley & Sons, Inc.
(Hoboken, NJ). 675–711. 2010. View Article : Google Scholar
|
|
6
|
Kubo T, Matsui Y, Naka N, Araki N, Myoui
A, Endo K, Yasui N, Ohtani O, Suzuki K, Kimura T, et al:
Specificity of fusion genes in adipocytic tumors. Anticancer Res.
30:661–664. 2010.PubMed/NCBI
|
|
7
|
Schoenmakers EF, Wanschura S, Mols R,
Bullerdiek J, Van den Berghe H and Van de Ven WJ: Recurrent
rearrangements in the high mobility group protein gene, HMGI-C, in
benign mesenchymal tumours. Nat Genet. 10:436–444. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Mandahl N: Methods in solid tumors
cytogenetics. Human Cytogenetics: Malignancies and Acquired
Abnormalities. Rooney D: Oxford University Press. (New York, NY).
165–203. 2001.
|
|
9
|
Schaffer LG, Slovak ML and Campbell LJ:
ISCN 2009: An International System for Human Cytogenetic
Nomenclature. Karger Bibliotheken Stiftung. Basel: 603–604.
2009.
|
|
10
|
Agostini A, Panagopoulos I, Andersen HK,
Johannesen LE, Davidson B, Tropé CG, Heim S and Micci F: HMGA2
expression pattern and TERT mutations in tumors of the vulva. Oncol
Rep. 33:2675–2680. 2015.PubMed/NCBI
|
|
11
|
Korbie DJ and Mattick JS: Touchdown PCR
for increased specificity and sensitivity in PCR amplification. Nat
Protoc. 3:1452–1456. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cleynen I and Van de Ven WJ: The HMGA
proteins: A myriad of functions (Review). Int J Oncol. 32:289–305.
2008.PubMed/NCBI
|
|
13
|
Chiappetta G, Avantaggiato V, Visconti R,
Fedele M, Battista S, Trapasso F, Merciai BM, Fidanza V, Giancotti
V, Santoro M, et al: High level expression of the HMGI (Y) gene
during embryonic development. Oncogene. 13:2439–2446.
1996.PubMed/NCBI
|
|
14
|
Rogalla P, Drechsler K, Frey G, Hennig Y,
Helmke B, Bonk U and Bullerdiek J: HMGI-C expression patterns in
human tissues. Implications for the genesis of frequent mesenchymal
tumors. Am J Pathol. 149:775–779. 1996.PubMed/NCBI
|
|
15
|
Fedele M and Fusco A: HMGA and cancer
(Review). Biochim Biophys Acta. 1799:48–54. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Morishita A, Zaidi MR, Mitoro A,
Sankarasharma D, Szabolcs M, Okada Y, D'Armiento J and Chada K:
HMGA2 is a driver of tumor metastasis. Cancer Res. 73:4289–4299.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Geurts JM, Schoenmakers EF and Van de Ven
WJ: Molecular characterization of a complex chromosomal
rearrangement in a pleomorphic salivary gland adenoma involving the
3′-UTR of HMGIC. Cancer Genet Cytogenet. 95:198–205. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mine N, Kurose K, Nagai H, Doi D, Ota Y,
Yoneyama K, Konishi H, Araki T and Emi M: Gene fusion involving
HMGIC is a frequent aberration in uterine leiomyomas. J Hum Genet.
46:408–412. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kazmierczak B, Meyer-Bolte K, Tran KH,
Wöckel W, Breightman I, Rosigkeit J, Bartnitzke S and Bullerdiek J:
A high frequency of tumors with rearrangements of genes of the HMGI
(Y) family in a series of 191 pulmonary chondroid hamartomas. Genes
Chromosomes Cancer. 26:125–133. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kolenda T, Przybyla W, Teresiak A,
Mackiewicz A and Lamperska KM: The mystery of let-7d-a small RNA
with great power. Contemp Oncol (Pozn). 18:293–301. 2014.PubMed/NCBI
|
|
21
|
Ayarpadikannan S and Kim HS: The impact of
transposable elements in genome evolution and genetic instability
and their implications in various diseases. Genomics Inform.
12:98–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ayarpadikannan S, Lee HE, Han K and Kim
HS: Transposable element-driven transcript diversification and its
relevance to genetic disorders. Gene. 558:187–194. 2015. View Article : Google Scholar : PubMed/NCBI
|