Introduction
Colorectal cancer (CRC) is the third most prevalent
cancer in males and the second most prevalent in females worldwide,
with ~55% of CRC cases occurring in developed countries (1). In the USA, there are ~132,700 new CRC
cases every year and the mortality rate remains significant at
~8.4% (2). In China, the incidence of
CRC has rapidly increased, and in 2011, it was the fifth leading
cause of cancer-associated mortality (3). Despite recent advances in treatment and
experimental oncology, the prognosis of patients diagnosed with CRC
remains poor; the 5-year survival rate for metastatic colorectal
cancer is <20% (2) A number of
molecular predictors have been systematically analyzed to determine
whether they may be used for the diagnosis and prognosis of CRC
(4). However, none are accurate
enough for clinical use, and thus have not been adopted by
practitioners.
Long non-coding RNAs (lncRNAs) are >200
nucleotides long and lack the ability to code proteins (5). Until recently, lncRNAs were not
considered to possess any useful biomedical functions (6). However, due to developments in
technology and genomics, an increasing number of studies have
demonstrated that lncRNAs are important in regulating gene
expression (7). Previous studies have
indicated that lncRNAs are associated with post-transcriptional
regulation (8), chromatin remodeling
(9), stem cell function (10), behavior of colonic epithelia (11) and cancer prognosis (12).
HOXA11-AS is a lncRNA located in the HOXA gene
cluster. This gene cluster consists of protein-coding genes and
genes for non-coding RNA, and certain HOXA lncRNAs may function as
biochemical markers in several forms of cancer (13,14).
HOXA11-AS was first discovered in a mouse embryonic cDNA library
using a probe from the sense HOXA11 cDNA sequence (15). The lncRNA is transcribed from the
opposite strand of the protein coding gene HOXA11; HOXA11-AS is
highly conserved and the 5.1 kb transcript is located on chromosome
7p15.2 (16).
It has been demonstrated that HOXA11-AS has the
ability to repress HOXA11 mRNA expression by transcriptional
interference, which is essential for embryo implantation and
endometrial development (17,18). Furthermore, previous studies have
indicated that HOXA11-AS may function as a biomarker for lung
cancer metastasis and poor patient prognosis (19), and that aberrant expression of
HOXA11-AS may be linked to the malignant characteristics of bladder
cancer (20).
However, the association between HOXA11-AS
expression and CRC development has not yet been thoroughly
investigated. Therefore, the present study aimed to measure
HOXA11-AS expression in CRC tissues, investigate the association
between HOXA11-AS levels and clinicopathological features, and
determine whether HOXA11-AS could serve as a viable biomarker for
CRC prognosis.
Materials and methods
Patient samples
All CRC tissues and adjacent non-cancerous tissues
were collected from 84 patients who underwent primary surgical
resection between January 2014 and January 2015 at the Department
of Colorectal and Anal Surgery, The First Affiliated Hospital of
Guangxi Medical University (Nanning, China). No patients received
radiotherapy or chemotherapy prior to surgery. The CRC and adjacent
healthy tissues were immediately preserved in liquid nitrogen and
stored at −80°C until total RNA extraction. The clinicopathological
factors of patients are presented in Table I. The present study was approved by
the Research Ethics Committee of the First Affiliated Hospital of
Guangxi Medical University and written informed consent was
obtained from all patients.
 | Table I.Association between HOXA11-AS
expression and clinicopathological features of colorectal
cancer. |
Table I.
Association between HOXA11-AS
expression and clinicopathological features of colorectal
cancer.
|
|
| HOXA11-AS
expression |
|
|---|
|
|
|
|
|
|---|
| Characteristics | N | Low | High | P-value |
|---|
| Gender |
|
|
| 0.943 |
| Male | 52 | 37 | 15 |
|
|
Female | 32 | 23 | 9 |
|
| Age, years |
|
|
| 0.685 |
|
<50 | 20 | 15 | 5 |
|
| ≥50 | 64 | 45 | 19 |
|
| Tumor diameter,
cm |
|
|
| 0.016a |
|
<5 | 38 | 18 | 14 |
|
| ≥5 | 46 | 42 | 10 |
|
| Tumor site |
|
|
| 0.437 |
|
Colon | 51 | 13 | 38 |
|
|
Rectum | 33 | 11 | 22 |
|
| Tumor
differentiation |
|
|
| 0.685 |
| Well and
moderate | 64 | 19 | 45 |
|
| Poor | 20 | 5 | 15 |
| Depth of
invasion |
|
|
| 0.890 |
|
T1-T2 | 50 | 36 | 14 |
|
|
T3-T4 | 34 | 24 | 10 |
|
| Lymphatic
metastasis |
|
|
| 0.035a |
|
Negative | 34 | 20 | 14 |
|
|
Positive | 50 | 40 | 10 |
|
| Venous
invasion |
|
|
| 0.754 |
|
Absent | 69 | 50 | 19 |
|
|
Present | 15 | 10 | 5 |
|
| Perineural
invasion |
|
|
| 1.000 |
|
Absent | 76 | 54 | 22 |
|
|
Present | 8 | 6 | 2 |
|
| TNM |
|
|
| 0.035a |
|
I–II | 34 | 20 | 14 |
|
|
III–IV | 50 | 40 | 10 |
|
| CEA, ng/ml |
|
|
| 0.015a |
|
<5 | 53 | 33 | 20 |
|
| ≥5 | 31 | 27 | 4 |
|
| CA199, U/ml |
|
|
| 0.770 |
|
<37 | 69 | 50 | 19 |
|
|
≥37 | 14 | 9 | 5 |
|
| Obstruction |
|
|
| 0.153 |
|
Absent | 12 | 6 | 6 |
|
|
Present | 72 | 54 | 18 |
|
Cell lines and culture conditions
Normal human colorectal CCD-18Co cells and human CRC
HCT8, HCT116 and RKO cells were purchased from the Shanghai
Institute of Biochemistry and Cell Biology (Shanghai, China). Cells
were cultured in Dulbecco's modified Eagle's medium (DMEM) with
high glucose (Wisent Biotechnology, Nanjing, China) supplemented
with 10% fetal bovine serum (Excell Bio, Shanghai, China), 100 U/ml
penicillin and 100 mg/ml streptomycin at 37°C in 5%
CO2.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the frozen tumor
tissues, adjacent non-cancerous tissues and cell lines using TRIzol
® reagent (Invitrogen; Thermo Fisher Scientific, Inc.,
Waltham, MA, USA) following the manufacturer's protocol. Next, a
total of 3 µg RNA for each sample was reverse transcribed into cDNA
using DNase I (#EN0521; Thermo Fisher Scientific, Inc.) and a
RevertAid First Strand cDNA Synthesis kit (#K1622; Thermo Fisher
Scientific, Inc.). qPCR was performed using the StepOnePlus™
Real-Time PCR system (ABI7500; Thermo Fisher Scientific, Inc.) and
SYBR ® Green PCR Master Mix (Roche Diagnostics GmbH,
Mannheim, Germany) according to the manufacturer's protocol. The
PCR cycles were as follows: 95°C for 10 min, followed by 40 cycles
of 95°C for 15 sec and 60°C for 1 min. All reactions were run in
triplicate using HOXA11-AS specific primers. β-actin expression
served as the endogenous control, and all samples were normalized
to human β-actin according to the 2− ΔΔ Cq method
(21). HOXA11-AS expression level was
determined by RT-qPCR using the following primer sequences:
HOXA11-AS, forward, 5′-TGCCAAGTTGTACTTACTACGTC-3′, and reverse,
5′-GTTGGAGGAGTAGGAGTATGTA-3′; β-actin, forward,
5′-GCACCACACCTTCTACAATGAGC-3′, and reverse,
5′-GGATAGCACAGCCTGGATAGCAAC-3′.
Statistical analysis
All data were analyzed by SPSS software v16.0 (SPSS,
Inc., Chicago, IL, USA) and GraphPad Prism v5.0 (GraphPad Software,
Inc., La Jolla, CA, USA). Comparisons between groups were performed
using the χ2 test or two-tailed Student's t-test.
Receiver operating characteristic (ROC) curves were constructed to
evaluate the diagnostic value of HOXA11-AS levels by plotting
sensitivity vs. 100% specificity. In all cases, P<0.05 was
considered to indicate a statistically significant difference.
Results
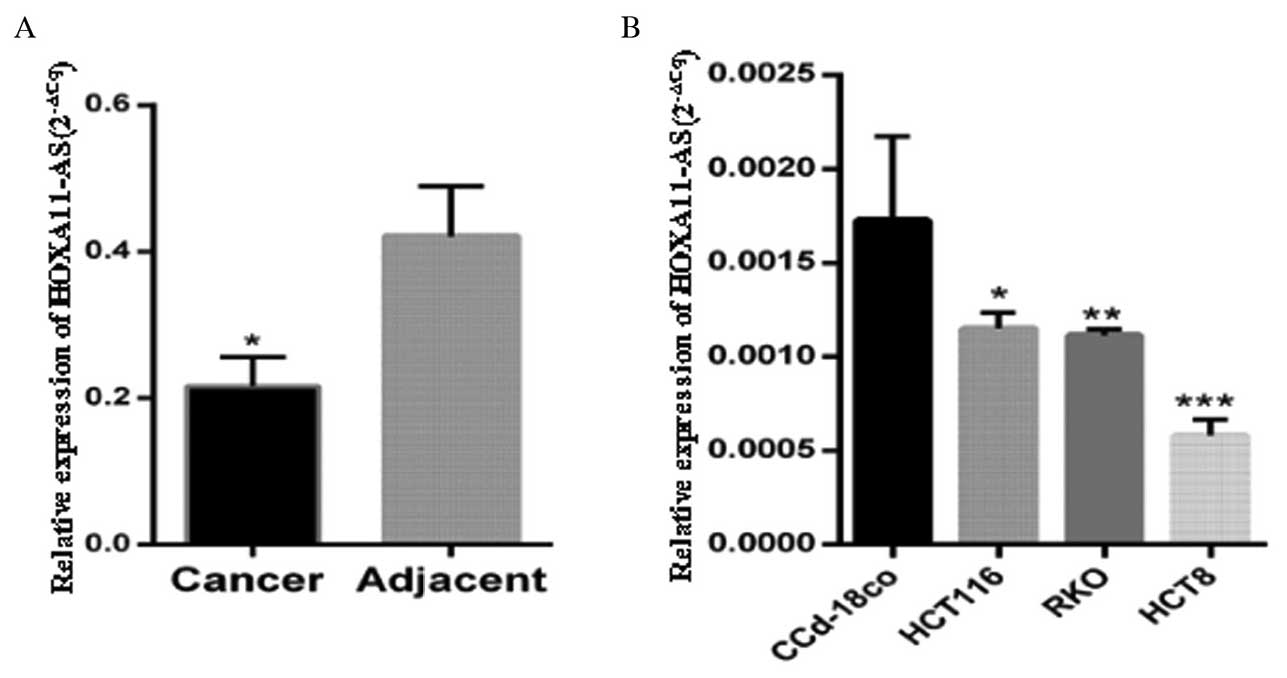
HOXA11-AS was downregulated in the CRC
tissues and cell lines
HOXA11-AS expression was measured in 84 paired CRC
and normal colonic mucosa tissues and observed to be significantly
lower in the CRC tissues compared with the paired normal colonic
mucosa tissues (P<0.05; Fig. 1A).
Furthermore, HOXA11-AS expression was measured in 3 CRC cell lines
(HCT8, RKO and HCT116) and 1 normal colorectal tissue cell line
(CCD-18Co), and was observed to be significantly lower in the CRC
cell lines compared with the normal colorectal tissue cells
(P<0.05; Fig. 1B).
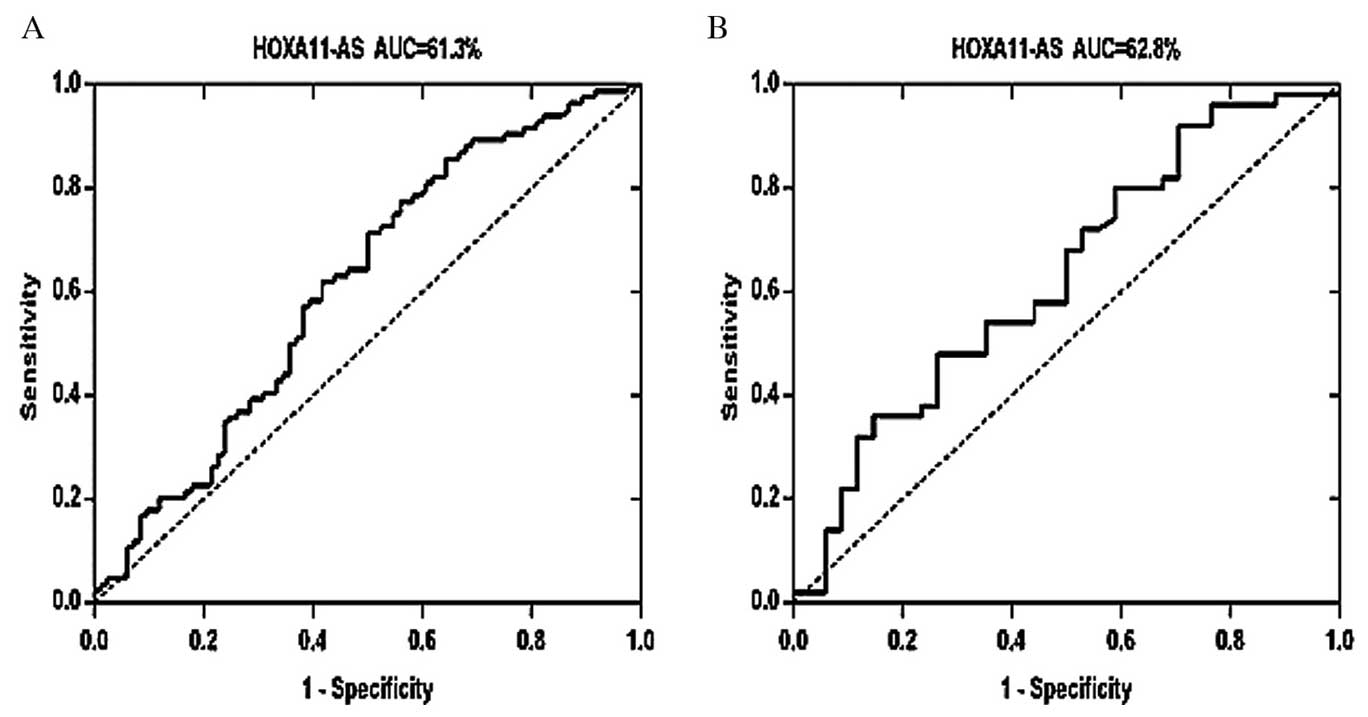
HOXA11-AS expression significantly
differed between the CRC tumor tissues and adjacent normal
tissues
A ROC curve was constructed by grouping all tumor
samples to determine whether HOXA11-AS was able to distinguish CRC
tissue from normal tissue, thus functioning as a biomarker for CRC.
As presented in Fig. 2A, the area
under the curve (AUC) of this ROC was 0.6130 [95% confidence
interval (CI), 0.5277–0.6983; P<0.05], and the cut-off value for
HOXA11-AS was 0.1908. Subsequently, a ROC was constructed using two
groups: CRC samples with lymphatic metastasis and CRC samples
without lymphatic metastasis. The AUC of this ROC was 0.628 (95%
CI, 0.5052–0.7507; P<0.05; Fig.
2B). Each ROC curve suggests that HOXA11-AS has potential
diagnostic value in CRC.
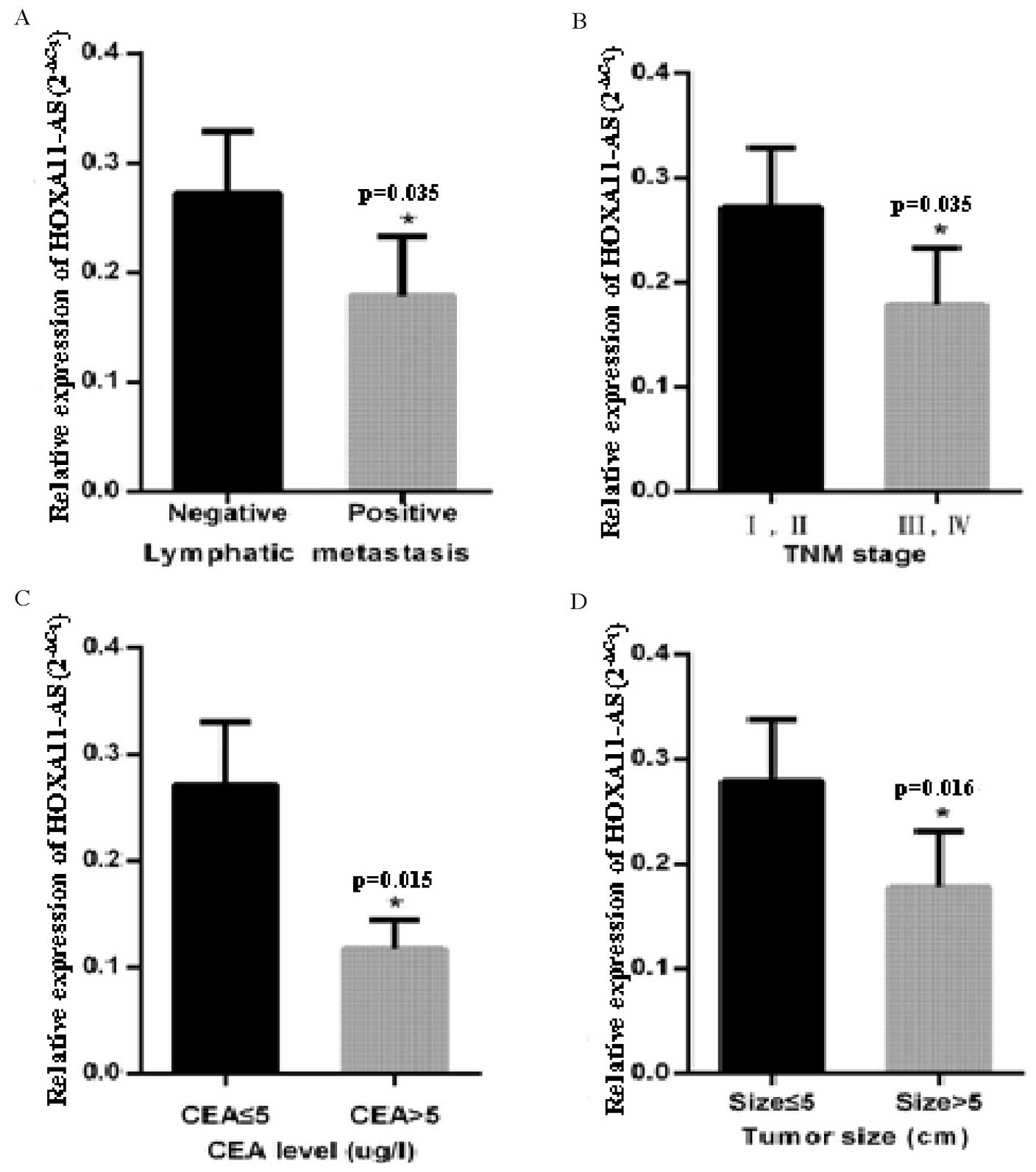
Association between HOXA11-AS
expression and clinicopathological factors in CRC
To investigate the possible correlation between
HOXA11-AS expression and clinicopathological factors, data from all
84 patients were collected and compared. As presented in Table I, decreased expression of HOXA11-AS
was observed in patients with advanced tumor-node-metastasis (TNM)
stage, lymphatic metastasis, large tumor size and a higher
carcinoembryonic antigen (CEA) level compared with other patients
in the corresponding groups (P<0.05). Moreover, Spearman's Rank
correlation coefficient demonstrated that HOXA11-AS expression
level was significantly correlated with advanced TNM stage
(r=−0.23; P<0.05), lymphatic metastasis (r=−0.23; P=0.035),
tumor size (r=−0.264; P<0.05) and CEA level (r=−0.265;
P<0.05) (Fig. 3). No significant
associations were observed between HOXA11-AS expression and other
patient clinicopathological features, including age, tumor
location, gender, histological grade, distant metastasis,
obstruction, depth of invasion, perineural invasion, venous
invasion and carbohydrate antigen 199 level (P>0.05; Table I).
Discussion
lncRNAs are non-coding RNAs that are important in
cancer metastasis and carcinogenesis (22). Previous studies have indicated that
the abnormal expression of certain well-defined lncRNAs, including
HOX transcript antisense RNA, maternally expressed 3 and metastasis
associated lung adenocarcinoma transcript 1, are strongly
correlated with CRC development and progression (23–25).
Therefore, the role of lncRNAs in the development and progression
of CRC is gaining increasing attention (26).
A number of previous studies have investigated how
HOXA11-AS expression affects different types of cancer. A gene
microarray analysis indicated that HOXA11-AS was downregulated in
bladder cancer, suggesting that it may function as a diagnostic
marker for patients with this disease. In addition, HOXA11-AS
expression has been demonstrated to be significantly lower in human
epithelial ovarian cancer (EOC) tissues compared with normal
ovarian tissues, and its upregulated expression may reduce cell
migration, invasion, survival and proliferation in EOC cells
(27). Therefore, such evidence
indicates that HOXA11-AS may serve an important role in
carcinogenesis.
In the current study, HOXA11-AS levels were
investigated in 84 pairs of CRC tissues and adjacent normal tissues
by RT-qPCR. The data obtained demonstrated that the relative
expression level of HOXA11-AS was markedly reduced in the tumor
tissues compared with the adjacent normal tissues. Furthermore,
HOXA11-AS expression was significantly reduced in the CRC HCT8,
HCT116 and RKO cells compared with the normal colon CCD-18Co cells.
The decreased expression of HOXA11-AS in the CRC tissues and
colorectal cells suggests that HOXA11-AS may function as a tumor
suppressor in CRC.
Previous studies have indicated that abnormal
expression of lncRNAs is associated with several
clinicopathological parameters of human cancer, including CEA
level, lymph node metastasis, tumor size, TNM stage and distant
metastasis (28–30). In the current study, the association
between HOXA11-AS expression and different clinicopathological
parameters of CRC tissues was investigated. The results
demonstrated that low HOXA11-AS expression was significantly
correlated with lymph node metastasis, advanced TNM stage, tumor
size and the CEA level of patients with CRC; however, HOXA11-AS
expression was not associated with any other clinicopathological
features. As lymph node metastasis indicates the likelihood of
tumor migration, this suggests that HOXA11-AS may be involved in
suppressing CRC tumorigenesis and metastasis.
However, the results of the current study did not
exhibit a correlation between HOXA11-AS expression and distant
metastasis or depth of invasion. This may be due to the lack of
samples available for subgroup analysis. A recent study of 29 pairs
of lung cancer samples indicated that HOXA11-AS expression levels
were higher in patients with lymph node metastasis and stage III
lung cancer (19), which is
inconsistent with the results of the present study. This may be
attributed to the small number of samples studied, or may be
explained by the notion that the progression of different tumors
may be regulated by different expression of the same lncRNA
(31,32). Further research is required, however,
taken together these results suggest that the function and
contribution of HOXA11-AS may be tumor dependent.
In the current study, a ROC curve was constructed to
distinguish CRC tissues from normal tissues. The AUC area
calculated from the ROC was 0.613, suggesting that HOXA11-AS
expression possesses promising diagnostic value in distinguishing
CRC tissues from normal tissues. HOXA11-AS expression levels also
differed between the CRC tissues with lymph node metastasis and the
CRC tissues without lymph node metastasis with an AUC area of
0.628, thus suggesting that the lncRNA may be used to distinguish
between the two groups. However, this was based on a limited number
of patients and future studies investigating larger patient samples
are required.
In conclusion, the results of the current study
demonstrated that HOXA11-AS expression was significantly decreased
in CRC tissues and certain CRC cell lines. Low HOXA11-AS expression
was significantly correlated with lymph node metastasis, advanced
TNM stage, tumor size and CEA level of patients with CRC. These
findings suggest that HOXA11-AS may function as a potential
prognostic indicator and an adjuvant therapy target in patients
with CRC. However, the molecular mechanisms of HOXA11-AS involved
in malignant phenotypes of CRC require further study.
Acknowledgements
The present study was supported by the Natural
Science Foundation, Guangxi, China (grant no. 2013GXNSFAA019153)
and the Universities Science Foundation, Guangxi, China (grant no.
2013ZD015).
References
|
1
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu S, Zheng R, Zhang M, Zhang S, Sun X
and Chen W: Incidence and mortality of colorectal cancer in China,
2011. Chin J Cancer Res. 27:22–28. 2015.PubMed/NCBI
|
|
4
|
Yan Y, Shen Z, Ye Y, Jiang K, Zhang H,
Shen C, Mustonen H, Puolakkainen P and Wang S: A novel molecular
marker of prognosis in colorectal cancer: Vasohibin-1. Med Oncol.
31:8162014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Derrien T, Johnson R, Bussotti G, Tanzer
A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG,
et al: The GENCODE v7 catalog of human long noncoding RNAs:
Analysis of their gene structure, evolution, and expression. Genome
Res. 22:1775–1789. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang KC and Chang HY: Molecular mechanisms
of long noncoding RNAs. Mol Cell. 43:904–914. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kornienko AE, Guenzl PM, Barlow DP and
Pauler FM: Gene regulation by the act of long non-coding RNA
transcription. BMC Biol. 11:592013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guttman M, Donaghey J, Carey BW, Garber M,
Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L, et al:
lincRNAs act in the circuitry controlling pluripotency and
differentiation. Nature. 477:295–300. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Franklin JL, Rankin CR, Levy S, Snoddy JR,
Zhang B, Washington MK, Thomson JM, Whitehead RH and Coffey RJ:
Malignant transformation of colonic epithelial cells by a
colon-derived long noncoding RNA. Biochem Biophys Res Commun.
440:99–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Jatkoe T, Zhang Y, Mutch MG,
Talantov D, Jiang J, McLeod HL and Atkins D: Gene expression
profiles and molecular markers to predict recurrence of Dukes' B
colon cancer. J Clin Oncol. 22:1564–1571. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu MX, Chen X, Chen G, Cui QH and Yan GY:
A computational framework to infer human disease-associated long
noncoding RNAs. PLoS One. 9:e844082014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu Y, Zhang L, Wang Y, Li H, Ren X, Wei F,
Yu W, Wang X, Zhang L, Yu J and Hao X: Long noncoding RNA HOTAIR
involvement in cancer. Tumour Biol. 35:9531–9538. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chau YM, Pando S and Taylor HS: HOXA11
silencing and endogenous HOXA11 antisense ribonucleic acid in the
uterine endometrium. J Clin Endocrinol Metab. 87:2674–2680. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Richards EJ, Permuth-Wey J, Li Y, Chen YA,
Coppola D, Reid BM, Lin HY, Teer JK, Berchuck A, Birrer MJ, et al:
A functional variant in HOXA11-AS, a novel long non-coding RNA,
inhibits the oncogenic phenotype of epithelial ovarian cancer.
Oncotarget. 6:34745–34757. 2015.PubMed/NCBI
|
|
17
|
Potter SS and Branford WW: Evolutionary
conservation and tissue-specific processing of Hoxa 11 antisense
transcripts. Mamm Genome. 9:799–806. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu H, Lindsay J, Feng ZP, Frankenberg S,
Hu Y, Carone D, Shaw G, Pask AJ, O'Neill R, Papenfuss AT and
Renfree MB: Evolution of coding and non-coding genes in HOX
clusters of a marsupial. BMC Genomics. 13:2512012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liang J, Lv J and Liu Z: Identification of
stage-specific biomarkers in lung adenocarcinoma based on RNA-seq
data. Tumour Biol. 36:6391–6399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo H, Zhao X, Wan X, Huang S and Wu D:
Gene microarray analysis of the lncRNA expression profile in human
urothelial carcinoma of the bladder. Int J Clin Exp Med.
7:1244–1254. 2014.PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Svoboda M, Slyskova J, Schneiderova M,
Makovicky P, Bielik L, Levy M, Lipska L, Hemmelova B, Kala Z,
Protivankova M, et al: HOTAIR long non-coding RNA is a negative
prognostic factor not only in primary tumors, but also in the blood
of colorectal cancer patients. Carcinogenesis. 35:1510–1515. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yin DD, Liu ZJ, Zhang E, Kong R, Zhang ZH
and Guo RH: Decreased expression of long noncoding RNA MEG3 affects
cell proliferation and predicts a poor prognosis in patients with
colorectal cancer. Tumour Biol. 36:4851–4859. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yang MH, Hu ZY, Xu C, Xie LY, Wang XY,
Chen SY and Li ZG: MALAT1 promotes colorectal cancer cell
proliferation/migration/invasion via PRKA kinase anchor protein 9.
Biochim Biophys Acta. 1852:166–174. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ma H, Hao Y, Dong X, Gong Q, Chen J, Zhang
J and Tian W: Molecular mechanisms and function prediction of long
noncoding RNA. ScientificWorldJournal. 2012:5417862012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Matei D, Fang F, Shen C, Schilder J,
Arnold A, Zeng Y, Berry WA, Huang T and Nephew KP: Epigenetic
resensitization to platinum in ovarian cancer. Cancer Res.
72:2197–2205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ye Z, Zhou M, Tian B, Wu B and Li J:
Expression of lncRNA-CCAT1, E-cadherin and N-cadherin in colorectal
cancer and its clinical significance. Int J Clin Exp Med.
8:3707–3715. 2015.PubMed/NCBI
|
|
29
|
Shi D, Zheng H, Zhuo C, Peng J, Li D, Xu
Y, Li X, Cai G and Cai S: Low expression of novel lncRNA
RP11-462C24.1 suggests a biomarker of poor prognosis in colorectal
cancer. Med Oncol. 31:312014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Qi P, Xu MD, Ni SJ, Huang D, Wei P, Tan C,
Zhou XY and Du X: Low expression of LOC285194 is associated with
poor prognosis in colorectal cancer. J Transl Med. 11:1222013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang EB, Yin DD, Sun M, Kong R, Liu XH,
You LH, Han L, Xia R, Wang KM, Yang JS, et al: P53-regulated long
non-coding RNA TUG1 affects cell proliferation in human non-small
cell lung cancer, partly through epigenetically regulating HOXB7
expression. Cell Death Dis. 5:e12432014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang MD, Chen WM, Qi FZ, Sun M, Xu TP, Ma
P and Shu YQ: Long non-coding RNA TUG1 is up-regulated in
hepatocellular carcinoma and promotes cell growth and apoptosis by
epigenetically silencing of KLF2. Mol Cancer. 14:1652015.
View Article : Google Scholar : PubMed/NCBI
|