Introduction
Squamous cell carcinoma (SCC) of the oral tongue is
one of the most common types of head and neck cancer (1,2). The
incidence peaks in the sixth decade of life (1), and is clearly associated with ethylism
and smoking (3). Previous studies
suggest that, although the overall incidence of head and neck
cancers is decreasing in the USA, the incidence of oral tongue
carcinoma specifically is increasing in young patients (2,4–7), particularly in white women (8). The factors responsible for this shift
remain unclear. Unlike oropharyngeal cancer (malignancy of the
tonsils or the base of tongue), whose rise in young patients has
been attributed to malignant transformation subsequent to human
papilloma virus infection (9,10), no such association has been observed
for oral tongue SCC.
The prognostic significance of age at diagnosis of
oral tongue SCC is controversial. Garavello et al (11) demonstrated that patients younger than
40 years are at higher risk of recurrence and mortality than
matched older patients. Other studies revealed that the prognostic
disadvantage of young age may be limited to individuals with
advanced, as opposed to early, disease at presentation (12) or with recurrent, as opposed to
new-onset, disease (13).
Accordingly, we previously reported that tongue carcinoma in
patients <30 years old is more advanced at presentation, with
significantly high rates of regional metastases and an increased
risk of distant failure compared with patients >30 years
(14). In addition, recurrent disease
is aggressive, with a fatality rate of 100% (14). However, other authors noticed that
clinical outcomes in young patients were similar to (15–18) or
even better than (19–21) those in older patients.
MicroRNAs (miRNAs or miRs) are short, non-coding RNA
molecules that regulate gene expression by controlling messenger
(m)RNA translation (22). They are
considered to play a role in normal biological processes, from cell
growth to cell differentiation and death (22). miRNAs are dysregulated in various
malignancies, and can act as both oncogenes and tumor suppressors
(22,23). In the head and neck, specific patterns
of miRNA expression have been detected in cell lines and tumoral
tissues from the hypopharynx, oral cavity, larynx, nasopharynx and
salivary glands (23). Specific
miRNAs that have been implicated in oral SCC cell lines and tumoral
tissues include miR-211 (22),
miR-184 (24), miR-137, miR-193a
(25), miR-133a, miR-133b (26), miR-98 (27), Homo sapiens (hsa)-miR-15a (28), miR-126 (29) and miR-155 (30). Certain patterns of miRNA expression
can also serve as prognostic markers. For example, high expression
of miR-211 (22) and miR-21 (31,32) was
associated with poor prognosis in patients with oral carcinoma.
The aim of the present study was to identify
specific miRNAs that are dysregulated in SCC of the oral tongue in
young patients and to determine if these aberrations resemble known
miRNA profiles in older patients.
Materials and methods
Study design and patient
characteristics
The present study was approved by the Institutional
Review Board of Rabin Medical Center (Petah Tikva, Israel), with
waiver of informed consent.
The study group included 12 patients aged ≤30 years
with SCC of the oral tongue who underwent partial glossectomy with
neck dissection at Rabin Medical Center between January 1996 and
December 2013. The mean age of the patients was 22.5 years (range,
15–30 years), and 7 (58%) patients were females. Two patients were
smokers (1 and 6 packs/year, respectively), and none consumed
alcohol. The mean follow-up time was 56 months.
At the time of diagnosis, 10 patients had advanced
disease (stage 3–4), 9 had regional lymph node involvement and none
had systemic metastases. All patients with advanced disease were
also treated with radiotherapy, and 8 received adjuvant
chemotherapy.
Five patients had a recurrence of the disease. The
mean time from diagnosis to recurrence was 11.4 months (range, 5–16
months). All patients succumbed to the disease within a mean time
of 18.4 months (range, 14–28 months). These patients were
categorized as having aggressive disease, and the other 7 patients,
without recurrence, were categorized as having non-aggressive
disease. The mean follow-up time of the group with non-aggressive
disease was 83 months (range, 24–132 months).
Sample collection
All tumors were diagnosed following partial
glossectomy. For the present study, all available hematoxylin and
eosin-stained slides were reviewed by a head and neck pathologist,
and paraffin-embedded blocks of the most representative slides were
used for RNA extraction. Samples of tumoral tissue and peritumoral
normal tongue tissue were collected for each patient.
Total RNA was extracted from formalin-fixed,
paraffin-embedded samples using RecoverAll Total Nucleic Acid
Isolation kit, according to the manufacturer's protocol (Ambion,
Life Technologies; Thermo Fisher Scientific, Inc., Waltham, MA,
USA), and further purified with phenol-chloroform, followed by
ethanol precipitation. The final RNA concentration and purity were
measured with an ND-1000 spectrophotometer (NanoDrop Technologies;
Thermo Fisher Scientific, Inc., Wilmington, DE, USA).
miRNA profiling
In total, >800 human miRNAs were profiled with
the multiplexed nCounter® miRNA Expression Assay kit
(NanoString Technologies, Seattle, WA, USA), according to the
manufacturer's protocol. Briefly, 100 ng of total RNA was used as
input material with 3 µl of 3-fold-diluted sample. A specific DNA
tag was ligated onto the 3′ end of each mature miRNA for exclusive
identification for each of the miRNA species present in the sample.
Tagging was performed in a multiplexed ligation reaction utilizing
reverse complementary bridge oligonucleotides (NanoString
Technologies). All hybridization reaction mixtures were incubated
at 64°C for 18 h. Excess tags were then removed, and the resulting
material was hybridized with a panel of fluorescently labeled,
bar-coded reporter probes, which were specific to the miRNA of
interest. miRNAs abundance was quantified with the
nCounter® Prep Station (NanoString Technologies) by
counting the individual fluorescent barcodes and identifying the
target miRNA molecules present in each sample. Each sample was
normalized to the geometric mean of the top 100 highest expressed
miRNAs. The mean value of the negative controls was set as the
lower threshold for each sample; thus, when ≥50% of the value was
equal to or lower than the lower threshold, the miRNA was excluded.
Using these criteria, 93 miRNAs were identified to be eligible for
subsequent analyses.
Quantitative polymerase chain reaction
(qPCR)
qPCR was performed to validate the top candidates
obtained with the nCounter® miRNA Expression Assay kit,
according to the manufacturer's protocol (NanoString Technologies).
The TaqMan Universal PCR Master Mix, Nno AmpErase UNG (Applied
Biosystems; Thermo Fisher Scientific, Inc.) was used to test miRNA
expression. PCR amplification and reading were performed with the
StepOne™ Real-Time PCR System (Life Technologies; Thermo Fisher
Scientific, Inc.) under the following conditions: 2 min at 50°C and
10 min at 95°C, followed by 40 cycles of 95°C for 30 sec and 60°C
for 1 min. Expression values were calculated using the comparative
threshold cycle method (33), with
normalization according to the total cellular RNA content.
Statistical analysis
Following data pre-processing and normalization,
differential expression analysis was performed using nSolver™
Analysis Software (version 2.0; NanoString Technologies) and
EXpression Analyzer and DisplayER version 6.06
(//acgt.cs.tau.ac.il/expander/). Student's t-test was used
for statistical analysis. P<0.05 was considered to indicate a
statistically significant difference. P-values were adjusted
according to the false-discovery rate (FDR).
Results
Comparisons
A total of 24 samples were analyzed, one from tumor
and one from adjacent healthy tissue, for each patient. The miRNA
results were compared between tumor and normal tissues, and between
aggressive and non-aggressive tumors. No significant differences
were identified in demographic characteristics or tumor stage
between patients with aggressive and non-aggressive tumors (P=0.87
and P=0.47, respectively). Table I
contains the significant differences in miRNA expression between
the groups.
 | Table I.miRNA expression by the studied
groups. |
Table I.
miRNA expression by the studied
groups.
| Comparison | miRNA | Fold-change | P-value |
|---|
| Tumors vs. healthy
tissue |
|
|
|
|
Upregulation | let-7f-5p | 1.54 | 0.015a |
|
| miR-30b-5p | 1.49 | 0.039 |
|
| let-7e-5p | 1.27 | 0.040 |
|
| miR-26a-5p | 1.51 | 0.086 |
|
Downregulation | miR-185-5p | 0.78 | 0.060 |
| Aggressive tumors
vs. non-aggressive tumors |
|
|
|
|
Upregulation | let-7c | 1.85 | 0.005a |
|
| miR-130a-3p | 1.39 | 0.008 |
|
| miR-361-5p | 1.47 | 0.021 |
|
| let-7d-5p | 1.45 | 0.022 |
|
| miR-99a-5p | 1.67 | 0.025 |
|
| miR-29c-3p | 2.29 | 0.043 |
|
| let-7f-5p | 1.49 | 0.051 |
|
Downregulation | miR-181a-5p | 0.46 | 0.069 |
|
| let-7i-5p | 0.67 | 0.075 |
| Normal tissue
adjacent to aggressive vs. non-aggressive tumors |
|
|
|
|
Upregulation | miR-107 | 1.86 | 0.010a |
|
| let-7f-5p | 1.72 | 0.016 |
|
| miR-30b-5p | 1.40 | 0.062 |
|
Downregulation | miR-34a-5p | 0.67 | 0.015 |
Tumors vs. adjacent normal
tissues
Comparisons of the 12 tumor samples (aggressive and
non-aggressive combined) with the 12 normal-tissue samples revealed
an upregulation of let-7f-5p, miR-30b-5p and let-7e-5p in the
tumors (P<0.05); the difference for let-7f-5p remained
significant following FDR correction. Stratification by disease
aggressiveness yielded an upregulation of let-7f-5p in
non-aggressive tumors and of let-7e-5p in aggressive tumors,
compared with the corresponding healthy-tissue samples.
Aggressive vs. non-aggressive
tumors
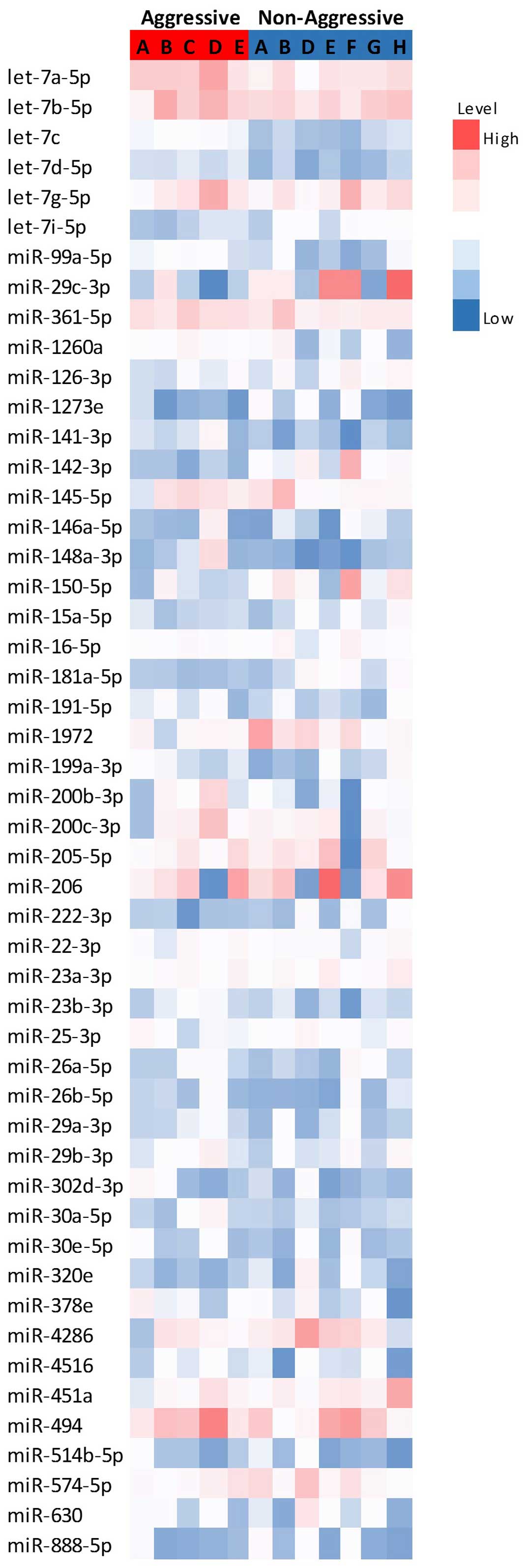
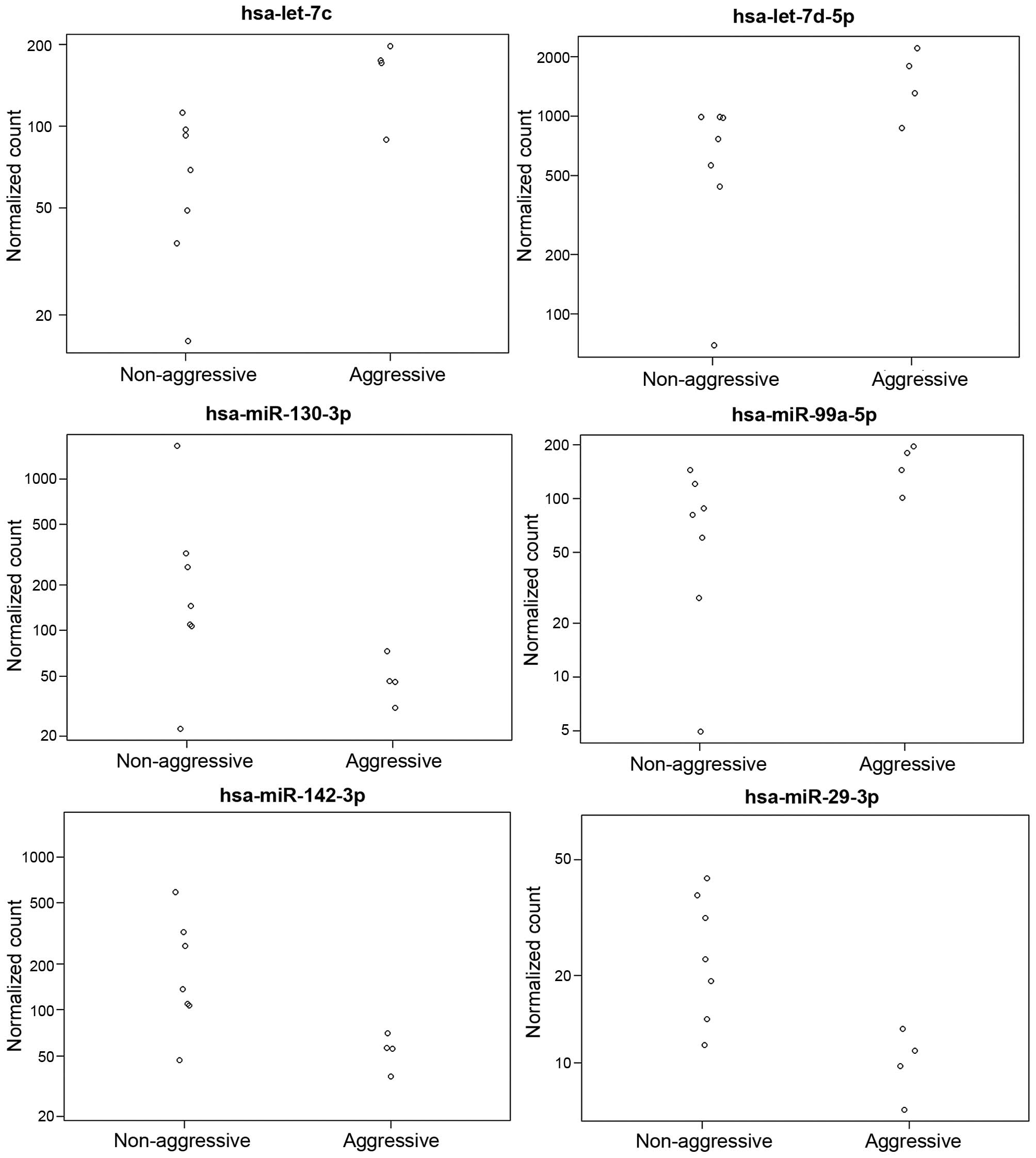
Fig. 1 represents the
differential expression of the miRNAs with the highest expression
levels, while Fig. 2 represents the
expression of the significantly altered miRNAs in aggressive vs.
non-aggressive tumors. There was a significant upregulation of
let-7c, miR-130a-3p, miR-361-5p, miR-99a-5p, miR-29c-3p and
let-7d-5p in the aggressive tumors (P<0.05); the difference for
let-7c remained significant after FDR correction. Additionally, the
aggressive tumors exhibited downregulation of miR-181a-5p and
let-7i-5p (P=0.06).
Normal tissues adjacent to aggressive
vs. non-aggressive tumors
Normal tissues adjacent to aggressive tumors
exhibited an upregulation of miR-107 and let-7f-5p (P<0.05); the
difference for miR-107 remained significant after FDR correction.
Normal tissues adjacent to non-aggressive tumors displayed a
downregulation of miR-34a-5p (P<0.05).
NanoString counts were validated by qPCR (Fig. 2). A total of 4 samples were tested for
let-7c expression and 4 were evaluated for miR-29c expression. A
correlation of 0.93 was observed.
Discussion
The rising incidence of oral tongue SCC among young
patients, combined with its weak association with known risk
factors of other head and neck cancers (including smoking, ethylism
and human papilloma virus infection) and its aggressiveness, have
prompted researchers to seek molecular aberrations specific to the
tumor in this age group (2,4–7). A high
incidence of DNA ploidy abnormalities and microsatellite
instability have been reported in young patients with tongue
carcinoma (34,35). Molecular markers observed to be
similar in young and old patients include p53, p21, retinoblastoma
protein and mouse double minute 2 homolog expression; presence of
mismatch repair genes such as human mutL homolog 1 and human mutS
homolog 2; loss of heterozygosity at the 3, 9 and 17p regions; and
high cell proliferation and angiogenesis index (34,36–39).
Numerous studies have reported that miRNA expression
differs between cancer cells and healthy cells (22,28,40–45),
although the exact mechanism by which each miRNA affects
carcinogenesis is unclear. The present study sought, for the first
time, to examine miRNA expression in oral cavity SCC in patients
<30 years old. Comparisons of the tumor tissue with adjacent
normal tissue revealed a significant upregulation of let-7f in
tumor tissues, which remained significant after FDR correction.
This increase was attributed mainly to overexpression of let-7f in
non-aggressive tumors and overexpression of let-7e in aggressive
tumors compared with normal tissues.
The tumoral upregulation of let-7f is in line with
previous reports in older patients. Christensen et al
(46) and Hui et al (40) demonstrated overexpression of let-7f in
oropharyngeal cancer compared with oral cavity cancer and benign
tonsillar tissue, and Cui et al (28) demonstrated overexpression of let-7f in
oral cavity cancer compared with normal tissue. Tran et al
(23) reported similar results in
cell lines of head and neck SCC.
Given the aggressiveness of recurrent oral cavity
cancer in young patients with a high mortality rate and increased
risk of distant metastases (14), the
present study analyzed molecular markers separately in aggressive
and non-aggressive tumors. The results revealed an overexpression
of let-7e in aggressive tumors compared with adjacent normal
tissues (paired analysis) that was not evident in non-aggressive
tumors. No dysregulation of let-7e was observed in any of the other
studies of oral cavity cancer or other head and neck cancers,
possibly suggesting a unique biology of aggressive tongue SCC in
young patients.
A second (non-paired) evaluation of aggressive vs.
non-aggressive tumors demonstrated an upregulation of let-7c,
let-7d-5p, miR-99a-5p, miR-29c-3p, miR-130a-3p and miR-361-5p in
the aggressive tumors, with let-7c remaining significant after FDR
correction. Previous reports of miRNA dysregulation in oral cavity
cancer that used normal tissue as the control demonstrated no
aberration in let-7c and downregulation of let-7 d in tumors
(28,41–43). By
contrast, in the present study, both markers were upregulated.
Additionally, our findings for mir-99 differed from a previous
study, in which forced expression of miR-99 family members in head
and neck SCC cell lines led to a reduction in cell proliferation
and cell migration, and an increase in apoptosis (47). In addition, mir-29 was downregulated
in that study (47) and in another
study of nasopharyngeal carcinoma associated with miR-29c
expression with increased sensitivity to cisplatin and inhibition
of invasion and metastatic spread (48), in contrast to the present
findings.
The literature on the role of miR-130a is
inconsistent. Studies of ovarian and hepatocellular carcinoma
tumors and head and neck SCC cell lines reported an association
between miR-130a downregulation with reduced response/resistance to
cisplatin (49) and docetaxel
(50), respectively, and conversely,
an association between miR-130a upregulation and increased
cisplatin resistance (51). In liver
cancer, downregulation of miR-130a was associated with poor
prognosis (52). However, studies of
cervical and esophageal cancer noticed that miR-130a upregulation
was associated with tumor development and poor prognosis (49,53), in
line with our results in tumor vs. healthy cells. Similarly, in a
study of 107 samples of oral cavity SCC, the copy number of mir-30b
was frequently increased in advanced tumors, in correlation with
hsa-miR-30b overexpression (54).
Previous studies evaluating let-7 expression in head
and neck malignancies are summarized in Table II. The let-7 family of miRNAs are
commonly considered tumor suppressors. They were originally
discovered in the roundworm Caenorhabditis elegans, a
popular model in biology, where they were observed to be
responsible for the development to the adult stage (55). let-7 miRNAs are dysregulated in lung,
colorectal and gastric cancers (56–58). In
lung cancer, reduced expression of let-7a was significantly
associated with shortened postoperative survival, while its
overexpression was significantly associated with inhibition of
cancer cell growth (59). let-7
expression has also been associated with disease severity and
survival in patients with head and neck cancer (56,60).
 | Table II.let-7 dysregulation in head and neck
carcinoma. |
Table II.
let-7 dysregulation in head and neck
carcinoma.
| Study, author
(ref) | Year | Collected
samples | Tissue
examined | n | Control | n | let-7 | Expression | Age,
yearsa |
|---|
| Chang et al
(22) | 2008 | Primary tumors | HNSCC | 4 | Healthy tissue
(UPPP patients) | 4 | let-7i | Upregulated | 54 |
| Tran et al
(23) | 2010 | Cell lines | HNSCC | 9 | NA |
| let-7a | Upregulated | NA |
|
|
|
|
|
|
|
| let-7b | Upregulated |
|
|
|
|
|
|
|
|
| let-7c | Upregulated |
|
|
|
|
|
|
|
|
| let-7d | Upregulated |
|
|
|
|
|
|
|
|
| let-7f | Upregulated |
|
| Cui et al
(28) | 2014 | NA | OCSCC | 17 | Healthy
controls | 3 | let-7a | Upregulated | NA |
|
|
|
|
|
|
|
| let-7f | Upregulated |
|
| Christensen et
al (46) | 2009 | Whole blood and
buccal cells | OCSCC | 10 | Pharyngeal
cancer | 4 | let-7a | Downregulated | NA |
|
|
|
|
|
|
|
| let-7c | Downregulated |
|
|
|
|
|
|
|
|
| let-7d | Downregulated |
|
|
|
|
|
|
|
|
| let-7f | Downregulated |
|
| Hui et al
(40) | 2013 | FFPE | Oropharyngeal
SCC | 88 | Tonsillectomy | 7 | let-7f | Upregulated | 55±11 |
|
|
|
|
|
|
|
| let-7g | Upregulated |
|
| Ricieri Brito et
al (41) | 2010 | Primary tumors and
whole blood | OCSCC | 20 |
Self-control-healthy tissue |
| let-7a | Downregulated
(n=18) | 57 (37–90) |
|
|
|
|
|
|
|
| let-7a | Upregulated
(n=13) |
|
| Chang et al
(42) | 2011 | Cell lines | OCSCC | NA | Human normal
keratinocyte cells | NA | let-7d | Downregulated | NA |
| Maclellan et
al (43) | 2012 | Serum samples | CIS+OCSCC | 30 | Non-cancer patients
serum | 26 | let-7a | Upregulated | 50–93 |
|
|
|
|
|
|
|
| let-7b | Upregulated |
|
|
|
|
|
|
|
|
| let7-d | Downregulated |
|
| Jakymiw et
al (44) | 2010 | Cell lines | HNSCC | 6 | Healthy gingival
cells | NA | let-7b | Downregulated | NA |
| Yu et al
(45) | 2011 | Primary tumors | Head and neck
cancer metastases | 107 |
Self-control-healthy tissue |
| let-7a | Downregulated | NA |
|
|
|
|
| 50 | No metastases | 57 | let-7a | Downregulated |
|
| Current study | 2015 | Primary tumors | OCSCC | 12 |
Self-control-healthy tissue | 12 | let-7f | Upregulated | 15–30 |
|
|
|
|
|
|
| let-7e | Upregulated |
|
|
|
|
| Aggressive | 5 | Non aggressive | 7 | let-7c | Upregulated |
|
|
|
|
| OCSCC |
| OCSCC |
| let-7d | Upregulated |
|
Jakymiw et al (44) hypothesized that the let-7 family
affects cancer biology by regulating the expression of Dicer, an
RNase III-type enzyme present in almost all eukaryotes. Dicer is
essential for RNA interference and miRNA pathway maturation
(61). Dicer was observed to be
overexpressed in prostate and lung adenocarcinomas (62,63) and
cervical cancers (53). Its
overexpression (by 24-fold) in cell lines of head and neck cancer
was associated with significantly reduced levels of let-7b. When
the cancer cell lines were transfected with chemically synthesized
let-7b, cell proliferation was inhibited by ≤83% (44).
Findings of reduced let-7 levels in the presence of
increased expression of the RAS protein (59) have led to the hypothesis that let-7
may inhibit cancer growth via its effect on the RAS protein.
Accordingly, in head and neck SCC, the presence of the RAS protein
was significantly associated with poor prognosis and increased
tumor size (46,64). The inhibitory effect of let-7a in
cancer development was further supported by findings of decreased
let-7a levels in head and neck cancer and lymph node metastases
(45).
Previous studies were inconsistent regarding the
expression profiles of let-7a, let-7b, let-7c, let-7d and let-7f,
with certain studies reporting overexpression while others
reporting reduced expression in cancer (22–23,28,40–46).
While the reduced expression of these miRNAs has provided a
theoretical explanation to the mechanism by which cancer cells
affect the RAS gene and protein, Dicer and HNC-TICS, no evidence
has supported the upregulation of the let-7 family, which may
explain the observed cell proliferation in head and neck
cancers.
The majority of relevant studies conducted to date
made no reference to patient age as a risk/contributory factor. Yu
et al (45) compared miRNA
expression in patients under and over the age of 50 years, and
observed no difference between the groups.
The present study is affected by certain
limitations. Oral cavity cancer in patients younger than 30 years
of age is very uncommon (1,2,4–7). Consequently, our samples are small and
the statistical power is limited. The present study evaluated miRNA
profiles in young patients and compared the results with published
data, rather than comparing them directly with samples from older
patients. Thus, methodological differences between the current
study and previous studies may affect our assumptions.
To conclude, the current study presented the miRNA
profile of oral cavity SCC in patients younger than 30 years of
age. Our finding of overexpression of let-7f in oral cavity cancer
in young patients compared with adjacent normal tissue is in line
with previous studies in older patients. However, further
stratification of the tumors by aggressiveness revealed that let-7f
was overexpressed in non-aggressive tumors, whereas the aggressive
tumors had a distinctive miRNA expression pattern, with
upregulation of let-7e (compared with normal tissue) as well as
let-7c and let-7d (compared with non-aggressive tumors). These
findings may aid the prognosis of oral cavity tumors in young
patients, which may be very aggressive and exhibit a high mortality
rate. Further studies with direct comparison with tissues from
older patients are warranted to validate the prognostic role of
these miRNA aberrations in oral cavity SCC.
Acknowledgements
The present study was supported by a research grant
from the Schauder Foundation, Sackler School of Medicine, Tel Aviv
University (Tel Aviv, Israel; grant no. 32003156000). The present
study was based on an abstract presented at The Annual Meeting of
The Israeli Society of Otolaryngology, Head and Neck Surgery on
March 9–12, 2016 in Eilat, Israel (available at www.entisrael.com/wp
content/uploads/2016/01/ENT2016_BookProgram.pdf).
References
|
1
|
Horner MJ, Ries LAG, Krapcho M, Neyman N,
Aminou R, Howlader N, et al: SEER Cancer Statistics Review,
1975–2006. National Cancer Institute; Bethesda, MD: simpleseer.cancer.gov/csr/1975_2006/based on November
2008 SEER data submission, posted to the SEER web site, 2009.
Accessed on June 27, 2016.
|
|
2
|
CancerResearch UK: Oral cancer - UK
incidence statistics. https://.cancerresearchuk.org/health-professional/cancer-statistics/statistics-by-cancer-type/oral-cancer/incidenceAccessed
on. June 27–2016
|
|
3
|
Mashberg A, Boffetta P, Winkelman R and
Garfinkel L: Tobacco smoking, alcohol drinking, and cancer of the
oral cavity and oropharynx among U.S. veterans. Cancer.
72:1369–1375. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Myers JN, Elkins T, Roberts D and Byers
RM: Squamous cell carcinoma of the tongue in young adults:
Increasing incidence and factors that predict treatment outcomes.
Otolaryngol Head Neck Surg. 122:44–51. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Annertz K, Anderson H, Biörklund A, Möller
T, Kantola S, Mork J, Olsen JH and Wennerberg J: Incidence and
survival of squamous cell carcinoma of the tongue in Scandinavia,
with special reference to young adults. Int J Cancer. 101:95–99.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shiboski CH, Schmidt BL and Jordan RC:
Tongue and tonsil carcinoma: Increasing trends in the U.S.
population ages 20–44 years. Cancer. 103:1843–1849. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Brown LM, Check DP and Devesa SS: Oral
cavity and pharynx cancer incidence trends by subsite in the United
States: Changing gender patterns. J Oncol. 2012:6494982012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Patel SC, Carpenter WR, Tyree S, Couch ME,
Weissler M, Hackman T, Hayes DN, Shores C and Chera BS: Increasing
incidence of oral tongue squamous cell carcinoma in young white
women, age 18 to 44 years. J Clin Oncol. 29:1488–1494. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mork J, Lie AK, Glattre E, Hallmans G,
Jellum E, Koskela P, Møller B, Pukkala E, Schiller JT, Youngman L,
et al: Human papillomavirus infection is a risk factor for
squamous-cell carcinoma of the head and neck. N Engl J Med.
344:1125–1131. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pintos J, Black MJ, Sadeghi N, Ghadirian
P, Zeitouni AG, Viscidi RP, Herrero R, Coutlée F and Franco EL:
Human papillomavirus infection and oral cancer: A case-control
study in Montreal, Canada. Oral Oncol. 44:242–250. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Garavello W, Spreafico R and Gaini RM:
Oral tongue cancer in young patients: A matched analysis. Oral
Oncol. 43:894–897. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Park JO, Sun DI, Cho KJ, Joo YH, Yoo HJ
and Kim MS: Clinical outcome of squamous cell carcinoma of the
tongue in young patients: A stage-matched comparative analysis.
Clin Exp Otorhinolaryngol. 3:161–165. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mallet Y, Avalos N, Le Ridant AM, Gangloff
P, Moriniere S, Rame JP, Poissonnet G, Makeieff M, Cosmidis A,
Babin E, et al: Head and neck cancer in young people: A series of
52 SCCs of the oral tongue in patients aged 35 years or less. Acta
Otolaryngol. 129:1503–1508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hilly O, Shkedy Y, Hod R, Soudry E,
Mizrachi A, Hamzany Y, Bachar G and Shpitzer T: Carcinoma of the
oral tongue in patients younger than 30 years: Comparison with
patients older than 60 years. Oral Oncol. 49:987–990. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Friedlander PL, Schantz SP, Shaha AR, Yu G
and Shah JP: Squamous cell carcinoma of the tongue in young
patients: A matched-pair analysis. Head Neck. 20:363–368. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
SiegelmannDanieli N, Hanlon A, Ridge JA,
Padmore R, Fein DA and Langer CJ: Oral tongue cancer in patients
less than 45 years old: Institutional experience and comparison
with older patients. J Clin Oncol. 16:745–753. 1998.PubMed/NCBI
|
|
17
|
Pitman KT, Johnson JT, Wagner RL and Myers
EN: Cancer of the tongue in patients less than forty. Head Neck.
22:297–302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Popovtzer A, Shpitzer T, Bahar G, Marshak
G, Ulanovski D and Feinmesser R: Squamous cell carcinoma of the
oral tongue in young patients. Laryngoscope. 114:915–917. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Davidson BJ, Root WA and Trock BJ: Age and
survival from squamous cell carcinoma of the oral tongue. Head
Neck. 23:273–279. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lee CC, Ho HC, Chen HL, Hsiao SH, Hwang JH
and Hung SK: Squamous cell carcinoma of the oral tongue in young
patients: A matched-pair analysis. Acta Otolaryngol. 127:1214–1217.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Udeabor SE, Rana M, Wegener G, Gellrich NC
and Eckardt AM: Squamous cell carcinoma of the oral cavity and the
oropharynx in patients less than 40 years of age: A 20-year
analysis. Head Neck Oncol. 4:282012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chang KW, Liu CJ, Chu TH, Cheng HW, Hung
PS, Hu WY and Lin SC: Association between high miR-211 microRNA
expression and the poor prognosis of oral carcinoma. J Dent Res.
87:1063–1068. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tran N, O'Brien CJ, Clark J and Rose B:
Potential role of micro-RNAs in head and neck tumorigenesis. Head
Neck. 32:1099–1111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wong TS, Liu XB, Chung-Wai Ho A, Po-Wing
Yuen A, Wai-Man Ng R and Ignace Wei W: Identification of pyrovate
kinase type M2 as potential oncoprotein in squamous cell carcinoma
of tongue through microRNA profiling. Int J Cancer. 123:251–257.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hebert C, Norris K, Scheper MA, Nikitakis
N and Sauk JJ: High mobility group A2 is a target for miRNA-98 in
head and neck squamous cell carcinoma. Mol Cancer. 6:52007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cui J, Li D, Zhang W, Shen L and Xu X:
Bioinformatics analyses combined microarray identify the
deregulated microRNAs in oral cancer. Oncol Lett. 8:218–222.
2014.PubMed/NCBI
|
|
29
|
Yang X, Wu H and Ling T: Suppressive
effect of microRNA-126 on oral squamous cell carcinoma in vitro.
Mol Med Rep. 10:125–130. 2014.PubMed/NCBI
|
|
30
|
Shi LJ, Zhang CY, Zhou ZT, Ma JY, Liu Y,
Bao ZX and Jiang WW: MicroRNA-155 in oral squamous cell carcinoma:
Overexpression, localization, and prognostic potential. Head Neck.
37:970–976. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li J, Huang H, Sun L, Yang M, Pan C, Chen
W, Wu D, Lin Z, Zeng C, Yao Y, et al: MiR-21 indicates poor
prognosis in tongue squamous cell carcinoma as an apoptosis
inhibitor. Clin Cancer Res. 15:3998–4008. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hedbäck N, Jensen DH, Specht L, Fiehn AM,
Therkildsen MH, Friis-Hansen L, Dabelsteen E and von Buchwald C:
MiR-21 expression in the tumor stroma of oral squamous cell
carcinoma: An independent biomarker of disease free survival. PLoS
One. 9:e951932014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kanno J, Aisaki KI, Igarashi K, Nakatsu N,
Ono A, Kodama Y and Nagao T: "Per cell" normalization method for
mRNA measurement by quantitative PCR and microarrays. BMC Genomics.
7:642006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang Y, Irish J, MacMillan C, Brown D,
Xuan Y, Boyington C, Gullane P and Kamel-Reid S: High frequency of
microsatellite instability in young patients with head-and-neck
squamous-cell carcinoma: Lack of involvement of the mismatch repair
genes hMLH1 AND hMSH2. Int J Cancer. 93:353–360. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
SantosSilva AR, Ribeiro AC, Soubhia AM,
Miyahara GI, Carlos R, Speight PM, Hunter KD, TorresRendon A,
Vargas PA and Lopes MA: High incidences of DNA ploidy abnormalities
in tongue squamous cell carcinoma of young patients: An
international collaborative study. Histopathology. 58:1127–1135.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Atula S, Grénman R, Laippala P and
Syrjänen S: Cancer of the tongue in patients younger than 40 years.
A distinct entity? Arch Otolaryngol Head Neck Surg. 122:1313–1319.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jin YT, Myers J, Tsai ST, Goepfert H,
Batsakis JG and el-Naggar AK: Genetic alterations in oral squamous
cell carcinoma of young adults. Oral Oncol. 35:251–256. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Regezi JA, Dekker NP, McMillan A,
RamirezAmador V, MenesesGarcia A, Ruiz-Godoy Rivera LM, Chrysomali
E and Ng IO: p53, p21, Rb, and MDM2 proteins in tongue carcinoma
from patients <35 versus >75 years. Oral Oncol. 35:379–383.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Benevenuto TG, Nonaka CF, Pinto LP and de
Souza LB: Immunohistochemical comparative analysis of cell
proliferation and angiogenic index in squamous cell carcinomas of
the tongue between young and older patients. Appl Immunohistochem
Mol Morphol. 20:291–297. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hui AB, Lin A, Xu W, Waldron L,
PerezOrdonez B, Weinreb I, Shi W, Bruce J, Huang SH, O'Sullivan B,
et al: Potentially prognostic miRNAs in HPV-associated
oropharyngeal carcinoma. Clin Cancer Res. 19:2154–2162. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
RicieriBrito JA, Gomes CC, Santos Pimenta
FJ, Barbosa AA, Prado MA, Prado VF, Gomez MV and Gomez RS: Reduced
expression of mir15a in the blood of patients with oral squamous
cell carcinoma is associated with tumor staging. Exp Ther Med.
1:217–221. 2010.PubMed/NCBI
|
|
42
|
Chang CJ, Hsu CC, Chang CH, Tsai LL, Chang
YC, Lu SW, Yu CH, Huang HS, Wang JJ, Tsai CH, et al: Let-7d
functions as novel regulator of epithelial-mesenchymal transition
and chemoresistant property in oral cancer. Oncol Rep.
26:1003–1010. 2011.PubMed/NCBI
|
|
43
|
Maclellan SA, Lawson J, Baik J, Guillaud
M, Poh CF and Garnis C: Differential expression of miRNAs in the
serum of patients with high-risk oral lesions. Cancer Med.
1:268–274. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jakymiw A, Patel RS, Deming N,
Bhattacharyya I, Shah P, Lamont RJ, Stewart CM, Cohen DM and Chan
EK: Overexpression of dicer as a result of reduced let-7 MicroRNA
levels contributes to increased cell proliferation of oral cancer
cells. Genes Chromosomes Cancer. 49:549–559. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yu CC, Chen YW, Chiou GY, Tsai LL, Huang
PI, Chang CY, Tseng LM, Chiou SH, Yen SH, Chou MY, et al: MicroRNA
let-7a represses chemoresistance and tumorigenicity in head and
neck cancer via stem-like properties ablation. Oral Oncol.
47:202–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Christensen BC, Moyer BJ, Avissar M,
Ouellet LG, Plaza SL, McClean MD, Marsit CJ and Kelsey KT: A let-7
microRNA-binding site polymorphism in the KRAS 3′UTR is associated
with reduced survival in oral cancers. Carcinogenesis.
30:1003–1007. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chen Z, Jin Y, Yu D, Wang A, Mahjabeen I,
Wang C, Liu X and Zhou X: Down-regulation of the microRNA-99 family
members in head and neck squamous cell carcinoma. Oral Oncol.
48:686–691. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhang JX, Qian D, Wang FW, Liao DZ, Wei
JH, Tong ZT, Fu J, Huang XX, Liao YJ, Deng HX, et al: MicroRNA-29c
enhances the sensitivities of human nasopharyngeal carcinoma to
cisplatin-based chemotherapy and radiotherapy. Cancer Lett.
329:91–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhang X, Huang L, Zhao Y and Tan W:
Downregulation of miR-130a contributes to cisplatin resistance in
ovarian cancer cells by targeting X-linked inhibitor of apoptosis
(XIAP) directly. Acta Biochim Biophys Sin (Shanghai). 45:995–1001.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dai Y, Xie CH, Neis JP, Fan CY, Vural E
and Spring PM: MicroRNA expression profiles of head and neck
squamous cell carcinoma with docetaxel-induced multidrug
resistance. Head Neck. 33:786–791. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu N, Shen C, Luo Y, Xia L, Xue F, Xia Q
and Zhang J: Upregulated miR-130a increases drug resistance by
regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell.
Biochem Biophys Res Commun. 425:468–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yang J, Han S, Huang W, Chen T, Liu Y, Pan
S and Li S: A meta-analysis of microRNA expression in liver cancer.
PLoS One. 9:e1145332014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
He L, Wang HY, Zhang L, Huang L, Li JD,
Xiong Y, Zhang MY, Jia WH, Yun JP, Luo RZ and Zheng M: Prognostic
significance of low DICER expression regulated by miR-130a in
cervical cancer. Cell Death Dis. 5:e12052014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Shao C, Yu Y, Yu L, Pei Y, Feng Q, Chu F,
Fang Z and Zhou Y: Amplification and up-regulation of microRNA-30b
in oral squamous cell cancers. Arch Oral Biol. 57:1012–1017. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Takamizawa J, Konishi H, Yanagizawa K,
Tomida S, Osada H, Endoh H, Harano T, Yatabe Y, Nagino M, Nimura Y,
et al: Reduced expression of the let-7 microRNAs in human lung
cancers in association with shortened postoperative survival.
Cancer Res. 64:3753–3756. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Akao Y, Nakagawa Y and Naoe T: Let-7
microRNA functions as a potential growth suppressor in human colon
cancer cells. Biol Pharm Bull. 29:903–906. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Zhang B, Pan X, Cobb GP and Anderson TA:
MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 302:1–12.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Lu J, Getz G, Miska EA, AlvarezSaavedra E,
Lamb J, Peck D, SweetCordero A, Ebert BL, Mak RH, Ferrando AA, et
al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 MicroRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Rana TM: Illuminating the silence:
Understanding the structure and function of small RNAs. Nat Rev Mol
Cell Biol. 8:23–36. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Chiosea S, Jelezcova E, Chandran U,
Acquafondata M, McHale T, Sobol RW and Dhir R: Up-regulation of
dicer, a component of the MicroRNA machinery, in prostate
adenocarcinoma. Am J Pathol. 169:1812–1820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Chiosea S, Jelezcova E, Chandran U, Luo J,
Mantha G, Sobol RW and Dacic S: Overexpression of Dicer in
precursor lesions of lung adenocarcinoma. Cancer Res. 67:2345–2350.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Perez-Ordoñez B, Beauchemin M and Jordan
RC: Molecular biology of squamous cell carcinoma of the head and
neck. J Clin Pathol. 59:445–453. 2006. View Article : Google Scholar : PubMed/NCBI
|