Introduction
MicroRNAs (miRs) are a group of endogenous RNAs
(containing ~23 nucleotides) that have significant gene expression
regulatory roles by pairing to the mRNAs of protein-coding genes to
direct their post-transcriptional repression (1). Approximately 50% of miR genes are
located in cancer-associated genomic regions or in fragile sites,
and a number of them have been demonstrated to exhibit tumor
suppressor activity, while others have been reported to act as
oncogenes in various tissues and environments (2). Rescued expression of downregulated or
functionally-deficient miRs and/or inhibition of overexpressed miRs
may contribute to rebalancing the expression of large gene clusters
implicated in cell differentiation, apoptosis, metabolism,
immunity, oncogenesis and cancer (3).
Previously, it has been demonstrated that the miR
clusters miR-143 and miR-145 at the chromosome 5q32 region are
downregulated in various human tumors and are able to suppress
tumor growth in cancer of the urogenital (4,5), digestive
(6) and respiratory systems (3), as well as in osteosarcoma (7), breast cancer (8) and leukemia (9). Furthermore, numerous types of human
cancer cell exhibit a markedly reduced expression of miR-143 and
miR-145 compared to normal tissues (10). Accumulating evidence indicates that
miR-143 may act as an anti-oncomir in a number of types of cancer
(11).
Cervical cancer exhibits an aberrant cellular miR
expression pattern (12). miR-143
expression has been reported to be downregulated in human cervical
cancer tissues (13). Furthermore,
previous studies have revealed that miR-143 is downregulated in
HPV-induced pre-neoplastic lesions, which suggests that miR-143 may
have a significant role in the early stages of cervical cancer
development (13,14). However, the mechanism of miR-143 in
apoptosis and cell cycle progression during cervical cancer and the
mechanisms underlying these processes remain to be elucidated.
miRs are able to regulate multiple target genes
simultaneously (15). miR-143 has
been demonstrated to target Kirsten rat sarcoma viral oncogene
homolog (KRAS) (16), matrix
metalloproteinase-13 (7),
epithelial-mesenchymal transition (17), cyclooxygenase-2 (6) and cluster of differentiation 44v3
(3), and is able to suppress cell
growth and metastasis in vitro and in vivo in several
types of tumor (13). Among the
target genes regulated by miR-143, in silico screening of
gene targets for miR-143 was performed using TargetScan (www.targetscan.org). Extracellular-signal-regulated
kinase 5 (ERK5), which is an upstream gene of mitogen-activated
protein kinase (MAPK), has been reported to be a potential target
of miR-143 and to be closely associated with tumorigenesis
(18). There is clinical evidence
that an increase in ERK5 signaling may be associated with cancer
progression. For example, miR-143 targeting by ERK5 was
demonstrated in prostate cancer (19), bladder cancer (10), gut tumors (20), colon carcinoma (21) and DLD-1 cells (22). This may be due to the fact that ERK5
is able to phosphorylate c-Fos, which is a highly inducible and
unstable transcription factor, and has a variety of functions in
cell proliferation, differentiation and transformation regulation
(23). The activity and stability of
c-Fos is affected by several kinases, including ERK1/2, ribosomal
s6 kinase, c-Mos, ERK5 and p38, via phosphorylation (23). Therefore, this suggests that targeted
therapies against ERK5 may have a more widespread clinical
application in numerous types of cancer.
In the present study, the effect of miR-143
overexpression was evaluated in HeLa cervical cancer cells. miR-143
expression in the transfectants was assessed by northern blotting.
The results indicated that miR-143 overexpression reduced HeLa cell
viability in a dose- and time-dependent manner compared to control
cells, via cell counting and
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. The present study also identified that the mechanism of
miR-143 inhibition of migration and invasion of HeLa cells may be
via targeting ERK5 and its downstream oncoprotein c-Fos.
Materials and methods
Cell culture, cell viability and
morphological study
Human cervical cancer HeLa cells (obtained from the
cell bank of the Institutes for Biological Sciences, Shanghai,
China) were cultured in RPMI-1640 (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) containing 10% (v/v) heat-inactivated fetal
bovine serum (Gibco; Thermo Fisher Scientific, Inc.), 2 mM
L-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin
(Sigma-Aldrich; EMD Millipore, Billerica, MA, USA), under an
atmosphere of 95% air and 5% CO2 at 37°C. Cell viability
was determined by cell counting and MTT assays. Briefly, the medium
in each well was replaced with 250 µl of fresh medium containing
0.5 mg/ml MTT (Sigma-Aldrich; EMD Millipore) and incubated for 4 h
at 37°C. Following removal of the medium and MTT, the remaining
crystals were dissolved in 150 µl dimethyl sulfoxide
(Sigma-Aldrich; EMD Millipore) and the plate was agitated for 5 min
in the dark. The absorbance at 490 nm was measured using an ELx800
enzyme immunoassay analyzer (BioTek Instruments, Inc. Winooski, VT,
USA). In a cell counting assay, 50 µl of cells was added to 450 µl
trypan blue (1:10 dilution) and the calls were counted using a
hemocytometer. The results are presented as the mean ± standard
error of quadruplicates of a representative experiment.
Transient transfection
A total of 20 or 40 nM of precursor-miR-143
(Pre-miR-143; 5′-UGAGAUGAAGCACUGUAGCUC-3′) or random sequence
negative control (5′-UUCUCCGAACGUGUCACGUTT-3′) (Ambion; Thermo
Fisher Scientific, Inc.) were transfected into 5×105
HeLa cells using Lipofectamine® 2000 transfection
reagent (Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. A second cell transfection was performed
at 48 h after the first transection using the same transfection
method for cell viability analyzed by cell counting. To confirm the
efficiency of transfection, northern blotting was performed
following transient transfection.
Northern blotting
Total RNA for reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis was extracted using
TRIzol (Invitrogen; Thermo Fisher Scientific, Inc.) as described
previously (24) and separated on a
10% polyacrylamide TBE-urea mini-gel (Invitrogen; Thermo Fisher
Scientific. Inc.) for miR analysis, followed by electroblotting
onto a Hybond N nylon filter (GE Healthcare Life Sciences,
Chalfont, UK). The membrane was hybridized with an end-labeled
oligonucleotide probe (Promega Corporation, Madison, WI, USA) for
miR-143 (5′-TGAGCTACAGTGCTTCATCTCA-3′) for 2 h at 42°C and washed 3
times with Hank's balanced salt solution buffer. The hybridization
was performed in Rapid-Hyb buffer (GE Healthcare Life Sciences).
The blot was probed for U6 as a control for equal loading (25,26). Data
were analyzed using a Storm 860 PhosphorImager (GE Healthcare Life
Sciences). Densitometric analysis was performed by Healthcare
ImageQuant TL 7.0 Image Analysis Software (GE Healthcare Life
Sciences), as described previously (24).
Target genes
The gene targets for miR-143 were predicted using
TargetScan (www.targetscan.org) and the potential
target of interest, ERK5, was screened as previously described
(19).
RT-qPCR
A total of 100 ng RNA was extracted with the
miRNeasy kit (Qiagen GmbH, Hilden, Germany) and was further
subjected to RT using a reverse transcription kit (New England
BioLabs, Inc., Ipswich, MA, USA) according to the manufacturer's
protocol. qPCR was performed using a SYBR Green Real-Time PCR
Master Mix (Bio-Rad Laboratories, Inc., Hercules, CA, USA) on an
iCycler (Bio-Rad Laboratories, Inc.). The PCR reaction consisted of
40 cycles (94°C for 45 sec, 55°C for 30 sec and 72°C for 30 sec)
following an initial denaturation step (95°C for 20 sec). The
sequences of the primers used in the present study were as follows:
ERK5 forward, 5′-CCTTCGATGTGACCTTTGAC-3′ and reverse,
5′-TGACACCATTGATCTGACCC-3′; β-actin forward,
5′-ATCGTGCGTGACATTAAGGAGAAG–3′ and reverse,
5′-AGGAAGGAAGGCTGGAAGAGTG-3′. Expression of ERK5 relative to
β-actin was determined using the 2-ΔΔCq method (27).
Flow cytometric analysis of cell cycle
and apoptosis
Cells were harvested by trypsinization, centrifuged
at 400 × g for 5 min at 4°C, suspended in 0.1 ml of
phosphate-buffered saline (PBS) and subsequently fixed by addition
of 1.0 ml 70% cold ethanol. Following pelleting and removal of
ethanol, RNA was extracted using 1 unit of RNase A (Sigma-Aldrich;
EMD Millipore). The cells were stained with 50 µg/ml propidium
iodide (PI; Sigma-Aldrich; EMD Millipore) for 30 min at room
temperature. The DNA content was subsequently analyzed using
cytofluorometry and cell cycle analysis was performed using FACScan
software (BD Biosciences, Franklin Lakes, NJ, USA), as described
previously (19).
To analyze apoptosis, cells were washed in PBS and
centrifuged at 400 × g for 5 min at 4°C. Cells were resuspended in
binding buffer (Biolegend, Inc., San Diego, CA, USA), and 5 µl of
fluorochrome-conjugated Annexin V-fluorescein isothiocyanate (FITC;
Roche Diagnostics, Meylan, France) and 10µl PI solution were added
and incubated with the cells for 10 min at 4°C. The percentage of
Annexin V-FITC-positive cells was immediately analyzed using a flow
cytometer and the data were analyzed using the Modfit LT software
for Windows version 3.2 (Verity Software House, Inc., Topsham, ME,
USA).
Western blotting
The cells were homogenized in chilled lysis buffer
comprising 10 mM Tris-HCl (pH 7.4), 1% NP-40, 0.1% deoxycholic
acid, 0.1% sodium dodecyl sulfate (SDS), 150 mM NaCl, 1 mM
ethylenediaminetetraacetic acid and 1% protease inhibitor cocktail
(Sigma-Aldrich; EMD Millipore) for 30 min on ice. Following
centrifugation at 16,000 × g for 20 min at 4°C, the supernatants
were collected as protein samples. Protein concentrations were
assessed with a DC protein assay kit (Bio-Rad Laboratories, Inc.).
A total of 10 µg of protein lysate was used for western blotting
and separated by 10% SDS-PAGE, followed by electroblotting onto a
polyvinylidene difluoride membrane (DuPont, Boston, MA, USA).
Following blockage of non-specific binding sites using
Tris-Buffered Saline and Tween 20 (TBST) containing 5% non-fat milk
for 1 h, the membrane was incubated with rabbit anti-human ERK5
monoclonal antibody (1:1,000 dilution; clone, D3I5V; catalog no.
12950; Cell Signaling Technology, Inc., Danvers, MA, USA) or mouse
anti-human c-Fos monoclonal antibody (1:1,000 dilution; clone,
6–2H-2F; catalog no. sc-447; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) overnight at 4°C. The membranes were washed three
times with TBST and incubated further with horseradish
peroxidase-conjugated sheep anti-mouse immunoglobulin (Ig)G,
horeradish peroxidase (HRP)-linked whole antibody (catalog no.
NA931) or enhanced chemiluminescence donkey anti-rabbit IgG,
HRP-linked whole antibody (catalog no. NA934V) (GE Healthcare Life
Sciences) for 1 h at room temperature. The immunoblots were
visualized using an enhanced chemiluminescence detection kit (New
England BioLabs, Inc.) following washing three times with TBST.
Quantification was performed using Imaging J version 1.45 (National
Institutes of Health, Bethesda, MD, USA).
Statistical analysis
Each experiment was performed in triplicate, with
the exception of the MTT assay, which was performed in
quadruplicate. All statistical analyses were performed using SPSS
version 13.0 (SPSS, Inc., Chicago, IL, USA). Data are presented as
the mean ± standard error, and differences between groups were
calculated by analysis of variance or χ2. P<0.05 was
considered to indicate a statistically significant difference.
Results
miR-143 overexpression decreases
growth of HeLa cells
It has been demonstrated that transient transfection
of human cancer cells with Pre-miR may significantly increase the
expression of mature miR (28). The
present study initially confirmed the expression of miR-143
following Pre-miR-143 transfection of HeLa cells, and investigated
the effect of transient miR-143 overexpression on HeLa cell growth.
This was performed by transfecting Pre-miR-143 into HeLa cells,
with a random sequence as a negative control. Northern blotting
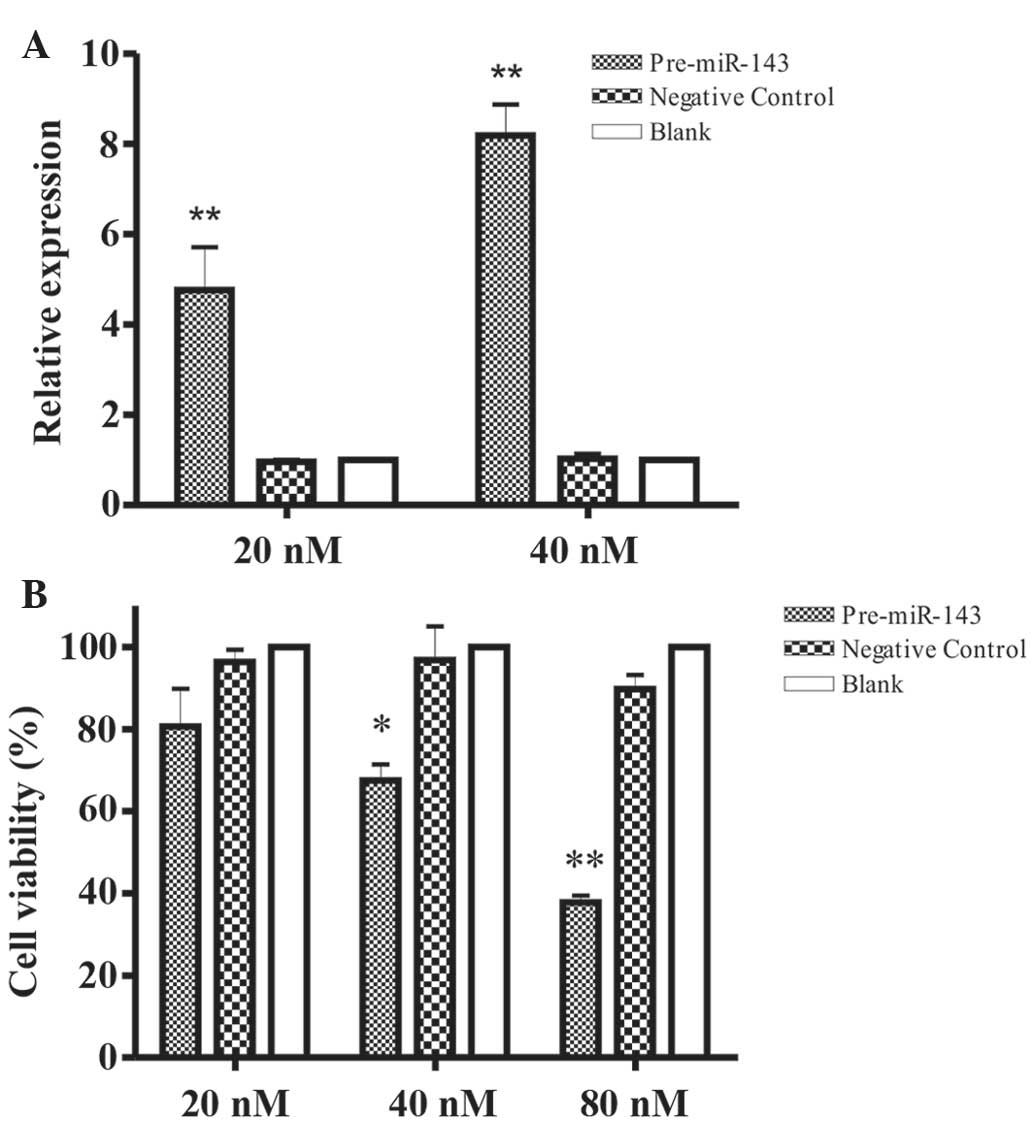
results clearly revealed that miR-143 expression was significantly
increased in 20 or 40 nM pre-miR-143 transfected HeLa cells
(Fig. 1A), as compared to negative
control transfected cells and blank buffer following normalization
to U6 expression (P<0.05).
To evaluate the impact of miR-143 overexpression on
the growth of HeLa cells, cell counting and MTT assays were
performed on HeLa cells following transient transfection with
Pre-miR-143 at 20, 40 and 80 nM respectively. The increased level
of Pre-miR-143 (40 and 80 nM) significantly inhibited HeLa cell
growth, compared to negative control transfected HeLa cell and
blank buffer groups (P=0.091, 0.008 and 0.002, for 20, 40 and 80
nM, respectively) as evaluated by cell counting (Fig. 1B). This inhibitory effect on cell
growth was not an immediate cell response; rather two consecutive
cell transfections at an interval of 48 h were required.
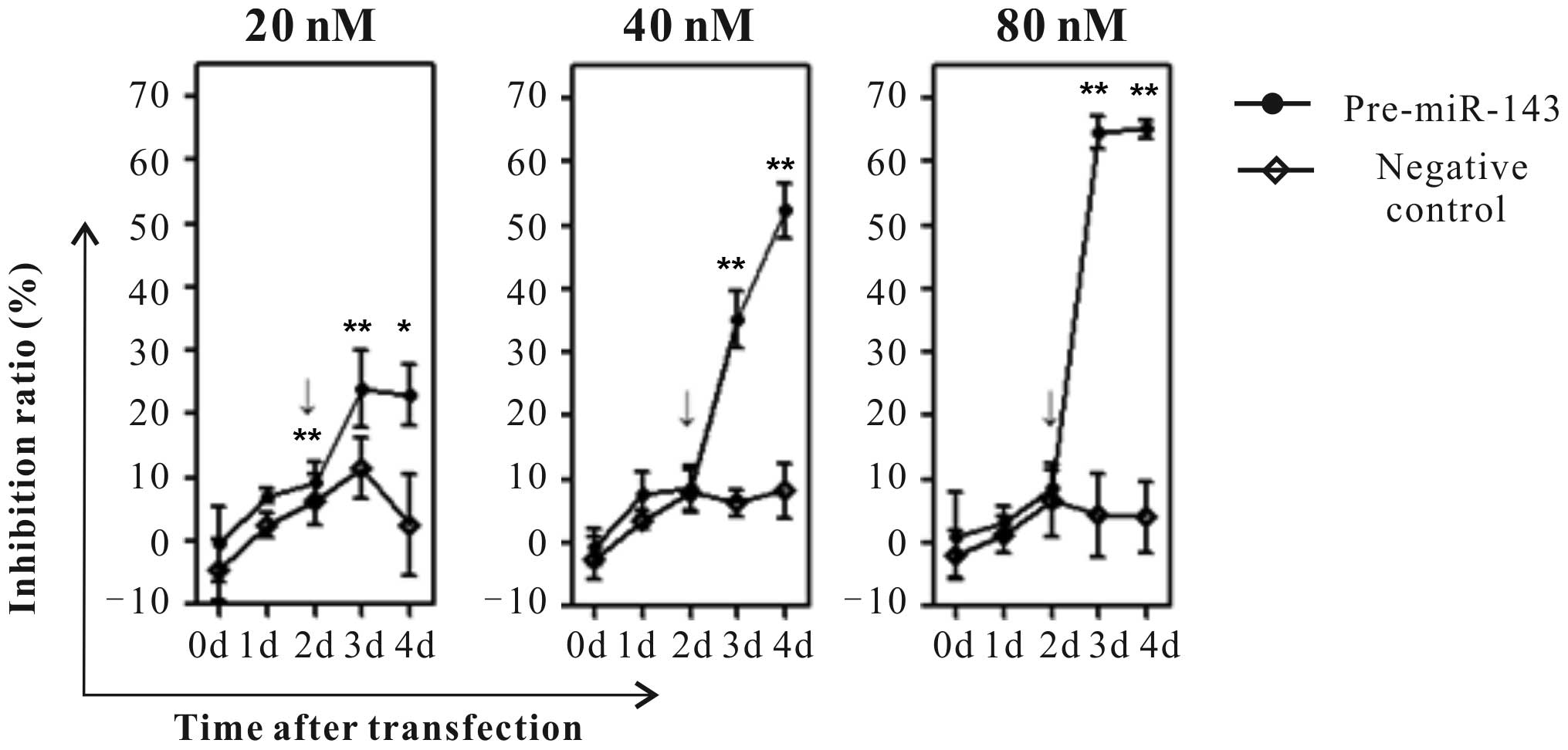
Furthermore, the cell viability was detected at 0, 1, 2, 3 and 4
days using MTT assays. Notably, the results of the present study
have shown that 40 and 80 nM Pre-miR-143 transfection significantly
suppresses HeLa cell growth in a dose- and time-dependent manner as
compared to the negative control transfected HeLa cells (Fig. 2; P<0.001).
miR-143 overexpression induces HeLa
cell apoptosis
Having validated the impact of miR-143 on the growth
rate of transfected HeLa cells, the present study additionally
characterized the effect of miR-143 overexpression on biological
responses, including the cell cycle and apoptosis, using flow
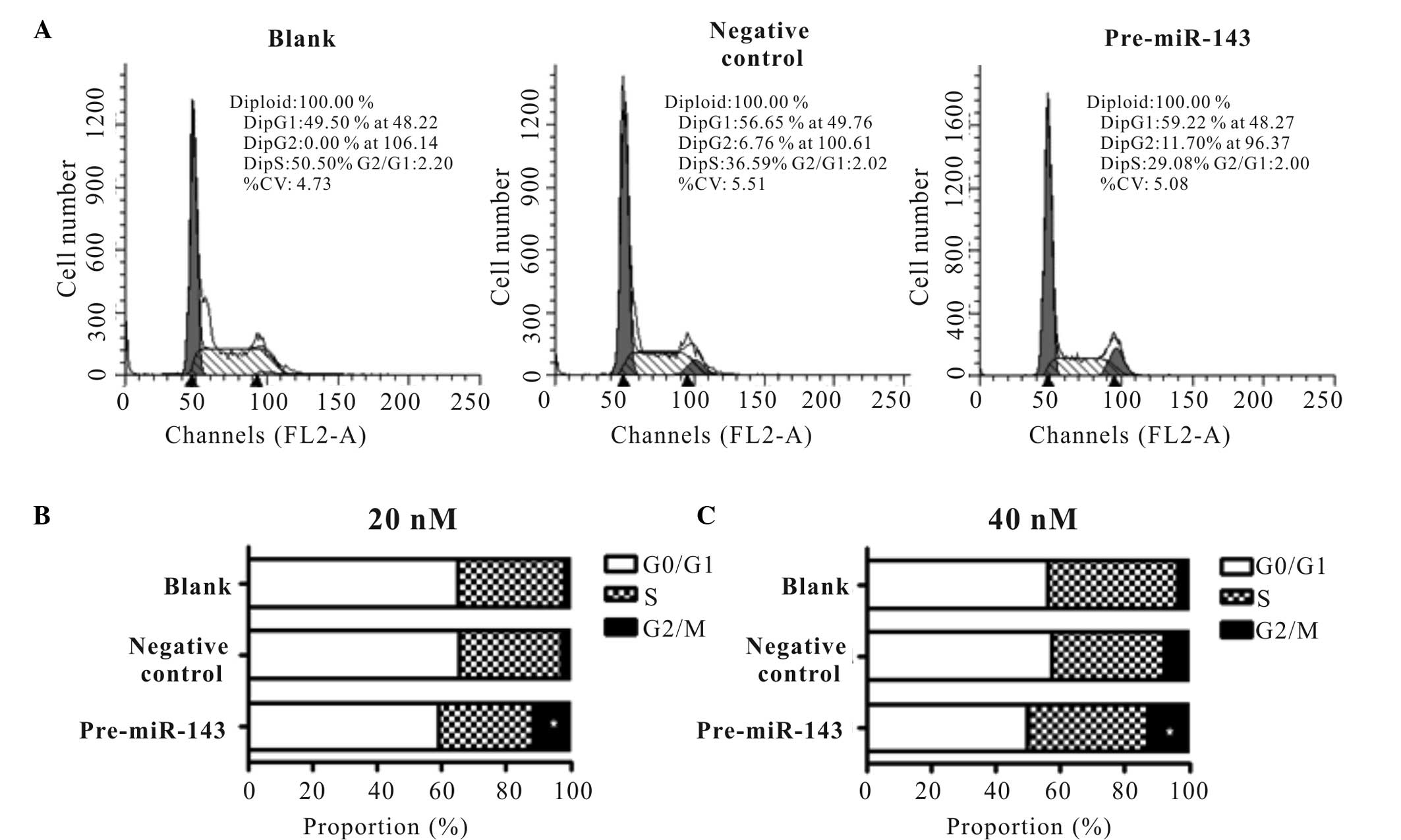
cytometry. The representative cell cycle distribution of confluent
is presented for 40 nM Pre-miR-143 transfected and negative control
transfected HeLa cells, as well as blank buffer (Fig. 3A). Notably, significant dose-dependent
increases in the apoptotic fraction and corresponding decreases in
the G0/G1 phase of the cell cycle were observed in the Pre-miR-143
transfected cells. By contrast, cells in G2/M phase of the cell
cycle were increased from 1.78 to 11.65% (P<0.05) following 20
nM Pre-miR-143 transfection and 3.5 to 12.99% following 40 nM
Pre-miR-143 transfection (P<0.001; Fig. 3B and C).
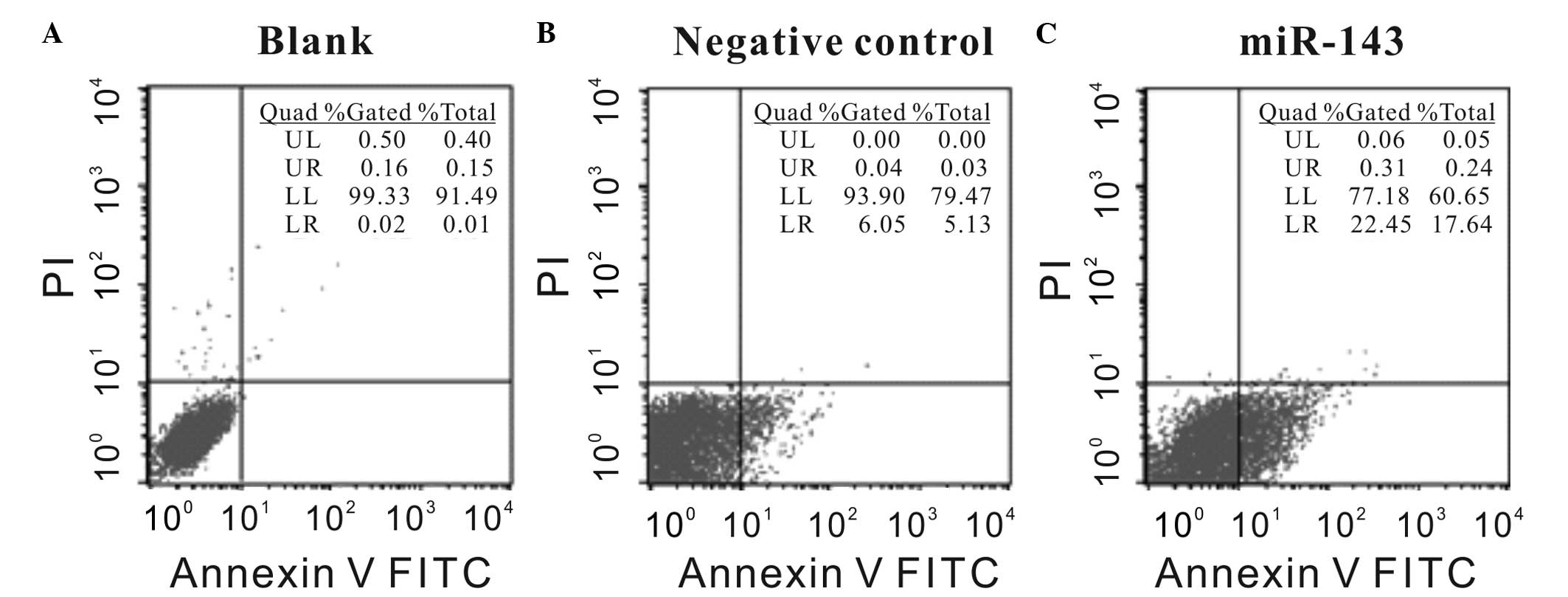
To additionally validate these results, the impact
of miR-143 overexpression on the apoptosis of HeLa cells was
evaluated by PI and Annexin V-FITC staining via flow cytometry. As
shown by representative figures, the Annexin V-FITC staining signal
of 40 nM Pre-miR-143 transfected cells showed a right shift in the
X-axis compared with the negative control transfected HeLa cells
and the blank cells. The apoptotic rate (37.51±0.032%) of miR-143
overexpressed HeLa cells was significantly increased compared to
the apoptotic rate of negative control transfected cells
(20.27±0.045%; P<0.05; Fig.
4).
miR-143 downregulates the expression
of ERK5 and c-Fos
In silico prediction of gene targets for
miR-143 was performed using TargetScan (www.targetscan.org) and ERK5 was screened as a
potential target. ERK5, also known as big MAPK, is activated by
oxidative stress, hyperosmolarity and certain growth factors
(29). Unlike other MAPK members,
ERK5 has a unique large C-terminal region, whose function has not
been fully elucidated (30). It has
been shown that ERK5 directly interacts with, phosphorylates and
activates a number of transcription factors, including c-Myc,
Sap1a, c-Fos, Fos-related antigen 1 and myocyte enhancer factor-2
family members (29). ERK5 is
important for promoting cell proliferation, differentiation and
neuronal survival (30). Its activity
has been previously considered to be dependent on its
phosphorylation by MEK5; however, post-transcriptional regulation
in this process is also considered to play a role, as has been
previously reported (19).
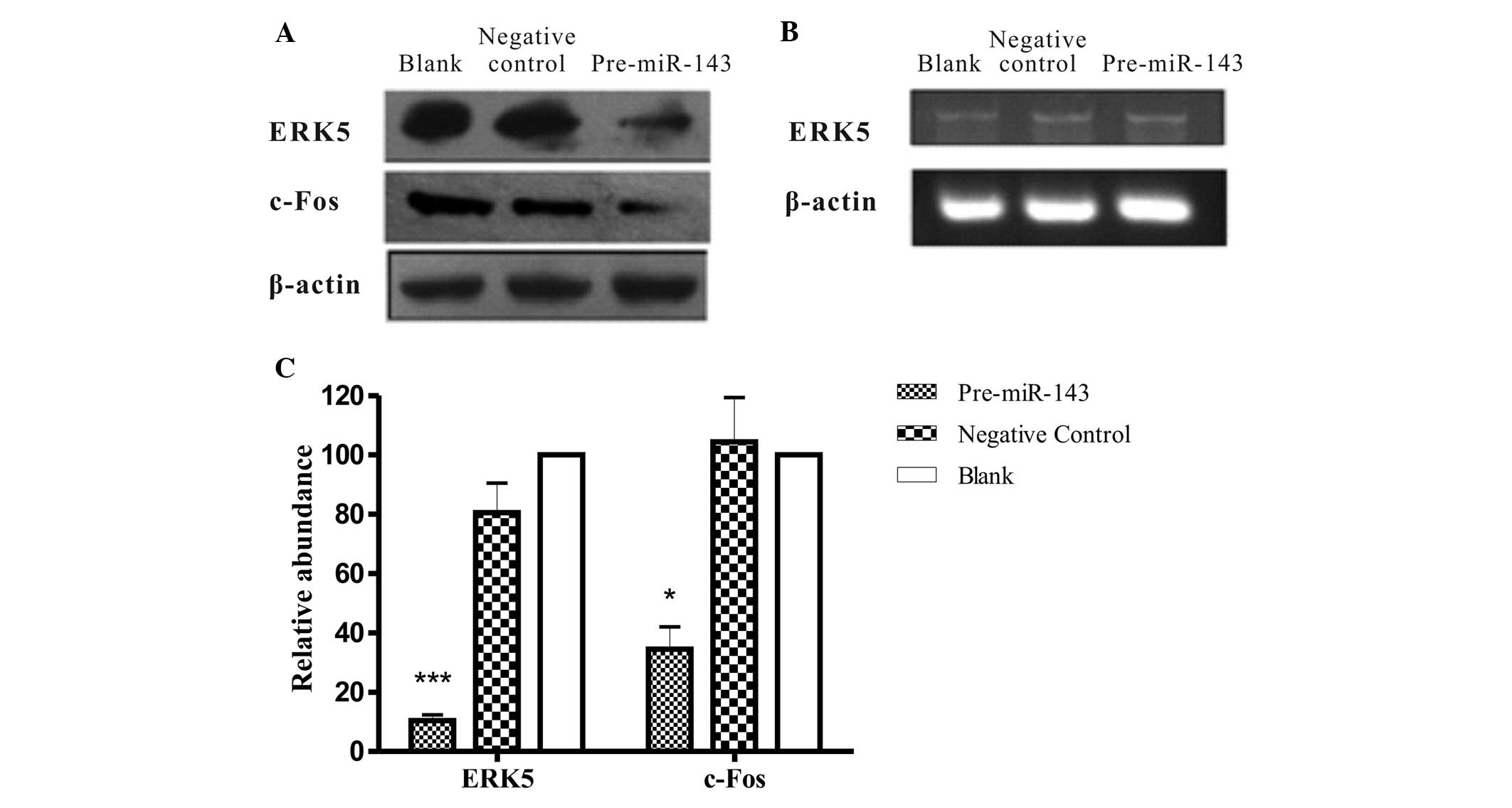
Overexpression of miR-143 has been demonstrated to downregulate
ERK5 expression in human bladder cancer cells (10), HCT116 human colon carcinoma cells
(21) and LNCaP prostate cancer cell
lines (19). To experimentally
validate regulation of miR-143 on its potential target ERK5, the
present study analyzed ERK5 expression in HeLa cells following
transient transfection with Pre-miR-143. Notably, ERK5 expression
was inversely correlated with miR-143 expression in HeLa cells as
demonstrated by western blot analysis. In a representative image
shown as Fig. 5A, the ERK5 protein
level in the HeLa cells transfected with Pre-miR-143 was decreased.
Whereas the mRNA level of ERK5 was almost unaffected, as confirmed
by RT-qPCR (Fig. 5B), when compared
with the negative control and blank cells, the ERK5 protein
expression in Pre-miR-143 transfectant was significantly
downregulated (P<0.001; Fig.5C).
Furthermore, activated ERK5 modulates cell differentiation and
proliferation via c-Fos, c-Myc, cyclin D1 and nuclear factor-кB
activation. Additionally, c-Fos has been demonstrated to have a
regulatory effect via a signaling pathway involving ERK5 (23,26).
Therefore, the transcription level and the expression level of
c-Fos were also evaluated. Notably, the c-Fox protein expression
was reduced concurrently with the downregulation of ERK5
(P<0.05; Fig. 5C). This indicates
that ERK5 may be negatively regulated by miR-143 at the
post-transcriptional level, which additionally decreases downstream
c-Fos oncoprotein expression.
Discussion
miR-143 is one of the most prominent miRs implicated
in the genesis and progression of human cancer. It has been
implicated in the promotion of tumor growth, proliferation,
apoptosis resistance and resistance to gemcitabine-based
chemotherapy (31). Furthermore, a
number of studies have identified that miR-143 was downregulated in
various tumor types (32–35). The expression levels of miR-143 and
−145 were observed to be decreased in the majority of human gastric
cancer cases, and growth inhibition and increased sensitivity to
5-fluorouracil was reported following transfection with miR-143
(36). It has also been reported that
miR-143 expression is significantly downregulated in bladder tumor
tissues compared with normal adjacent tissues, and miR-143
transfection into EJ and T24 cells significantly inhibited cell
proliferation (37). Taken together,
the results of a number of studies, including the present, support
the hypothesis that miR-143 may be one of the most relevant tumor
suppressors among the class of miRs.
However, limited efforts have been made to elucidate
the role of miR-143 in cervical cancer. The molecular mechanism
modulated by miR-143, leading to cell growth inhibition and death
remains to be fully elucidated and the role of miR-143 in
regulation of the cell cycle and apoptosis is unclear. Lin et
al (38) used a direct sequencing
method to characterize the profiles of miRs and other small RNA
segments for six human cervical carcinoma cell lines and five
normal cervical samples, confirmed with a panel of 29 matched pairs
of human cervical cancer and normal cervical samples. Reduced
expression of miR-143 and increased expression of miR-21 were
observed in cervical cancer samples, suggesting the potential value
of these miRs as tumor markers (24,38).
The present study demonstrates that miR-143 is a
negative regulator of HeLa cell growth. It may additionally be
speculated that miR-143 can induce apoptosis, at least in part, via
negatively regulating ERK5. As in the present transfection
experiments ERK5 mRNA level was unaltered, contrasting to the
significant change in ERK5 protein levels, it may be proposed that
the main mechanism of miR-143-induced ERK5 suppression occurs at a
post-transcriptional level.
To the best of our knowledge, the results of the
present study comprise the first report of a causal association
between miR-143 and G2-M progression. Zen et al (39) reported that downregulation of MAPK7 by
small interfering RNA suppressed the growth of SNU449 cells, which
comprise the HCC cell line with the greatest amplification and
overexpression of MAPK7. ERK5, which is phosphorylated during the
G2/M phases of the cell cycle, may regulate entry into mitosis in
SNU449 cells (40). As a significant
step in determining its role in cervical tumorigenesis, the present
study identifies a novel function for miR-143/ERK5 in the
regulation of G2-M transition.
ERK5 has been demonstrated to inhibit cancer cell
viability, achieved in part by inhibiting the nuclear export of
c-Fos and disrupting the interaction of c-Fos with ubiquitin
protein ligase E3 component N-recognin 1 by phosphorylating Ser32
(23). Additional mechanisms and
targets of miR-143 besides ERK5 are likely to contribute to
miR-143-induced tumor cell growth inhibition. It has been reported
that miR-143 is significant in suppressing colorectal cancer cell
growth via inhibition of KRAS translation (41). Additionally, restoration of miR-143
expression in colon cell lines decreased tumor cell growth and
soft-agar colony formation, and downregulated DNA
(cytosine-5)-methyltransferase 3A (DNMT3A) expression in terms of
mRNA and protein levels. DNMT3A has been demonstrated to be a
direct target of miR-143 by luciferase reporter assay (42).
An improved understanding of the signaling pathway
investigated in the present study will bring us closer to
understanding the molecular mechanisms underlying cervical cancer
and may lead to the development of novel approaches for detection
and therapy. Taken together with the results of previous
correlational studies on miR-143 and ERK5, the results of the
present study indicate that strategies rescuing miR-143 expression,
enhancing the miR-143/ERK5 interaction, or inhibiting ERK5
expression may have a strong rationale for therapeutic applications
in the treatment of cancer.
Acknowledgements
The present study was financially supported by the
Fundamental Research Funds for the Central Universities (grant no.
XJJ2015056) and (grant no. 1191320048).
References
|
1
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ha M and Kim VN: Regulation of microRNA
biogenesis. Nat Rev Mol Cell Biol. 15:509–524. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma QP, Jiang Q, Pu Q, Zhang X, Yang W,
Wang Y, Ye S, Wu S, Zhong G, Ren J, et al: MicroRNA-143 inhibits
migration and invasion of human non-small-cell lung cancer and its
relative mechanism. Int J Biol Sci. 9:680–692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Song T, Zhang X, Wang C, Wu Y, Dong J, Gao
J, Cai W and Hong B: Expression of miR-143 reduces growth and
migration of human bladder carcinoma cells by targeting
cyclooxygenase-2. Asian Pac J Cancer Prev. 12:929–933.
2011.PubMed/NCBI
|
|
5
|
Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z,
Li P, Zhang W, Wu H, Feng N, et al: mir-143 decreases prostate
cancer cells proliferation and migration and enhances their
sensitivity to docetaxel through suppression of KRAS. Mol Cell
Biochem. 350:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu XL, Cheng B, Li PY, Huang HJ, Zhao Q,
Dan ZL, Tian DA and Zhang P: MicroRNA-143 suppresses gastric cancer
cell growth and induces apoptosis by targeting COX-2. World J
Gastroenterol. 19:7758–7765. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Osaki M, Takeshita F, Sugimoto Y, Kosaka
N, Yamamoto Y, Yoshioka Y, Kobayashi E, Yamada T, Kawai A, Inoue T,
et al: MicroRNA-143 regulates human osteosarcoma metastasis by
regulating matrix metalloprotease-13 expression. Mol Ther.
19:1123–1130. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chang YY, Kuo WH, Hung JH, Lee CY, Lee YH,
Chang YC, Lin WC, Shen CY, Huang CS, Hsieh FJ, et al: Deregulated
microRNAs in triple-negative breast cancer revealed by deep
sequencing. Mol Cancer. 14:362015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Akao Y, Nakagawa Y, Kitade Y, Kinoshita T
and Naoe T: Downregulation of microRNAs-143 and −145 in B-cell
malignancies. Cancer Sci. 98:1914–1920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Noguchi S, Yasui Y, Iwasaki J, Kumazaki M,
Yamada N, Naito S and Akao Y: Replacement treatment with
microRNA-143 and −145 induces synergistic inhibition of the growth
of human bladder cancer cells by regulating PI3K/Akt and MAPK
signaling pathways. Cancer Lett. 328:353–361. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ahmad I, Singh LB, Yang ZH, Kalna G,
Fleming J, Fisher G, Cooper C, Cuzick J, Berney DM, Møller H, et
al: Mir143 expression inversely correlates with nuclear ERK5
immunoreactivity in clinical prostate cancer. Br J Cancer.
108:149–154. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hou LJ and Zhai JJ: Aberrant expression
profile of translationally controlled tumor protein and
tumor-suppressive microRNAs in cervical cancer. J BUON.
20:1504–1509. 2015.PubMed/NCBI
|
|
13
|
Honegger A, Schilling D, Bastian S,
Sponagel J, Kuryshev V, Sültmann H, Scheffner M, Hoppe-Seyler K and
Hoppe-Seyler F: Dependence of intracellular and exosomal microRNAs
on viral E6/E7 oncogene expression in HPV-positive tumor cells.
PLoS Pathog. 11:e10047122015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang XH, Tang S, Le SY, Lu R, Rader JS,
Meyers C and Zheng ZM: Aberrant expression of oncogenic and
tumor-suppressive microRNAs in cervical cancer is required for
cancer cell growth. PLoS One. 3:e25572008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Pinweha P, Rattanapornsompong K,
Charoensawan V and Jitrapakdee S: MicroRNAs and oncogenic
transcriptional regulatory networks controlling metabolic
reprogramming in cancers. Comput Struct Biotechnol J. 14:223–233.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mekenkamp LJ, Tol J, Dijkstra JR, de
Krijger I, Vink-Börger ME, van Vliet S, Teerenstra S, Kamping E,
Verwiel E, Koopman M, et al: Beyond KRAS mutation status: Influence
of KRAS copy number status and microRNAs on clinical outcome to
cetuximab in metastatic colorectal cancer patients. BMC Cancer.
12:2922012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Peng X, Guo W, Liu T, Wang X, Tu X, Xiong
D, Chen S, Lai Y, Du H, Chen G, et al: Identification of miRs-143
and-145 that is associated with bone metastasis of prostate cancer
and involved in the regulation of EMT. PLoS One. 6:e203412011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lochhead PA, Gilley R and Cook SJ: ERK5
and its role in tumour development. Biochem Soc Trans. 40:251–256.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Clapé C, Fritz V, Henriquet C, Apparailly
F, Fernandez PL, Iborra F, Avancès C, Villalba M, Culine S and
Fajas L: miR-143 interferes with ERK5 signaling, and abrogates
prostate cancer progression in mice. PLoS One. 4:e75422009.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Takaoka Y, Shimizu Y, Hasegawa H, Ouchi Y,
Qiao S, Nagahara M, Ichihara M, Lee JD, Adachi K, Hamaguchi M and
Iwamoto T: Forced expression of miR-143 represses ERK5/c-Myc and
p68/p72 signaling in concert with miR-145 in gut tumors of Apc(Min)
mice. PLoS One. 7:e421372012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Borralho PM, Simões AE, Gomes SE, Lima RT,
Carvalho T, Ferreira DM, Vasconcelos MH, Castro RE and Rodrigues
CM: miR-143 overexpression impairs growth of human colon carcinoma
xenografts in mice with induction of apoptosis and inhibition of
proliferation. PLoS One. 6:e237872011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakagawa Y, Iinuma M, Naoe T, Nozawa Y and
Akao Y: Characterized mechanism of alpha-mangostin-induced cell
death: Caspase-independent apoptosis with release of endonuclease-G
from mitochondria and increased miR-143 expression in human
colorectal cancer DLD-1 cells. Bioorg Med Chem. 15:5620–5628. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sasaki T, Kojima H, Kishimoto R, Ikeda A,
Kunimoto H and Nakajima K: Spatiotemporal regulation of c-Fos by
ERK5 and the E3 ubiquitin ligase UBR1, and its biological role. Mol
Cell. 24:63–75. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kajimoto K, Naraba H and Iwai N: MicroRNA
and 3T3-L1 pre-adipocyte differentiation. RNA. 12:1626–1632. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Corney DC, Flesken-Nikitin A, Godwin AK,
Wang W and Nikitin AY: MicroRNA-34b and MicroRNA-34c are targets of
p53 and cooperate in control of cell proliferation and
adhesion-independent growth. Cancer Res. 67:8433–8438. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Peltier HJ and Latham GJ: Normalization of
microRNA expression levels in quantitative RT-PCR assays:
Identification of suitable reference RNA targets in normal and
cancerous human solid tissues. RNA. 14:844–852. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak and Schmittgen, . Analysis of
relative gene expression data using real-time quantitative PCR and
the 2-ΔΔCt method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu L, Yu X, Guo X, Tian Z, Su M, Long Y,
Huang C, Zhou F, Liu M, Wu X and Wang X: miR-143 is downregulated
in cervical cancer and promotes apoptosis and inhibits tumor
formation by targeting Bcl-2. Mol Med Rep. 5:753–760.
2012.PubMed/NCBI
|
|
29
|
Simões AE, Rodrigues CM and Borralho PM:
The MEK5/ERK5 signalling pathway in cancer: A promising novel
therapeutic target. Drug Discov Today. S1359-S6446(16): 30228–8.
2016.
|
|
30
|
Nithianandarajah-Jones GN, Wilm B,
Goldring CE, Müller J and Cross MJ: ERK5: Structure, regulation and
function. Cell Signal. 24:2187–2196. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen B, Li H, Zeng X, Yang P, Liu X, Zhao
X and Liang S: Roles of microRNA on cancer cell metabolism. J
Transl Med. 10:2282012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Truta A, Popon TA, Saraci G, Ghervan L and
Pop IV: Novel non invasive diagnostic strategies in bladder cancer.
Clujul Med. 89:187–192. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Goto Y, Kurozumi A, Enokida H, Ichikawa T
and Seki N: Functional significance of aberrantly expressed
microRNAs in prostate cancer. Int J Urol. 22:242–252. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Banno K, Iida M, Yanokura M, Kisu I, Iwata
T, Tominaga E, Tanaka K and Aoki D: MicroRNA in cervical cancer:
OncomiRs and tumor suppressor miRs in diagnosis and treatment.
Scientific World Journal. 2014:1780752014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pan Y, Meng M, Zhang G, Han H and Zhou Q:
Oncogenic microRNAs in the genesis of leukemia and lymphoma. Curr
Pharm Des. 20:5260–5267. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Bai L, Mao R, Wang J, Ding L, Jiang S, Gao
C, Kang H, Chen X, Sun X and Xu J: ERK1/2 promoted proliferation
and inhibited apoptosis of human cervical cancer cells and
regulated the expression of c-Fos and c-Jun proteins. Med Oncol.
32:572015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ng EK, Tsang WP, Ng SS, Jin HC, Yu J, Li
JJ, Röcken C, Ebert MP, Kwok TT and Sung JJ: MicroRNA-143 targets
DNA methyltransferases 3A in colorectal cancer. Br J Cancer.
101:699–706. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lin T, Dong W, Huang J, Pan Q, Fan X,
Zhang C and Huang L: MicroRNA-143 as a tumor suppressor for bladder
cancer. J Urol. 181:1372–1380. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zen K, Yasui K, Nakajima T, Zen Y, Zen K,
Gen Y, Mitsuyoshi H, Minami M, Mitsufuji S, Tanaka S, et al: ERK5
is a target for gene amplification at 17p11 and promotes cell
growth in hepatocellular carcinoma by regulating mitotic entry.
Genes Chromosomes Cancer. 48:109–120. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lui WO, Pourmand N, Patterson BK and Fire
A: Patterns of known and novel small RNAs in human cervical cancer.
Cancer Res. 67:6031–6043. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Deftereos G, Corrie SR, Feng QH, Morihara
J, Stern J, Hawes SE and Kiviat NB: Expression of mir-21 and
mir-143 in cervical specimens ranging from histologically normal
through to invasive cervical cancer. PLoS One. 6:e284232011.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cude K, Wang YP, Choi HJ, Hsuan SL, Zhang
H, Wang CY and Xia Z: Regulation of the G2-M cell cycle progression
by the ERK5-NFkappa B signaling pathway. J Cell Biol. 177:253–264.
2007. View Article : Google Scholar : PubMed/NCBI
|