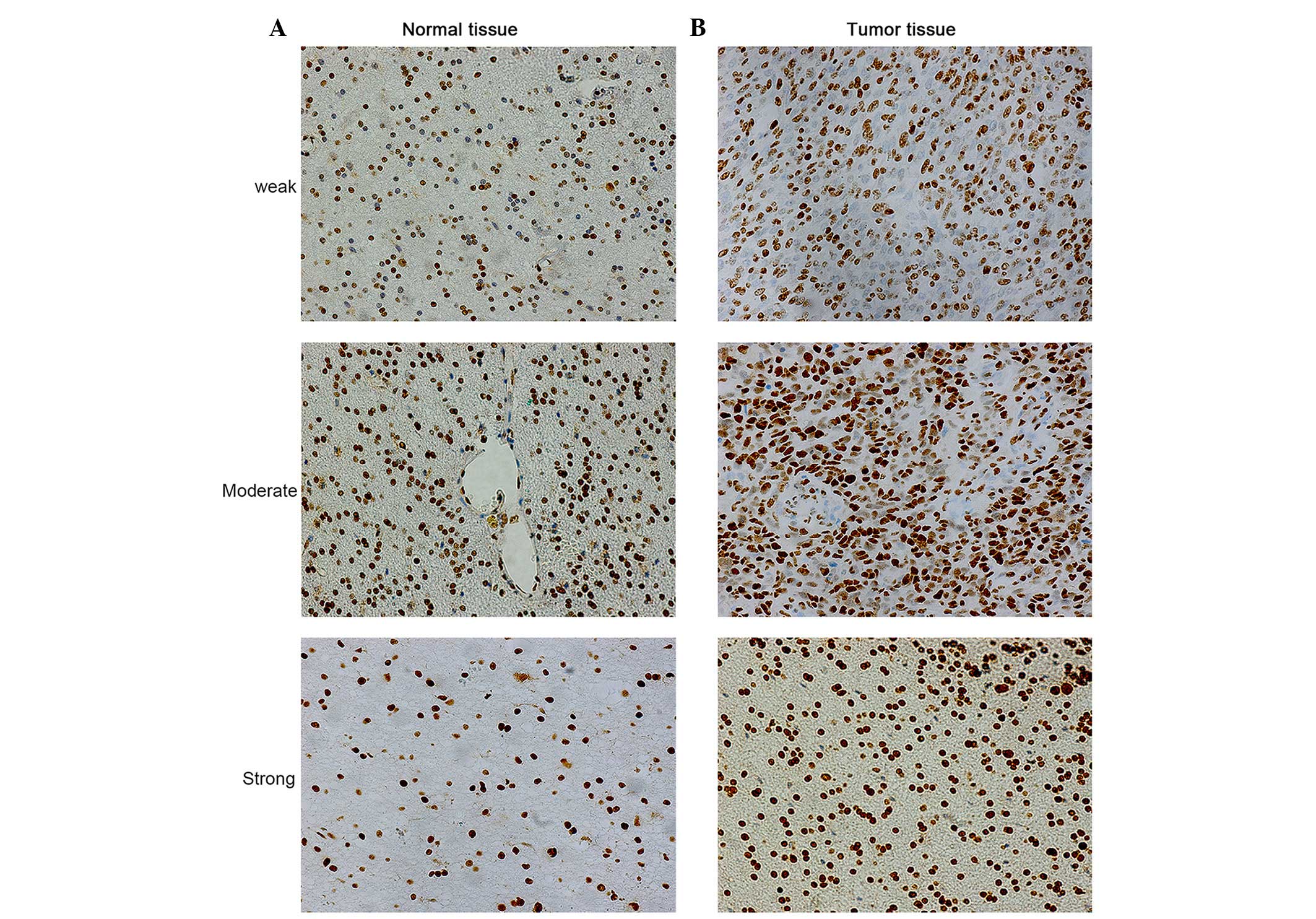
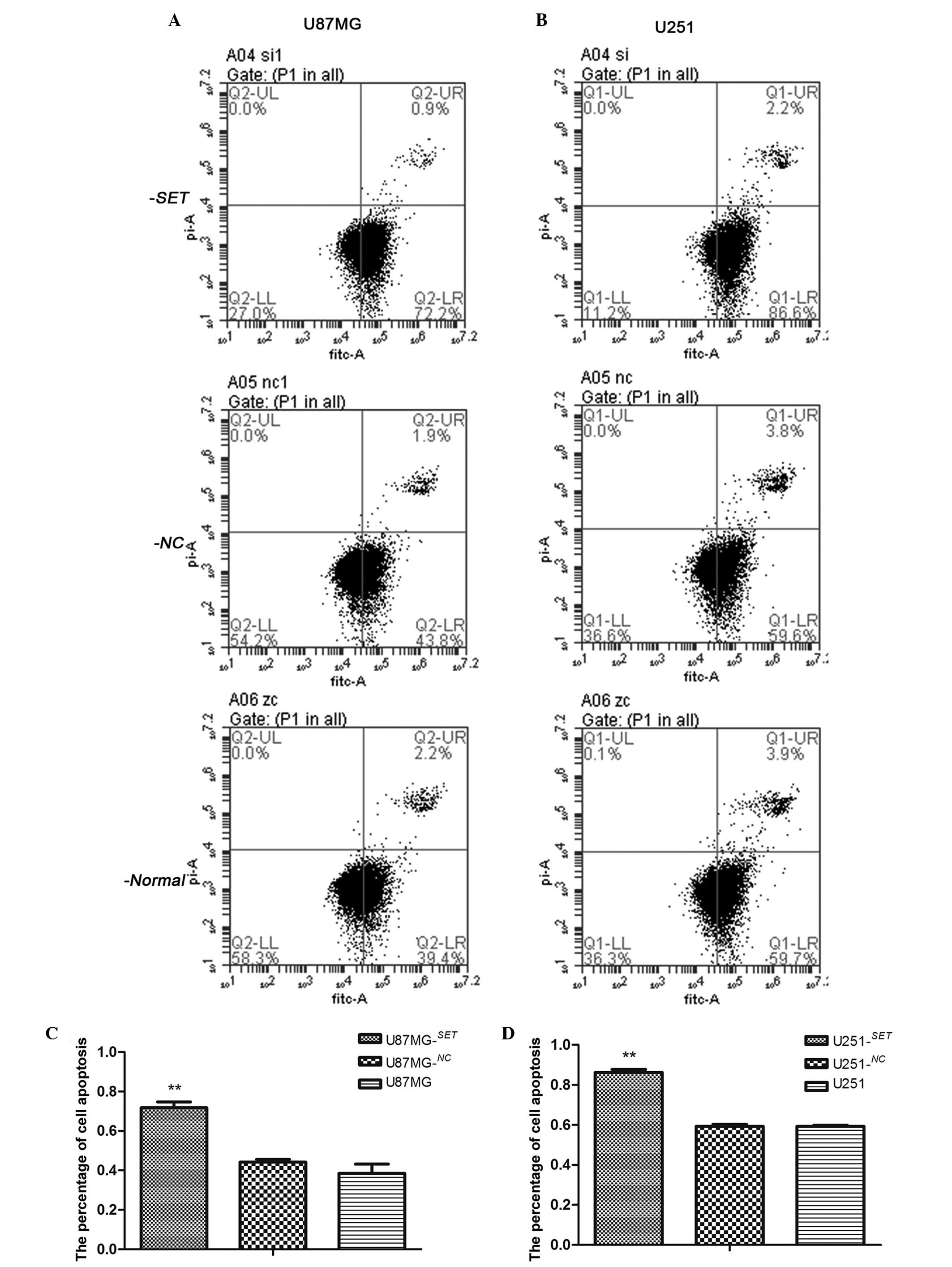
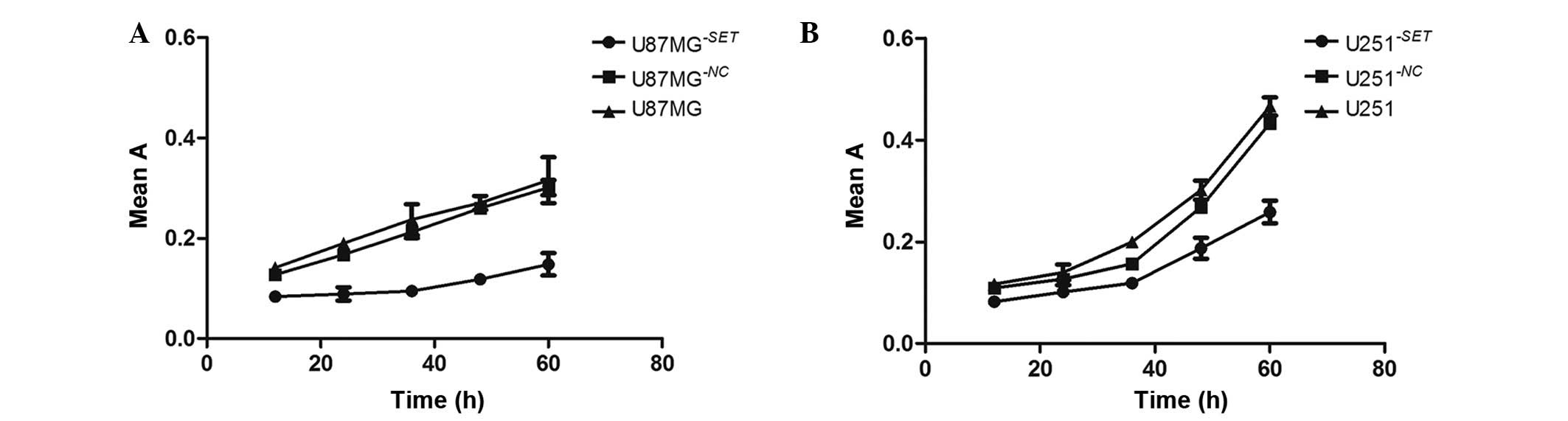
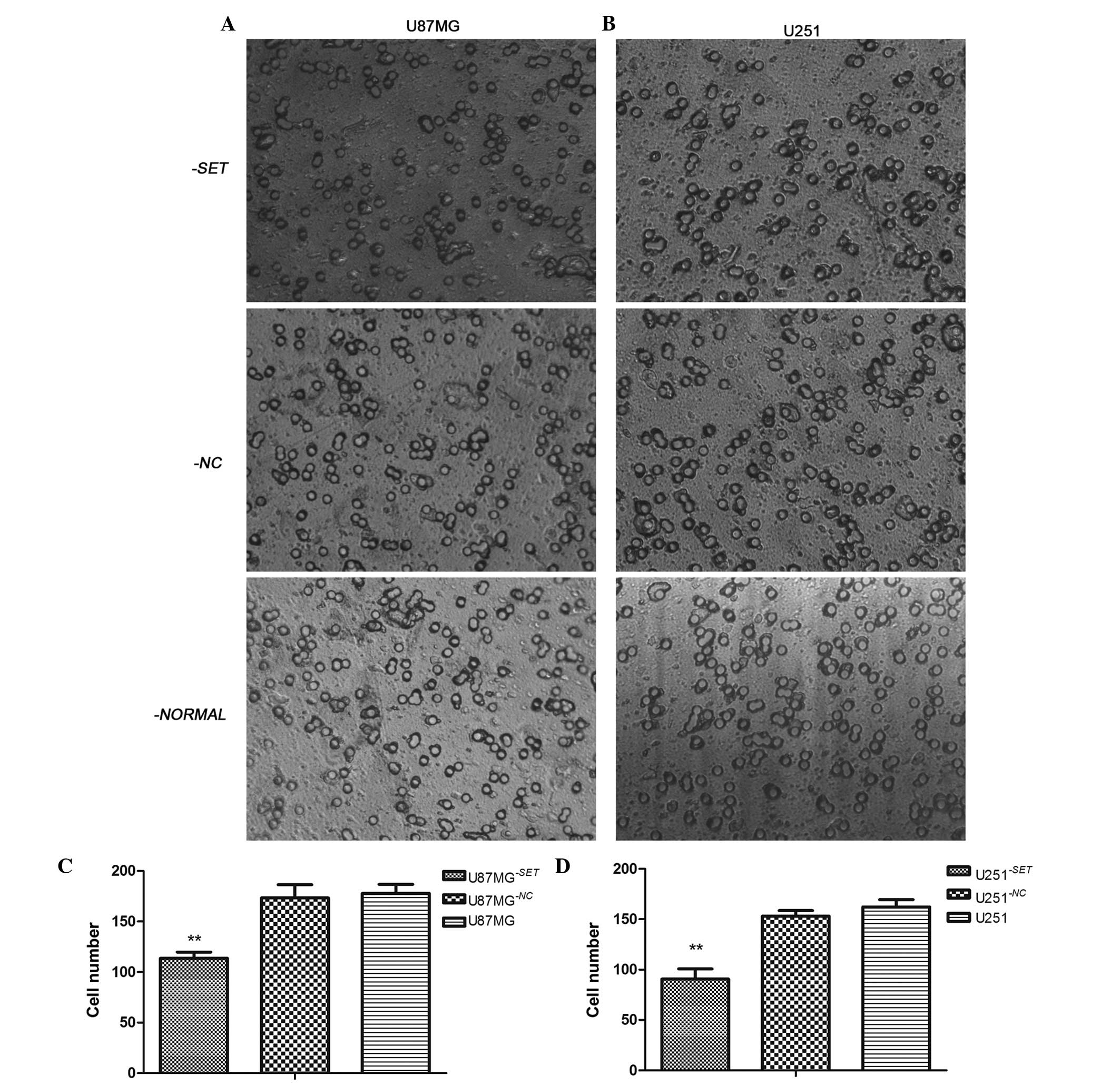
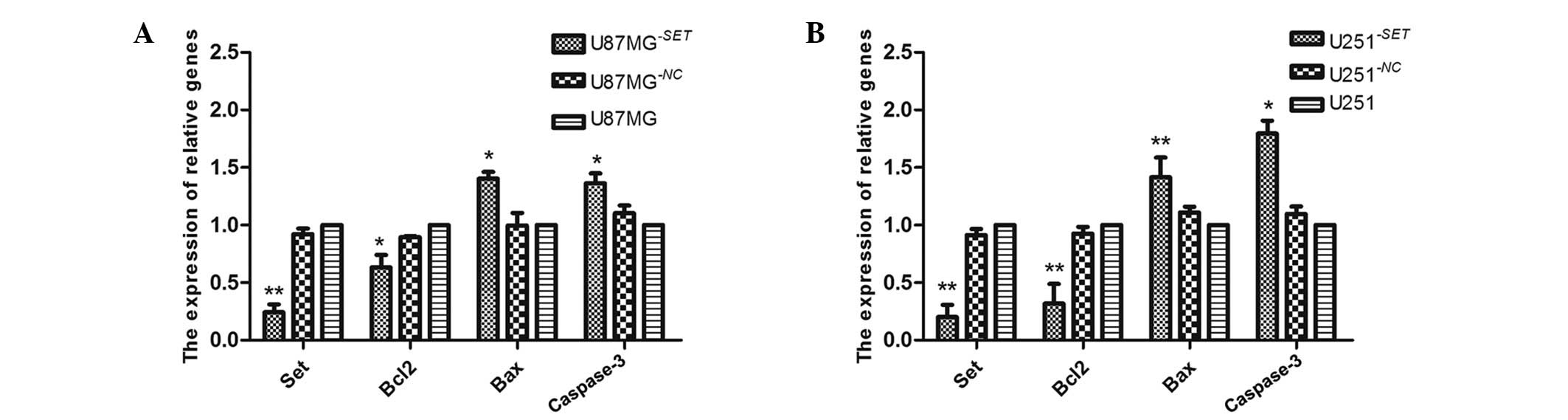
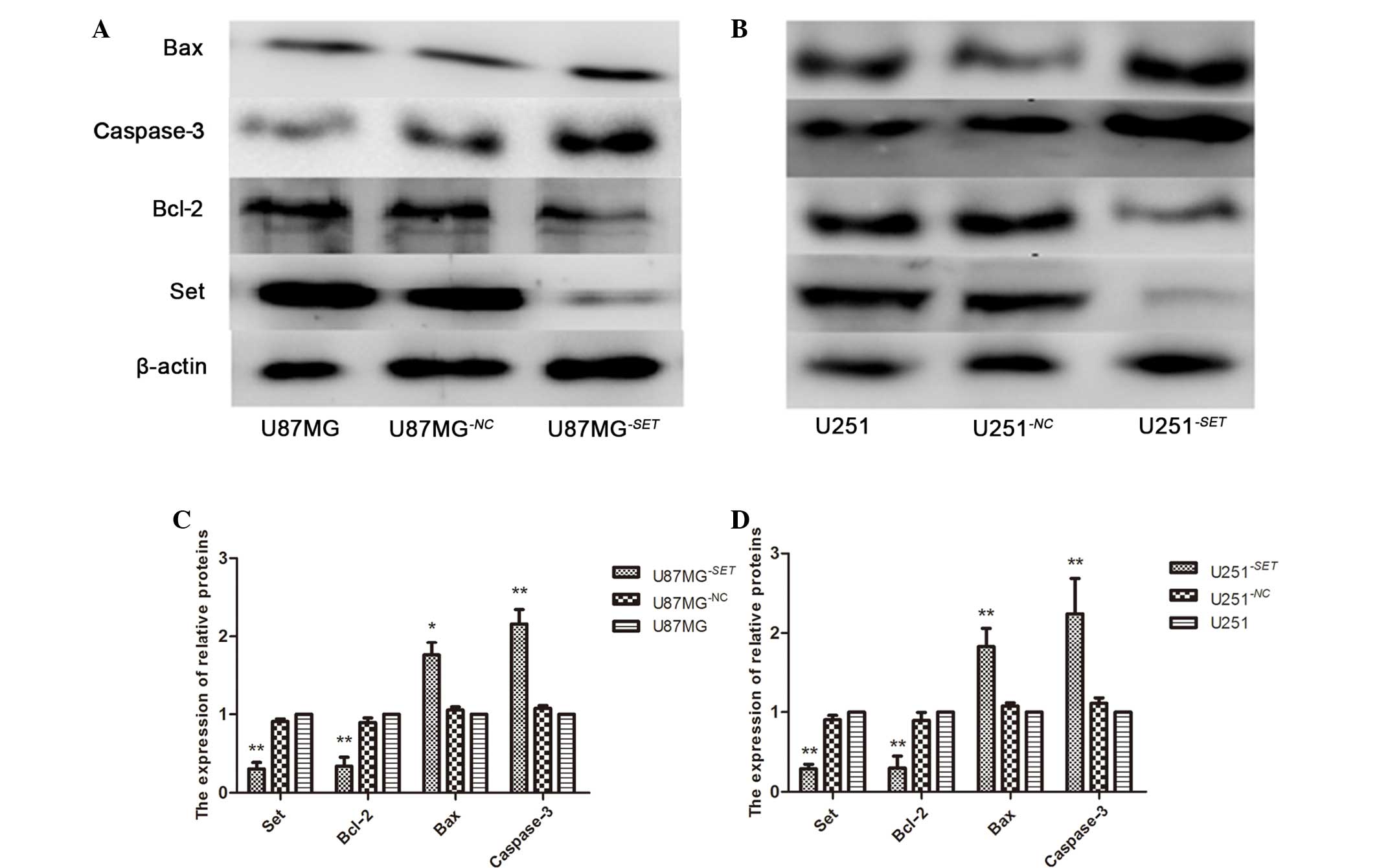
|
1
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Liu SY
and Zhou CF: Peritumoral edema shown by MRI predicts poor clinical
outcome in glioblastoma. World J Surg Oncol. 13:972015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thakkar JP, Dolecek TA, Horbinski C,
Ostrom QT, Lightner DD, BarnholtzSloan JS and Villano JL:
Epidemiologic and molecular prognostic review of glioblastoma.
Cancer Epidemiol Biomarkers Prev. 23:1985–1996. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang L, Wang H, Ding K and Xu J: FTY720
induces autophagy-related apoptosis and necroptosis in human
glioblastoma cells. Toxicol Lett. 236:43–59. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Koo S, Martin G and Toussaint LG:
MicroRNA-145 promotes the phenotype of human glioblastoma cells
selected for invasion. Anticancer Res. 35:3209–3215.
2015.PubMed/NCBI
|
|
5
|
Chakrabarti M and Ray SK: Direct
transfection of miR-137 mimics is more effective than DNA
demethylation of miR-137 promoter to augment anti-tumor mechanisms
of delphinidin in human glioblastoma U87MG and LN18 cells. Gene.
573:141–152. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang G, Wang J, Zhao H, Wang J and Tony To
SS: The role of Myc and let-7a in glioblastoma, glucose metabolism
and response to therapy. Arch Biochem Biophys. 580:84–92. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cimino PJ, Bredemeyer A, Abel HJ and
Duncavage EJ: A wide spectrum of EGFR mutations in glioblastoma is
detected by a single clinical oncology targeted next-generation
sequencing panel. Exp Mol Pathol. 98:568–573. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yan J, Kong LY, Hu J, Gabrusiewicz K,
Dibra D, Xia X, Heimberger AB and Li S: FGL2 as a multimodality
regulator of Tumor-mediated immune suppression and therapeutic
target in gliomas. J Natl Cancer Inst. 107:pii.djv1372015.
View Article : Google Scholar
|
|
9
|
Karetsou Z, Emmanouilidou A, Sanidas I,
Liokatis S, Nikolakaki E, Politou AS and Papamarcaki T:
Identification of distinct SET/TAF-Ibeta domains required for core
histone binding and quantitative characterisation of the
interaction. BMC Biochem. 10:102009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Irie A, Harada K, Araki N and Nishimura Y:
Phosphorylation of SET protein at Ser171 by protein kinase D2
diminishes its inhibitory effect on protein phosphatase 2A. PLoS
One. 7:e512422012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li M, Makkinje A and Damuni Z: The myeloid
leukemia-associated protein SET Is a potent inhibitor of protein
phosphatase 2A. J Biol Chem. 271:11059–11062. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Almeida LO, Garcia CB, MatosSilva FA,
Curti C and Leopoldino AM: Accumulated SET protein up-regulates and
interacts with hnRNPK, increasing its binding to nucleic acids, the
Bcl-xS repression, and cellular proliferation. Biochem Biophys Res
Commun. 445:196–202. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Xu Z, Yang W, Shi N, Gao Y, Teng M and Niu
L: Cloning, purification, crystallization and preliminary X-ray
crystallographic analysis of SET/TAF-Iß δN from Homo sapiens. Acta
Crystallogr Sect F Struct Biol Cryst Commun. 66:926–928. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lam BD, Anthony EC and Hordijk PL:
Cytoplasmic targeting of the proto-oncogene SET promotes cell
spreading and migration. FEBS Lett. 587:111–119. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cristóbal I, Garcia-Orti L, Cirauqui C,
Cortes-Lavaud X, García-Sánchez MA, Calasanz MJ and Odero MD:
Overexpression of SET is a recurrent event associated with poor
outcome and contributes to protein phosphatase 2A inhibition in
acute myeloid leukemia. Haematologica. 97:543–550. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mukhopadhyay A, Tabanor K, Chaguturu R and
Aldrich JV: Targeting inhibitor 2 of protein phosphatase 2A as a
therapeutic strategy for prostate cancer treatment. Cancer Biol
Ther. 14:962–972. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gonzales MF: Grading of gliomas. J Clin
Neurosci. 4:16–18. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kontos CK, Fendri A, Khabir A,
MokdadGargouri R and Scorilas A: Quantitative expression analysis
and prognostic significance of the BCL2-associated X gene in
nasopharyngeal carcinoma: A retrospective cohort study. BMC Cancer.
13:2932013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Manoochehri M, Karbasi A, Bandehpour M and
Kazemi B: Down-regulation of BAX gene during carcinogenesis and
acquisition of resistance to 5-FU in colorectal cancer. Pathol
Oncol Res. 20:301–307. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Croci DO, Cogno IS, Vittar NB, Salvatierra
E, Trajtenberg F, Podhajcer OL, Osinaga E, Rabinovich GA and
Rivarola VA: Silencing survivin gene expression promotes apoptosis
of human breast cancer cells through a caspase-independent pathway.
J Cell Biochem. 105:381–390. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Choi JE, Kang SH, Lee SJ and Bae YK:
Prognostic significance of Bcl-2 expression in non-basal
triple-negative breast cancer patients treated with
anthracycline-based chemotherapy. Tumour Biol. 35:12255–12263.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gómez-Crisóstomo NP, López-Marure R,
Zapata E, Zazueta C and Martinez-Abundis E: Bax induces cytochrome
c release by multiple mechanisms in mitochondria from MCF7 cells. J
Bioenerg Biomembr. 45:441–448. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kavanagh E, Rodhe J, Burguillos MA, Venero
JL and Joseph B: Regulation of caspase-3 processing by cIAP2
controls the switch between pro-inflammatory activation and cell
death in microglia. Cell Death Dis. 5:e15652014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu X, He Y, Li F, Huang Q, Kato TA, Hall
RP and Li CY: caspase-3 promotes genetic instability and
carcinogenesis. Mol Cell. 58:284–296. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Amar SK, Goyal S, Mujtaba SF, Dwivedi A,
Kushwaha HN, Verma A, Chopra D, Chaturvedi RK and Ray RS: Role of
type I & type II reactions in DNA damage and activation of
caspase 3 via mitochondrial pathway induced by photosensitized
benzophenone. Toxicol Lett. 235:84–95. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wen X, Zhu J, Dong L and Chen Y: The role
of c2orf68 and PI3K/Akt/mTOR pathway in human colorectal cancer.
Med Oncol. 31:922014. View Article : Google Scholar : PubMed/NCBI
|