Introduction
Colorectal cancer (CRC) is the third most frequent
cancer and the fourth most common cause of cancer-associated
mortality globally, with an annual incidence of 17.2/100,000 people
and an age-standardized mortality rate of 8.4/100,000 people
worldwide (1,2). The preferred treatment option is
complete surgical resection, yet in up to 30% of cases the cancer
recurs either locally or as distant metastasis (2). In order to reduce the incidence of
disease recurrences, chemotherapy or radiochemotherapy are
indicated, in addition to surgery in certain patients. It has been
shown that only patients with cancer that has spread to the local
lymph nodes benefit from these additional treatment options
(1). Therefore, appropriate pre- and
post-operative tumor-node-metastasis (TNM) staging is crucial for
the optimal treatment of CRC patients and for the estimation of
their prognosis (1,3). Follow-up for several years after
treatment is necessary for the early detection of disease
recurrence (2).
Since surgery is the treatment of choice in CRC, the
completeness of the surgical removal of the tumor is the most
important treatment-related prognostic factor in CRC patients. The
completeness of the surgical removal of the tumor is expressed in
terms of the R category: R0 meaning the complete removal of the
tumor, R1 meaning microscopic residual disease and R2 meaning
macroscopic residual disease (4,5).
The two most important prognostic factors (the stage
of the disease at diagnosis and residual disease after surgical
resection) are assessed by the pathohistological analysis of the
resected tumor specimen. Clinical decisions regarding adjuvant
treatment or follow-up after surgery are based on the
pathohistological analysis of the removed specimen and are based on
the probability of malignant tissue remaining in the patient
(6). A more direct and effective
method would be to introduce a biomarker that could measure the
disease burden, as well as the residual disease, directly in the
patient's body (7).
This idea has led to the search for a specific
molecular marker in the peripheral blood for the detection of
minimal residual disease after the primary treatment of CRC. The
majority of investigated molecular markers have been
tumor-associated membrane proteins, such as cytokeratin 20 (CK20),
and tumor-specific DNA mutations, such as KRAS proto-oncogene,
GTPase (KRAS) (8). The
detection of minimal residual disease in the peripheral blood
through the use of molecular markers has been shown to have an
impact on a patient's prognosis in a number of studies, although
the methods are not yet sufficiently standardized for clinical use
(9–11).
The present study was aimed at determining the
expression of CK20 and the presence of KRAS and B-Raf
proto-oncogene, serine/threonine kinase (BRAF)
tumor-specific mutations in DNA isolated from the peripheral blood
prior to and following surgery, in order to assess whether the
serological markers can be used to determine the stage of the
disease at diagnosis and the R status after surgery.
Materials and methods
Patients
The study was approved by the Republic of Slovenia
National Medical Ethics Committee (Ljubljana, Slovenia). All
patients made an informed decision to take part in the study and
provided written consent. Patients were informed that the results
of tests would not be disclosed to them, as they would not affect
the standard treatment provided, to which all patients
consented.
A total of 65 patients scheduled for elective
surgery at University Medical Centre Ljubljana (Ljubljana,
Slovenia) due to CRC or adenomas between July 2011 and January 2012
were enrolled into the study. The patients were staged into Union
for International Cancer Control (UICC) TNM categories and stages
according to the pre-operative staging results, intraoperative
findings and pathohistological results of the resected specimens,
as described previously (1). After
surgery, the pathohistological analysis of the resected specimens
was performed in a standard manner (3). The clearance of tumor tissue by surgery
was expressed by R category based on surgery reports and
pathohistological analysis of the resected specimen.
Samples
Formalin-fixed paraffin-embedded (FFPE) tumor tissue
and matched normal tissue samples were obtained from each patient.
The appropriate FFPE tumor and normal tissue block was selected by
the pathologist, who also evaluated the percentage of tumor cells
in the paraffin slides from the first and last hematoxylin and
eosin (HE)-stained sections. Six consecutive non-colored FFPE
sample cuts between the HE colored sections with the thickness of
10-µm were prepared in an Eppendorf tube by the pathologist.
Additionally, from each patient, the peripheral whole blood samples
were collected immediately prior to the surgery and 5–7 days after
the surgery. The whole blood samples were collected in 10-ml EDTA
tubes and stored at −80°C. The samples (1 ml) were centrifuged at
1,700 × g for 25 min at 4°C to obtain pellets (cells and
cellular debris). These pellets were then used for DNA and RNA
extraction.
DNA isolation
For the DNA extraction, a QIAamp DNA FFPE kit
(Qiagen, Hilden, Germany) was used according to the manufacturer's
protocols. The DNA from the blood samples was extracted using an
Arrow Blood DNA kit (NorDiag, Dublin, Ireland) according to the
manufacturer's protocols. After the isolation, the DNA
concentration was measured spectrophotometrically at 280/260 nm
using a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific,
Inc., Waltham, MA, USA).
RNA isolation
Total RNA was isolated from the blood using an
miRNeasy Mini kit (Qiagen) according to the manufacturer's
protocols. RNA was quantified using the NanoDrop spectrophotometer,
and cDNA was synthesized from 500 ng of the total RNA using the
High-Capacity cDNA Reverse Transcription kit with RNase Inhibitor
(Applied Biosystems; Thermo Fisher Scientific, Inc. Foster City,
CA, USA), according to the manufacturer's protocols.
Determination of KRAS and BRAF
mutations
Determination of KRAS and BRAF
mutation status was performed using the KRAS/BRAF Mutation Analysis
kit (EntroGen, Tarzana, CA, USA) for a quantitative polymerase
chain reaction (qPCR) assay based on allele-specific PCR. The assay
is designed to preferentially amplify mutant DNA even in samples
that have mostly wild-type DNA. In the assay, an endogenous control
gene is included in order to ensure that a sufficient amount of DNA
is available for amplification. The detection of the amplification
product is performed using fluorescent hydrolysis probes. The assay
was performed according to the manufacturer's protocols, with 25 ng
DNA per reaction using an ABI7900 system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). For data analysis, the
manufacturer's instructions were followed (EntroGen). The KRAS/BRAF
Mutation Analysis kit includes primers and probes for detection of
the 18 most common KRAS mutations in codons 12, 13, 61, 117
and 146, and a single BRAF mutation in codon 600 (Table I).
 | Table I.List of KRAS and BRAF
mutations covered by the KRAS/BRAF mutation analysis kit
(EntroGen). |
Table I.
List of KRAS and BRAF
mutations covered by the KRAS/BRAF mutation analysis kit
(EntroGen).
| Gene | Exon | Nt change | aa change |
|---|
| KRAS | 2 | c.34G>C | p.Gly12Arg |
|
|
| c.34G>T | p.Gly12Cys |
|
|
| c.34G>A | p.Gly12Ser |
|
|
| c.35G>T | p.Gly12Val |
|
|
| c.35G>C | p.Gly12Ala |
|
|
| c.35G>A | p.Gly12Asp |
|
|
| c.38G>A | p.Gly13Asp |
|
| 3 | c.182A>G | p.Gln61Arg |
|
|
| c.183A>C | p.Gln61His |
|
|
| c.183A>T | p.Gln61His |
|
|
| c.182A>T | p.Gln61Leu |
|
| 4 | c.351A>C |
p.Lys117Asn |
|
|
| c.351A>T |
p.Lys117Asn |
|
|
| c.350A>G |
p.Lys117Arg |
|
|
| c.349A>G |
p.Lys117Glu |
|
|
| c.436G>A |
p.Ala146Thr |
|
|
| c.436G>C |
p.Ala146Pro |
|
|
| c.437C>T |
p.Ala146Val |
| BRAF | 15 | c.1799T>A |
p.Val600Glu |
Determination of CK20 gene
expression
Reverse transcription-qPCR (RT-qPCR) was performed
using a LightCycler 480 instrument (Roche Applied Science,
Mannheim, Germany). The primers used for amplification of the
target gene, CK20, and a housekeeping gene, glyceraldehyde
3-phosphate dehydrogenase (GAPDH), were as follows: CK20 forward,
5′-CTGAATAAGGTCTTTGATGACC-3′ and reverse,
5′-ATGCTTGTGTAGGCCATCGA-3′; and GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse, 5′-GAAGATGGTGATGGGATTTC-3′,
as described previously by Shen et al (12). RT-qPCR was set up in triplicate in
384-well plates and performed in 10-µl reactions with 2X
LightCycler 480 SYBR Green I Master (Roche Applied Science), 10 µM
of each primer (Applied Biosystems) and 300 ng cDNA. The
thermocycling conditions were as follows: 95°C for 10 min, and 45
cycles of 95°C for 15 sec, 60°C for 45 sec and 72°C for 15 sec,
followed by a melting curve construction by increasing the
temperature from 65–95°C with a temperature transition rate of
0.1°C/sec. A melting curve analysis was performed to verify that
the product consisted of a single amplicon. In each experiment, the
control group consisted of a pool of RNA samples isolated from the
whole blood of healthy volunteers. The data were analyzed by Roche
LightCycler 480 software (Roche Applied Science) using the advanced
relative quantification method, according to the manufacturer
instructions, where the ΔCq values of a control group were used for
normalization.
Statistical analysis
Statistical analyses were performed using IBM SPSS
Statistics Version 20 (IBM SPSS, Armonk, NY, USA). As the dependent
variable could be assumed to be normally distributed, a
Jocheere-Terpstra test was used for testing the difference in the
relative CK20 levels between TNM stages. Fisher's exact test was
used for comparing the presence of specific tumor mutations in the
peripheral blood, vascular invasion and lymphatic infiltration
between TNM stages. A Mann-Whitney test was used for testing the
difference between the levels of CK20 in groups described as R0 and
R2 resections. Fisher's exact test was used for testing the
difference in the change patterns of tumor mutations between the
groups described as R0 and R2 resections. P<0.05 was considered
to indicate a statistically significant difference.
Results
Detection of tumor-specific DNA
mutations
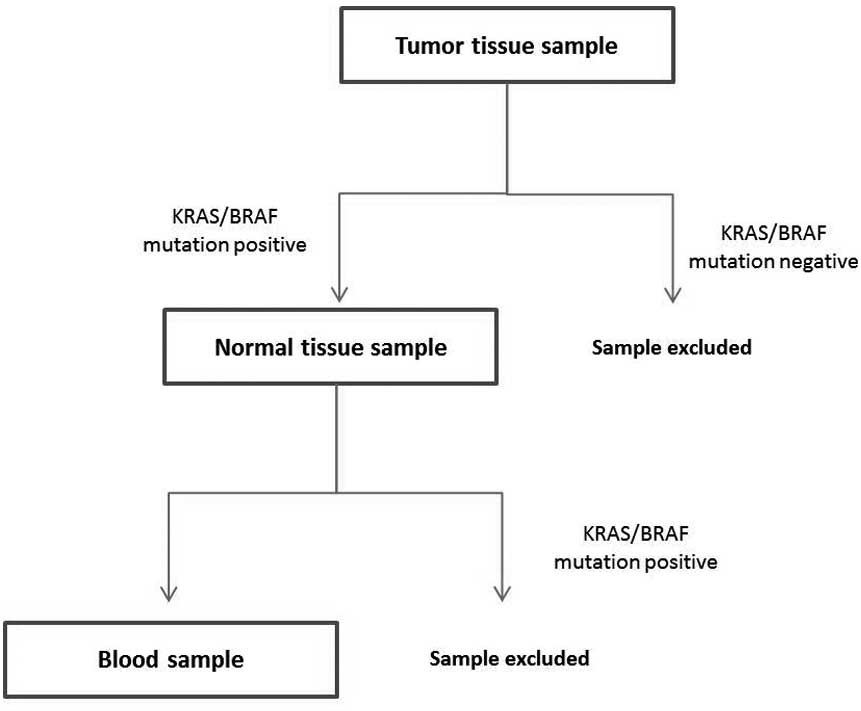
In 35/65 patients, the primary tumor tested positive
for the specific mutations (KRAS mutations in codons 12, 13,
61, 117 and 146, and BRAF mutations in codon 600) (Table II). In all these patients, the
specimen of normal bowel resected with the tumor was also tested
for the presence of the same mutations in order to exclude the
germ-line mutations. Only patients who tested positive for the
specific mutation in the primary tumor were included in further
analysis (Fig. 1).
 | Table II.Type and distribution of
KRAS/BRAF mutations in colorectal cancer
patients. |
Table II.
Type and distribution of
KRAS/BRAF mutations in colorectal cancer
patients.
| Sample | Tumor tissue | Normal tissue | Blood sample
collected prior to surgery | Blood sample
collected 5–7 days after surgery |
|---|
| 1 | p.Gln61Arg | wt | wt | wt |
| 2 | p.Gly12Asp | wt | wt | wt |
| 3 | p.Gly12Asp | wt | wt | wt |
| 4 | p.Gly12Arg | wt | wt | wt |
| 5 | p.Gly12Ser | wt | wt | wt |
| 6 | p.Gly12Asp | wt | wt | wt |
| 7 | p.Gly12Asp | wt | wt | wt |
| 8 | p.Gly12Asp | wt | p.Gly12Asp | wt |
| 9 | p.Gly12Val | wt | p.Gly12Val | wt |
| 10 | p.Gly12Val | wt | wt | wt |
| 11 | p.Val600Glu | wt | wt | wt |
| 12 | p.Val600Glu | wt | wt | wt |
| 13 | p.Gly13Asp | wt | wt | wt |
| 14 | p.Gln61His | wt | wt | p.Gln61His |
| 15 | p.Gly12Asp | wt | p.Gly12Asp | wt |
| 16 | p.Gly12Val | wt | p.Gly12Val | wt |
| 17 | p.Gly12Asp | wt | wt | wt |
| 18 | p.Gly12Asp | wt | wt | wt |
| 19 | p.Gly12Asp | wt | wt | ND |
| 20 | p.Gly12Cys | wt | p.Gly12Cys | p.Gly12Cys |
| 21 | p.Gly12Val | wt | p.Gly12Val | wt |
| 22 | p.Gly12Val | wt | p.Gly12Val | wt |
| 23 | p.Gly12Cys | wt | wt | wt |
| 24 | p.Val600Glu | wt | wt | wt |
| 25 | p.Gln61His | wt | wt | wt |
| 26 | p.Gln61His | wt | wt | wt |
| 27 | p.Gly12Ser | wt | wt | wt |
| 28 | p.Ala146x | wt | p.Ala146x | p.Ala146x |
| 29 | p.Val600Glu | wt | wt | wt |
| 30 | p.Gln61Leu | wt | wt | p.Gln61Leu |
| 31 | p.Gly12Asp | wt | p.Gly12Asp | p.Gly12Asp |
| 32 | p.Gln61His | wt | wt | wt |
| 33 | p.Gly12Alaq | wt | wt | wt |
| 34 | p.Gly12Asp | wt | p.Gly12Asp | wt |
| 35 | p.Ala146x | wt | p.Ala146x | p.Ala146x |
The mean age of the 35 mutation-positive patients
was 68 years (range, 43–83 years), and 21 (60%) of the patients
were male and 14 (40%) were female.
The stage distribution of these 35 patients was 4,
14, 8 and 5 patients in UICC stages I, II, III and IV,
respectively. In 4 patients, the resected specimen turned out to be
a non-malignant adenoma. A total of 14 tumors were located in the
right colon or hepatic flexure, 3 tumors were located in the
splenic flexure or left colon, 11 tumors were located in the
sigmoid colon or in the region of rectosigmoid junction, and 7
tumors were located in the rectum. Only one of the rectal tumors
was treated pre-operatively with 5×5 Gy short-course
radiotherapy.
Regarding the radicality of surgery, there were 30
R0 resections and 5 R2 resections. The R2 resections were all due
to non-resectable liver metastases. Overall, 8 (23%) of the
resections were performed laparoscopically and 27 (77%) were
performed using the classic surgical technique.
The same mutation as in the primary tumor was
detected in the first blood sample of 11 patients (31%). In the
second blood sample, the same mutation as in the tumor was detected
in 6 patients (17%). Additionally, 2 of the patients who were
negative for the mutation in the first blood sample were positive
for the mutation in the second blood sample (Table II).
Tumor stage at the time of
surgery
The median relative levels of CK20 were 1.91, 1.95,
2.05 and 2.17 for tumor stages I, II, III and IV, respectively. The
differences in relative CK20 levels in the peripheral blood between
the different UICC stages were not statistically significant
(P=0.689).
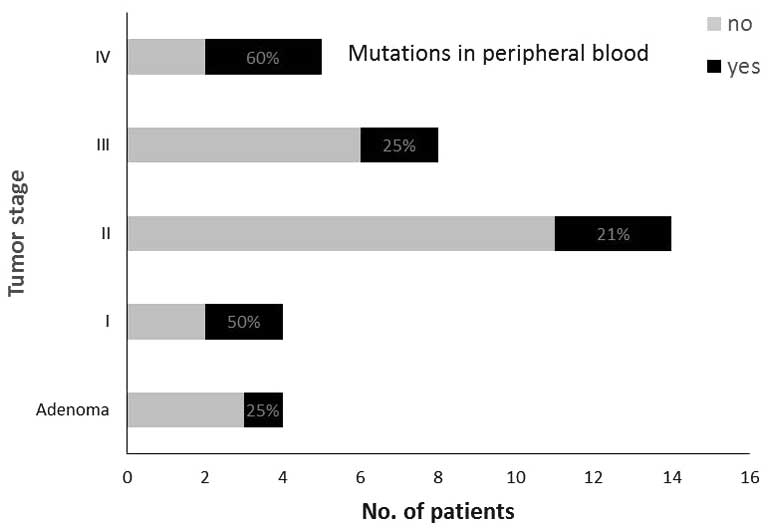
The same specific mutation as in the primary tumors
was found in the peripheral blood of 2 out of 4 (50%) patients in
stage I, in 3 out of 14 patients (21%) in stage II, in 2 out of 8
patients (25%) in stage III and in 3 out of 5 patients (60%) in
stage IV (Fig. 2). Vascular invasion
was reported in 1, 3, 2 and 3 tumor specimens of stages I, II, III
and IV, respectively. Lymphocyte infiltration was reported in 3, 7,
3 and 3 tumor specimens of stages I, II, III and IV, respectively.
The differences between UICC stages were not statistically
significant regarding the proportion of tumor-specific mutations
found in the peripheral blood, vascular invasion or lymphocyte
infiltration (P=0.491, P=0.435, and P=0.717, respectively)
(Table III).
 | Table III.Correlation of tumor-node-metastasis
stages with vascular invasion and lymphocyte infiltration in
primary tumors, as well as with the presence of specific DNA
mutations and CK20 in peripheral blood. |
Table III.
Correlation of tumor-node-metastasis
stages with vascular invasion and lymphocyte infiltration in
primary tumors, as well as with the presence of specific DNA
mutations and CK20 in peripheral blood.
|
| Stage of colorectal
cancer |
|---|
|
|
|
|---|
| Factor | I (n=4) | II (n=14) | III (n=8) | IV (n=5) | P-value |
|---|
| Vascular invasion,
n (%) | 1 (25) | 3 (21) | 2 (25) | 3 (60) | 0.435a |
| Lymphocyte
infiltration, n (%) | 3 (75) | 7 (50) | 3 (38) | 3 (60) | 0.717a |
| DNA mutation
detected | 2 (50) | 3 (21) | 2 (25) | 3 (60) | 0.491a |
| Median (range)
relative CK20 | 1.91
(1.57–3.14) | 1.95
(0.97–3.32) | 2.05
(1.09–3.73) | 2.17
(2.02–3.14) | 0.689b |
In the group of patients with adenomas, the specific
mutation was found in the peripheral blood of 1 out of 4 patients
(25%); the median level of CK20 was 1.68 (range, 1.28–1.84)
(Fig. 2). Information on vascular
invasion or lymphocyte infiltration was not provided in the
pathohistological report.
Residual disease after surgery
In the group of patients with R0 resections, the
specific mutations were found in the peripheral blood of 8/30
patients (27%) prior to surgery and in 3/30 patients (10%) after
surgery. The median levels of CK20 were 1.82 (range, 0.97–3.73)
prior to surgery and 2.34 (range, 1.11–11.29) after surgery. In the
R2 resection group, the specific mutations in the peripheral blood
were detected in 3/5 of the patients (60%) prior to and after
surgery. The median levels of CK20 were 2.16 (range, 2.02–3.14) and
2.60 (range, 2.42–2.87) prior to and after surgery,
respectively.
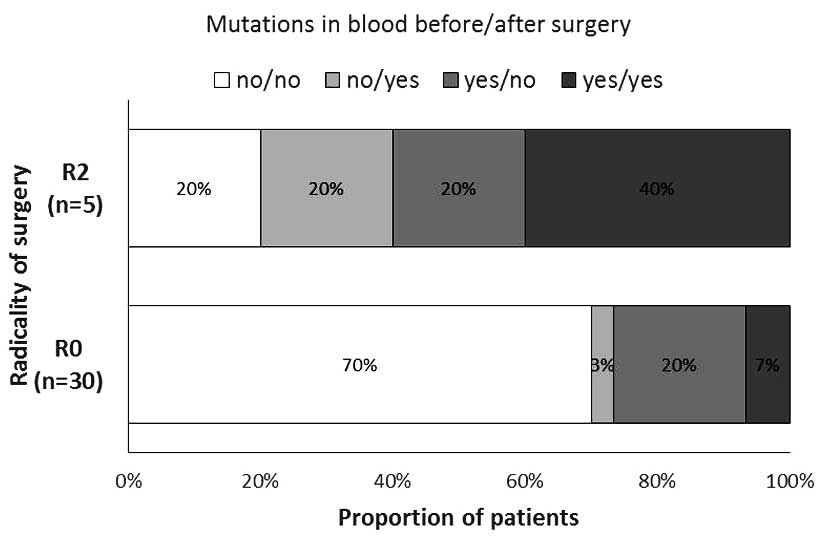
The proportion of patients with the various patterns
of tumor-specific mutations in the blood samples prior to and after
the surgery was significantly different between the groups of
patients described as R0 and R2 resections (P=0.038, Fig. 3). There was a higher proportion of
patients with detected specific tumor mutations in each blood
sample (collected prior to and after the surgery) in the R2
resection group.
The change in CK20 level was not statistically
significantly different between the groups of patients described as
R0 and R2 resection (P=0.671).
Discussion
According to the current understanding of tumor
biology, the dissemination of the primary tumor is a complex
process that occurs rather early in the course of the disease. It
is known that numerous tumor cells are shed daily from the tumor
even at early stages of the disease and that the majority of these
cells die off very quickly in the peripheral circulation (13,14).
Therefore, it was first thought that the detection of circulating
tumor cells (CTCs) in the peripheral blood would be the ultimate
tool for monitoring the activity of the disease. However, the
detection of CTCs turned out to be more complicated than expected.
CTCs are extremely rare cells hidden in the background of numerous
normal blood cells (15). From
studies with proven metastatic CRC, it is known that only a few
CTCs, if any at all, are found in standard blood probes (16). For the detection of CTCs, different
molecular markers have been used, yet only some of them are
specific for CTCs (17,18). It was previously shown that specific
DNA mutations found in tumors can mostly be found in metastases and
also in isolated CTCs (19,20). Therefore, it has been proposed that
circulating free tumor DNA in the peripheral blood may be a good
estimate of CTCs and of disease burden (21–23). It is
expected that there should be no tumor DNA present in the
circulation after the surgical removal of the whole tumor (24). The presence of tumor-specific DNA in
the peripheral blood following primary tumor removal would be an
indication of active metastatic sites in the patient's body, i.e.,
residual disease after surgery.
A few studies performed on CRC patients support the
concept of serological staging by reporting a statistically
significant association between CK20 levels or tumor-specific DNA
mutations in the DNA from the peripheral blood and the tumor stage
(12,25–27).
Therefore, the aim of the present study was to assess whether the
detection of the expression of CK20 and the presence of KRAS
and BRAF tumor-specific mutations in the peripheral blood
can be used to determine the stage of a CRC patient prior to
surgery and the R status after surgery.
However, no statistically significant correlation
could be found between the relative levels of CK20 or the presence
of tumor-specific KRAS or BRAF mutations in DNA from
the peripheral blood prior to surgery and the UICC disease stage.
These results are in concordance with the results of numerous other
studies that also failed to confirm the correlation between CK20,
the presence of KRAS mutated or other tumor-derived DNA in
the blood of CRC patients and the tumor stage (28–35). The
present study was also not able to confirm a statistically
significant correlation between residual disease after surgery, as
described by R category, and CK20 levels in the blood prior to and
following surgery.
However, statistically significant differences were
found with regard to the proportions of patients with detected
tumor-specific DNA mutations in the peripheral blood between the
groups of patients with R0 and R2 resections. Statistical
significance was associated with all patterns (combinations) of
mutation presence in the blood samples prior to and following
surgery. Tumor-specific DNA mutations in the peripheral blood were
more frequently detected in the patients with incomplete surgical
clearance of the tumor due to macroscopic residual disease (R2
resections). In the literature, only scant and indirect data exist
on the correlation of the radicality of surgery and the levels of
molecular markers in the blood in the post-operative period
(33). Studies mainly report solely
on the detection rate of different molecular markers in the blood
after surgery and focus on the tumor recurrence during the
follow-up period (29,32,35). A
difference in pre- and post-operative levels of different molecular
markers in the blood of patients operated upon for CRC has also
been reported, yet with only an indirect correlation to the
radicality of surgery (36,37).
In the present study, tumor-specific DNA showed
greater potential for residual disease detection than CK20. In
theory, tumor-specific DNA also appears to be a more promising
serological marker for tumor staging than non-specific epithelial
markers, such as CK20. The main problem with using the detection of
tumor-specific DNA in the peripheral blood for clinical use is the
fact that there is no single tumor-specific mutation that can be
found in all tumors of the same type (38). For CRC, for example, it is known that
only up to 50% of all tumors have mutated KRAS (39). Even in proven mutated tumors the
entire tumor cell population does not show specific mutations, but
only certain portions of the tumor (40,41). All
of this has to be taken into account when interpreting the
detection rates of specific tumor DNA mutations in the peripheral
blood.
Despite conflicting evidence from previous studies,
as well as the from results of the present study, the detection of
tumor-specific DNA in the peripheral blood remains a promising, but
highly challenging method for estimating the tumor burden at
diagnosis and the residual disease after resection (42). The possibility of serological staging
by the detection of tumor-specific DNA from the peripheral blood
would be of great clinical importance in situations in which no
tumor tissue is available for standard pathohistological staging.
The important clinical issues of the residual disease after
surgery, regression of the tumor after neoadjuvant or adjuvant
treatment and the follow-up on watch-and-wait strategy of CRC
patients with complete response after radiochemotherapy are all not
assessable by pathohistological analysis (43). This fact is also recognized by the
most recent, 7th edition, of the TNM classification, which moved
away from the purely pathological to the clinicopathological
staging in order to improve a number of the clinically apparent
shortcomings of previous editions (44). With evolving diagnostic tools and
treatment modalities, the concept of staging will inevitably drift
away from the pathohistological analysis of the specimens even more
in the future.
Overall, the present study concludes that the
follow-up of somatic KRAS- and BRAF-mutated DNA in
the peripheral blood of CRC patients may be useful in assessing the
surgical clearance of the disease. Regardless of all the
difficulties associated with the improvement of blood-specific
DNA-based staging and the follow-up of CRC, the concept of
serological staging may gain importance in the near future. Until
then, good clinical and pathohistological staging and R category
determination should remain the gold standard upon which all novel
techniques are to be measured.
References
|
1
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Pita-Fernández S, Alhayek-Aí M,
González-Martín C, López-Calviño B, Seoane-Pillado T and
Pértega-Díaz S: Intensive follow-up strategies improve outcomes in
nonmetastatic colorectal cancer patients after curative surgery: A
systematic review and meta-analysis. Ann Oncol. 26:644–656. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Compton CC: Colorectal carcinoma:
Diagnostic, prognostic, and molecular features. Mod Pathol.
16:376–388. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wittekind C, Compton CC, Greene FL and
Sobin LH: TNM residual tumor classification revisited. Cancer.
94:2511–2516. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wittekind C: Problems with residual tumor
classification, particularly R1. Chirurg. 78:785–791. 2007.(In
German). View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greene FL: Cancer staging in outcomes
assessment. J Surg Oncol. 110:616–620. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nicholls J: Commentary. Colorectal Dis.
14:1074–1075. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tsouma A, Aggeli C, Pissimissis N,
Lembessis P, Zografos GN and Koutsilieris M: Circulating tumor
cells in colorectal cancer: Detection methods and clinical
significance. Anticancer Res. 28:3945–3960. 2008.PubMed/NCBI
|
|
9
|
Sergeant G, Penninckx F and Topal B:
Quantitative RT-PCR detection of colorectal tumor cells in
peripheral blood-a systematic review. J Surg Res. 150:144–152.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Peach G, Kim C, Zacharakis E, Purkayastha
S and Ziprin P: Prognostic significance of circulating tumour cells
following surgical resection of colorectal cancers: A systematic
review. Br J Cancer. 102:1327–1334. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rahbari NN, Aigner M, Thorlund K, Mollberg
N, Motschall E, Jensen K, Diener MK, Büchler MW, Koch M and Weitz
J: Meta-analysis shows that detection of circulating tumor cells
indicates poor prognosis in patients with colorectal cancer.
Gastroenterology. 138:1714–1726. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen CX, Hu LH, Xia L and Li YR:
Quantitative real-time RT-PCR detection for survivin, CK20 and CEA
in peripheral blood of colorectal cancer patients. Jpn J Clin
Oncol. 38:770–776. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weinberg RA: Leaving home early:
Reexamination of the canonical models of tumor progression. Cancer
Cell. 14:283–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Klein CA: Cancer. The metastasis cascade.
Science. 321:1785–1787. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Allan AL and Keeney M: Circulating tumor
cell analysis: Technical and statistical considerations for
application to the clinic. J Oncol. 2010:4262182010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cohen SJ, Punt CJ, Iannotti N, Saidman BH,
Sabbath KD, Gabrail NY, Picus J, Morse MA, Mitchell E, Miller MC,
et al: Prognostic significance of circulating tumor cells in
patients with metastatic colorectal cancer. Ann Oncol.
20:1223–1229. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Dotan E, Cohen SJ, Alpaugh KR and Meropol
NJ: Circulating tumor cells: Evolving evidence and future
challenges. Oncologist. 14:1070–1082. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Maheswaran S and Haber DA: Circulating
tumor cells: A window into cancer biology and metastasis. Curr Opin
Genet Dev. 20:96–99. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Knijn N, Mekenkamp LJ, Klomp M,
Vink-Börger ME, Tol J, Teerenstra S, Meijer JW, Tebar M, Riemersma
S, van Krieken JH, et al: KRAS mutation analysis: A comparison
between primary tumours and matched liver metastases in 305
colorectal cancer patients. Br J Cancer. 104:1020–1026. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mostert B, Jiang Y, Sieuwerts AM, Wang H,
Bolt-de Vries J, Biermann K, Kraan J, Lalmahomed Z, van Galen A, de
Weerd V, et al: KRAS and BRAF mutation status in circulating
colorectal tumor cells and their correlation with primary and
metastatic tumor tissue. Int J Cancer. 133:130–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pantel K and Alix-Panabières C: Real-time
liquid biopsy in cancer patients: Fact or fiction? Cancer Res.
73:6384–6388. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bidard FC, Weigelt B and Reis-Filho JS:
Going with the flow: From circulating tumor cells to DNA. Sci
Transl Med. 5:207ps142013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bettegowda C, Sausen M, Leary RJ, Kinde I,
Wang Y, Agrawal N, Bartlett BR, Wang H, Luber B, Alani RM, et al:
Detection of circulating tumor DNA in early- and late-stage human
malignancies. Sci Transl Med. 6:224ra242014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Diehl F, Schmidt K, Choti MA, Romans K,
Goodman S, Li M, Thornton K, Agrawal N, Sokoll L, Szabo SA, et al:
Circulating mutant DNA to assess tumor dynamics. Nat Med.
14:985–990. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
25
|
Iinuma H, Okinaga K, Egami H, Mimori K,
Hayashi N, Nishida K, Adachi M, Mori M and Sasako M: Usefulness and
clinical significance of quantitative real-time RT-PCR to detect
isolated tumor cells in the peripheral blood and tumor drainage
blood of patients with colorectal cancer. Int J Oncol. 28:297–306.
2006.PubMed/NCBI
|
|
26
|
Tsouma A, Aggeli C, Lembessis P, Zografos
GN, Korkolis DP, Pectasides D, Skondra M, Pissimissis N, Tzonou A
and Koutsilieris M: Multiplex RT-PCR-based detections of CEA, CK20
and EGFR in colorectal cancer patients. World J Gastroenterol.
16:5965–5974. 2010.PubMed/NCBI
|
|
27
|
Lin JK, Lin PC, Lin CH, Jiang JK, Yang SH,
Liang WY, Chen WS and Chang SC: Clinical relevance of alterations
in quantity and quality of plasma DNA in colorectal cancer
patients: Based on the mutation spectra detected in primary tumors.
Ann Surg Oncol. 21(Suppl 4): S680–S686. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang JY, Wu CH, Lu CY, Hsieh JS, Wu DC,
Huang SY and Lin SR: Molecular detection of circulating tumor cells
in the peripheral blood of patients with colorectal cancer using
RT-PCR: Significance of the prediction of postoperative metastasis.
World J Surg. 30:1007–1013. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Uen YH, Lu CY, Tsai HL, Yu FJ, Huang MY,
Cheng TL, Lin SR and Wang JY: Persistent presence of postoperative
circulating tumor cells is a poor prognostic factor for patients
with stage I–III colorectal cancer after curative resection. Ann
Surg Oncol. 15:2120–2128. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ryan BM, Lefort F, McManus R, Daly J,
Keeling PW, Weir DG and Kelleher D: A prospective study of
circulating mutant KRAS2 in the serum of patients with colorectal
neoplasia: Strong prognostic indicator in postoperative follow up.
Gut. 52:101–108. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hsieh JS, Lin SR, Chang MY, Chen FM, Lu
CY, Huang TJ, Huang YS, Huang CJ and Wang JY: APC, K-ras, and p53
gene mutations in colorectal cancer patients: Correlation to
clinicopathologic features and postoperative surveillance. Am Surg.
71:336–343. 2005.PubMed/NCBI
|
|
32
|
Frattini M, Gallino G, Signoroni S,
Balestra D, Lusa L, Battaglia L, Sozzi G, Bertario L, Leo E,
Pilotti S and Pierotti MA: Quantitative and qualitative
characterization of plasma DNA identifies primary and recurrent
colorectal cancer. Cancer Lett. 263:170–181. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lecomte T, Ceze N, Dorval E and
Laurent-Puig P: Circulating free tumor DNA and colorectal cancer.
Gastroenterol Clin Biol. 34:662–681. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kopreski MS, Benko FA, Borys DJ, Khan A,
McGarrity TJ and Gocke CD: Somatic mutation screening:
Identification of individuals harboring K-ras mutations with the
use of plasma DNA. J Natl Cancer Inst. 92:918–923. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lu CY, Uen YH, Tsai HL, Chuang SC, Hou MF,
Wu DC, Juo SH, Lin SR and Wang JY: Molecular detection of
persistent postoperative circulating tumour cells in stages II and
III colon cancer patients via multiple blood sampling: Prognostic
significance of detection for early relapse. Br J Cancer.
104:1178–1184. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Allen-Mersh TG, McCullough TK, Patel H,
Wharton RQ, Glover C and Jonas SK: Role of circulating tumour cells
in predicting recurrence after excision of primary colorectal
carcinoma. Br J Surg. 94:96–105. 2007. View
Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lindforss U, Zetterquist H,
Papadogiannakis N and Olivecrona H: Persistence of K-ras mutations
in plasma after colorectal tumor resection. Anticancer Res.
25:657–661. 2005.PubMed/NCBI
|
|
38
|
Wood LD, Parsons DW, Jones S, Lin J,
Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al: The
genomic landscapes of human breast and colorectal cancers. Science.
318:1108–1113. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ličar A, Cerkovnik P and Novaković S:
Distribution of some activating KRAS and BRAF mutations in Slovene
patients with colorectal cancer. Med Oncol. 28:1048–1053. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gerlinger M, Rowan AJ, Horswell S, Larkin
J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A,
Tarpey P, et al: Intratumor heterogeneity and branched evolution
revealed by multiregion sequencing. N Engl J Med. 366:883–892.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bork U, Grützmann R, Rahbari NN, Schölch
S, Distler M, Reissfelder C, Koch M and Weitz J: Prognostic
relevance of minimal residual disease in colorectal cancer. World J
Gastroenterol. 20:10296–10304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Yong E: Cancer biomarkers: Written in
blood. Nature. 511:524–526. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Chapuis PH, Bokey L, Chan C and Dent OF:
Colorectal cancer staging revisited: Time for critical evaluation?
Colorectal Dis. 14:1043–1044. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wittekind C and Oberschmid B: TNM
classification of malignant tumors 2010: General aspects and
amendments in the general section. Pathologe. 31:333–334, 336–338.
2010.(In German). View Article : Google Scholar : PubMed/NCBI
|