Introduction
Irinotecan, also termed Camptosar and CPT-11, is a
type of semi-artificial camptothecin compound that is one of the
effective cytotoxic antineoplastic agents used to treat colorectal
cancer (1). Irinotecan can be
activated to SN-38 subsequent to hydrolysis of carboxylesterase
in vivo, and derived into an inhibitor of topoisomerase I,
which can synchronously be inactivated into SN-38G by UDP
glucuronosyltransferase family 1 member A complex locus (UGT1A)
(2). Thus, irinotecan interrupts the
replication and transcription of DNA by preventing the relegation
of single-stranded DNA and causes the death of tumor cells
following a series of biotransformations. Irinotecan received
authorization from the US Food and Drug Administration (FDA) in
1998, and has been approved as the primary therapy of metastatic
adult colorectal cancer, combined with 5-fluorouracil (5-FU) and
leucovorin (LV) (3). However, severe
neutropenia and diarrhea, the primary toxicities of irinotecan,
have restrained the dose and clinical efficiency of irinotecan
(4). It has been reported that the
curative effect and negative toxicity are each dependent on
ethnicity (5–7).
The UGT1A gene family has 13 members in Homo
sapiens, and is located on Chr2 (2q37). Each member is composed
of a unique exon 1, followed by four common exons (exons 2–5).
Therefore, each functional protein of the UGT1A family has a
specific N-terminus, which is determined by the first exon
sequence, to recognize the corresponding substrate. In addition,
the last four mutual exons provide the identical C termini, which
consists of 245 amino acids and guarantee the combination with
uridine diphosphate glucuronic acid (8). UGT1A1, UGT1A7 and UGT1A9 have been shown
to be the major effective factors during the inactivation of SN-38
(9,10), and genetic polymorphism in the UGT1A
family has also been shown to affect the variability of irinotecan
toxicity during clinical application (11,12).
Considering the complex mechanism involved in the
association between the UGT1A family and toxicity of irinotecan,
the FDA has instructed the manufacturer of irinotecan to revise the
label and add the caution of relative toxicity and dose titration
in patients with the UGT1A1*28 polymorphism, as well as the advice
for patients to undergo allelic detection (13). For patients from the Asian population,
UGT1A1*6 is considered to be another potential risk factor that is
responsible for the toxicity among the Asian population in addition
to the UGT1A1*28 allele (11,12,14). The
Japanese Pharmaceuticals and Medical Devices Agency package insert
for irinotecan states that individuals who are homozygous for the
UGT1A1*6 or UCT1A1*28 alleles, or heterozygous with the
UGT1A1*6/*28 genotype, are at an increased risk of severe adverse
events, particularly neutropenia (15). The impact of UGT1A7 polymorphism on
irinotecan-induced toxicity has also been well-studied (16–18). In
addition, UGT1A9*22 has been reported to be associated with the
toxic potency of irinotecan (18–20).
Considering the ethnicity-specific distribution of genetic
polymorphisms, the predictive markers for severe toxicity in
Chinese patients treated with irinotecan-based regimens have not
been affirmatively reported by studies yet.
In the present study, direct sequencing was adopted
to avoid ethnic heterogeneity and to identify novel variations. The
first exon regions of UGT1A1, UGT1A7 and UGT1A9 were sequenced, and
comprehensive analysis of genetic polymorphisms was performed to
determine the association between inherited genetic variations and
irinotecan-induced toxicity. The major aim of the current study was
to identify toxicity predictable markers of Chinese metastatic
gastrointestinal cancer patients treated with irinotecan-based
regimens.
Patients and methods
Patients
In total, 70 patients with histologically-confirmed
metastatic gastrointestinal cancer were enrolled from the Cancer
Institute and Hospital, Chinese Academy of Medical Sciences
(Beijing, China) between January 2012 and December 2014. All
patients were treated with the irinotecan-based folinic acid,
fluorouracil (5-FU) and irinotecan (FOLFIRI) or capecitabine and
irinotecan (XELIRI) regimens. The study protocol was approved by
the Ethics Committee of Beijing Chao-Yang Sanhuan Cancer Hospital
(Beijing, China), which is affiliated with the Cancer Institute and
Hospital of the Chinese Academy of Medical Sciences. Written
informed consent was obtained from all patients prior to entering
the study.
Eligibility criteria
All patients were diagnosed with
histologically-confirmed gastrointestinal cancer and were
undergoing their first irinotecan-based chemotherapy. The patients
were aged >18 years and had an Eastern Cooperative Oncology
Group (ECOG) performance status (PS) of 0–2 and a life expectancy
of ≥3 months.
Chemotherapy
The patients were treated with one of the following
FOLFIRI-based regimens until progressive disease (PD), unacceptable
toxicity, patient refusal or a medical decision to discontinue
treatment. Patients treated with the FOLFIRI regimen received 180
mg/m2 irinotecan intravenously (IV), 400
mg/m2 LV IV and 400 mg/m2 5-FU bolus,
followed by 2,400 mg/m2 5-FU IV, all administered on day
1, every 2 weeks. Patients treated with the XELIRI regimen received
120 mg/m2 irinotecan IV on days 1 and 8, and 800
mg/m2 oral capecitabine twice per day on days 1–14 every
3 weeks.
Genotyping and genetic analysis
Peripheral venous blood samples were obtained from
70 patients with metastatic gastrointestinal cancer, who were
treated with irinotecan-based regimens (37 with FOLFIRI and 33 with
XELIRI) for isolation of genomic DNA. Genomic DNA was isolated
using the QIAamp DNA blood mini kit (Qiagen, Inc., Valencia, CA,
USA). The promoter (−1000 bp) and exon 1 regions of UGT1A1, UGT1A7
and UGT1A9 were resequenced to screen for single nucleotide
polymorphisms (SNPs), using the DYEnamic ET terminator cycle
sequencing kit (GE Healthcare, Chalfont St. Giles, UK) on the ABI
Prism 3730×l DNA analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). Polymerase chain reaction
(PCR) was performed in a final volume of 15 µl, containing 30 ng
genomic DNA, 1*PCR Takara buffer (Mg2+ plus; Takara
Biotechnology Co., Ltd., Dalian, China), 2.5 pmol of each primer,
25 pmol deoxynucleotide triphosphates and 1 U of Taq DNA polymerase
(Takara Biotechnology Co., Ltd.). Following pre-denaturation at
93°C for 3 min, amplification was performed under the following
conditions for 32 cycles: Denaturation at 95°C for 30 sec;
annealing at 58°C for 40 sec; and extension at 72°C for 2 min. The
primer sequences used are shown in Table
I (21).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Genes | Primer ID | Primer sequences |
|---|
| UGT1A1 | U1_E1AF |
TCGTCCTTCTTCCTCTCTGG |
|
| U1_E1AR |
GCAGTGCATGCAAGAAGAAT |
|
| U1_E1BF |
TGTCTGGCTGTTCCCACTTA |
|
| U1_E1BR |
CCAGAAGATGATGCCAAAGA |
|
| U1_PF |
GGTCATTCTCTACCCCAGCA |
|
| U1_PR |
AAAGCTGTCAGTCCACAAAGG |
| UGT1A7 | U7_E1AF |
AAACTCATATTGCAGCACAGG |
|
| U7_E1AR |
AAGTCAAAAATACCATTGGATGAA |
|
| U7_E1BF |
GGAAGATCACTGAATTGCACAG |
|
| U7_E1BR |
TTCCTCTGGGGGTAGTGTAGAA |
|
| U7_PF |
TCTTTCCGTCGAACATGAGA |
|
| U7_PR |
CACATTCACTGCCAATGATTTA |
| UGT1A9 | U9_E1AF |
CCAAGGCAAAGACCATAAGCTA |
|
| U9_E1AR |
CAAACTCCTGCAATTTGAAAAA |
|
| U9_E1BF |
CATATACCCTGGAGGATCTGGA |
|
| U9_E1BR |
CTGACGAGTACACGCATTGG |
|
| U9_PF |
CCTCTGACCTCAAGGAGTGC |
|
| U9_PR |
CAATGATTTACCCAAAAGAACAAG |
Base calling, quality assessment and polymorphism
determination from DNA sequencing were analyzed using Phred, Phrap,
Consed (http://www.phrap.org/phredphrapconsed.html) and
PolyPhred (http://droog.gs.washington.edu/polyphred/). Haploview
4.2 (Broad Institute of MIT and Harvard, Cambridge, MA, USA
(22) was used to estimate allele
frequencies, test the Hardy-Weinberg equilibrium, measure pairwise
linkage disequilibrium (LD), establish haplotypes, estimate
haplotype frequency, and analyze the association between the
haplotypes and toxicity. LD blocks were defined by the ‘solid spine
of LD’ algorithm, and LD structure was exhibited using GOLD heatmap
color scheme (23,24). The allele designations were defined
according to the UGT Allele Nomenclature Committee (25).
Statistical analysis
Patients underwent baseline evaluations, including
physical examination, complete medical history, ECOG PS, complete
blood count, and hepatic and renal function tests. Toxicity
evaluations were performed according to National Cancer Institute
Common Terminology Criteria for Adverse Events version 3.0
criteria. Associations between the clinical features and toxicity
were analyzed by two-tailed Fisher's exact test or Student's
t-test. In addition, associations between the alleles or genotypes
and toxicity were analyzed by two-tailed Fisher's exact test.
P<0.05 was considered to indicate a statistically significant
difference, as calculated by PLINK v1.07 (http://pngu.mgh.harvard.edu/~purcell/plink) (26). Due to the exploratory nature of the
present study, no adjustments were made for multiple
comparisons.
Results
In total, 70 patients with histologically-confirmed
metastatic gastrointestinal cancer, consisting of 37 patients with
metastatic colorectal cancer and 33 patients with metastatic
gastric cancer, were enrolled in the present study. The rates of
grade 3–4 toxicity were as follows: Neutropenia, 18.57% (n=13); and
diarrhea, 10% (n=7). These results are in accordance with a
previous study of Asian patients (6).
The clinical features of patients and the incidence of severe
toxicity of the irinotecan-based regimens FOLFIRI and XELIRI are
summarized in Table II.
 | Table II.Clinical features of 70 patients with
histologically-confirmed metastatic gastrointestinal cancer. |
Table II.
Clinical features of 70 patients with
histologically-confirmed metastatic gastrointestinal cancer.
| Features | Value, n (%) |
|---|
| Age, years |
|
|
Median | 54 |
|
Range | 25–77 |
| Gender |
|
|
Male | 48 (68.57) |
|
Female | 22 (31.43) |
| Tumor location |
|
|
Colon | 37 (52.86) |
|
Stomach | 33 (47.14) |
| ECOG PS |
|
| 0 | 19 (27.14) |
| 1 | 46 (65.71) |
| 2 | 5 (7.14) |
| TNM stage IV | 70
(100.00) |
| Regimen |
|
|
FOLFIRI | 37 (52.86) |
|
XELIRI | 33 (47.14) |
| Neutropenia grade
3–4 | 13 (18.57) |
| Diarrhea grade
3–4 | 7
(10.00) |
A total of 23 different genetic variants were
detected in the present study, all of the variants were consistent
with the Hardy-Weinberg equilibrium (P>0.05). Genetic variations
in the UGT1A1, UGT1A7 and UGT1A9 genes were compared with the
genomic reference sequence AF039138.1. The amino acids were located
in the reference sequences NP_000454.1 (UGT1A1), NP_061950.2
(UGTIA7) and NP_066307.1 (UGTIA9). Of these variants, 2 novel
polymorphisms were identified: M06 and M09, which are located in
the promoter regions of UGT1A9 and UGT1A7, respectively. UGT1A1*6,
which is found only in Asian populations, had the highest frequency
[minor allele frequency (MAF), 22.5%] of the SNPs in UGT1A1 in the
present study. The frequency of the UGT1A1*28 SNP (MAF, 9.3%) was
comparable to the frequency previously found in the Asian
population (MAF, 7.0%), and was much lower compared with the
frequency in Caucasian patients (MAF, 31.6%), as previously
reported (11,20). Other known UGT1A alleles, UGT1A1*80,
UGT1A1*81, UGT1A7*2, UGT1A7*3 and UGT1A9*1b (UGT1A9*22), were found
concurrently. Table III lists the
summary of identified polymorphisms in UGT1A1, UGT1A7 and
UGT1A9.
 | Table III.Summary of identified polymorphisms
in UGT1A1, UGT1A7 and UGT1A9. |
Table III.
Summary of identified polymorphisms
in UGT1A1, UGT1A7 and UGT1A9.
| ID | Location | db SNP ID | Change | Allele | Function | MAF | H-Wpval |
|---|
| M01 | UGT1A9
promoter | rs3806598 | A>C | – | – | 0.243 | 0.747 |
| M02 | UGT1A9
promoter | rs45440791 | T>C | – | – | 0.007 | 1.000 |
| M03 | UGT1A9
promoter | rs2741045 | C>T | – | – | 0.007 | 1.000 |
| M04 | UGT1A9
promoter | rs2741046 | T>A | – | – | 0.007 | 1.000 |
| M05 | UGT1A9
promoter | rs3832043 | T9>T10 | *1b | – | 0.616 | 0.818 |
| M06 | UGT1A9
promoter | – | C>T | – | – | 0.007 | 1.000 |
| M07 | UGT1A9 exon 1 | rs151216459 | G>T | – | Synonymous | 0.014 | 1.000 |
| M08 | UGT1A7
promoter | rs4530361 | A>G | – | – | 0.250 | 0.633 |
| M09 | UGT1A7
promoter | – | T>C | – | – | 0.007 | 1.000 |
| M10 | UGT1A7
promoter | rs28946877 | C>T | – | – | 0.143 | 1.000 |
| M11 | UGT1A7
promoter | rs7586110 | T>G | – | – | 0.246 | 0.296 |
| M12 | UGT1A7 exon 1 | rs7577677 | C>A | – | Synonymous | 0.243 | 0.314 |
| M13 | UGT1A7 exon 1 | rs17868323 | T>G | *2&*3 | N129K | 0.407 | 1.000 |
| M14 | UGT1A7 exon 1 | rs17863778 | C>A | *2&*3 | R131K | 0.407 | 1.000 |
| M15 | UGT1A7 exon 1 | rs17868324 | G>A | *2&*3 | R131Q | 0.407 | 1.000 |
| M16 | UGT1A7 exon 1 | rs139969318 | A>G | – | Synonymous | 0.007 | 1.000 |
| M17 | UGT1A7 exon 1 | rs11692021 | T>C | *3 | W208R | 0.25 | 0.633 |
| M18 | UGT1A7 exon 1 | rs45462096 | C>T | – | Synonymous | 0.014 | 1.000 |
| M19 | UGT1A7 exon 1 | rs17864686 | G>A | – | Synonymous | 0.157 | 1.000 |
| M20 | UGT1A1
promoter | rs887829 | C>T | *80 | – | 0.107 | 1.000 |
| M21 | UGT1A1
promoter | rs873478 | G>C | *81 | – | 0.036 | 1.000 |
| M22 | UGT1A1
promoter | rs3064744 | TA6>TA7 | *28 | – | 0.093 | 1.000 |
| M23 | UGT1A1 exon 1 | rs4148323 | G>A | *6 | G71R | 0.225 | 0.550 |
In total, 6 SNPs exhibited an association with grade
3–4 neutropenia, including M01 (P=0.009), M08 (P=0.011), M10
(P=0.025), M17 (P=0.011), M19 (P=0.014) and M23 (P=0.016) (Table IV). In addition, a significant
association was observed between the UGT1A9*1b polymorphism (M05)
and severe diarrhea (P= 0.045; Table
IV). Of these 7 SNPs, two are functional mutations, consisting
of UGT1A7*3 (P=0.011; OR, 3.391) and UGT1A1*6 (P=0.016; OR, 3.373).
The hotspot allele UGT1A1*28, which has been previously reported
(20), was not significantly
associated with grade 3–4 neutropenia or severe diarrhea (P=1.000
and 0.620, respectively; Table IV).
The present study established haplotypes of those 7 SNPs and
calculated the association with irinotecan-induced toxicity
(Table V). Of the 7 haplotypes that
were assessed, haplotypes H03, H04, H05 and H06 were observed to be
associated with a higher risk of grade 3–4 neutropenia (P=0.037,
P=0.043, P=0.097 and P=0.004, respectively). In addition, H04 was
found to be associated with grade 3–4 diarrhea (P=0.001).
 | Table IV.Association between UGT1A
polymorphisms and severe toxicity. |
Table IV.
Association between UGT1A
polymorphisms and severe toxicity.
|
|
| Neutropenia grade
3–4 | Diarrhea grade
3–4 |
|---|
|
|
|
|
|
|---|
| SNP ID | db SNP ID | P-value | OR | 95% CI | P-value | OR | 95% CI |
|---|
| M01 | rs3806598 | 0.009 | 3.584 | 1.457–8.821 | 0.745 | 1.280 | 0.374–4.379 |
| M02 | rs45440791 | 1.000 | 0.000 | NA | 1.000 | 0.000 | NA |
| M03 | rs2741045 | 1.000 | 0.000 | NA | 0.100 | NA | NA |
| M04 | rs2741046 | 1.000 | 0.000 | NA | 0.100 | NA | NA |
| M05 | rs3832043 | 1.000 | 1.003 | 0.417–2.411 | 0.045 | 3.273 | 1.033–10.37 |
| M06 | – | 0.186 | NA | NA | 1.000 | 0.000 | NA |
| M07 | rs151216459 | 1.000 | 0.000 | NA | 1.000 | 0.000 | NA |
| M08 | rs4530361 | 0.011 | 3.391 | 1.384–8.312 | 0.339 | 1.778 | 0.553–5.714 |
| M09 | – | 1.000 | 0.000 | NA | 0.100 | NA | NA |
| M10 | rs28946877 | 0.025 | 0.000 | NA | 1.000 | 1.000 | 0.206–4.845 |
| M11 | rs7586110 | 0.081 | 2.292 | 0.923–5.693 | 1.000 | 0.818 | 0.214–3.124 |
| M12 | rs7577677 | 0.077 | 2.344 | 0.944–5.819 | 1.000 | 0.836 | 0.219–3.19 |
| M13 | rs17868323 | 0.659 | 1.314 | 0.558–3.099 | 0.084 | 2.925 | 0.925–9.245 |
| M14 | rs66534818 | 0.659 | 1.314 | 0.558–3.099 | 0.084 | 2.925 | 0.925–9.245 |
| M15 | rs17868324 | 0.659 | 1.314 | 0.558–3.099 | 0.084 | 2.925 | 0.925–9.245 |
| M16 | rs139969318 | 0.186 | NA | NA | 1.000 | 0.000 | NA |
| M17 | rs11692021 | 0.011 | 3.391 | 1.384–8.312 | 0.339 | 1.778 | 0.553–5.714 |
| M18 | rs45462096 | 1.000 | 0.000 | NA | 1.000 | 0.000 | NA |
| M19 | rs17864686 | 0.014 | 0.000 | NA | 0.236 | 2.400 | 0.679–8.481 |
| M20 | rs887829 | 0.737 | 0.647 | 0.137–3.062 | 0.647 | 1.449 | 0.292–7.199 |
| M21 | rs873478 | 0.584 | 0.000 | NA | 0.414 | 2.346 | 0.244–22.59 |
| M22 | rs3064744 | 1.000 | 0.780 | 0.162–3.754 | 0.620 | 1.742 | 0.345–8.802 |
| M23 | rs4148323 | 0.016 | 3.373 | 1.350–8.431 | 0.191 | 0.241 | 0.03–1.92 |
 | Table V.Correlation between UGT1A haplotypes
and severe toxicity. |
Table V.
Correlation between UGT1A haplotypes
and severe toxicity.
|
| UGT1A
haplotype |
| P-value |
|---|
|
|
|
|
|
|---|
| Haplotype ID | M01 | M05 | M08 | M10 | M17 | M19 | M23 | Frequency | Neutropenia grade
3–4 | Diarrhea grade
3–4 |
|---|
| H01 | A | 10T | A | C | T | G | G | 0.555 | 0.269 | 0.114 |
| H02 | C | 9T | G | C | C | G | A | 0.176 | 0.183 | 0.274 |
| H03 | A | 9T | A | T | T | A | G | 0.120 | 0.037 | 0.783 |
| H04 | C | 9T | G | C | C | G | G | 0.044 | 0.043 | 0.001 |
| H05 | A | 10T | A | C | T | G | A | 0.030 | 0.097 | 0.502 |
| H06 | C | 10T | G | C | C | G | A | 0.015 | 0.004 | 0.629 |
| H07 | A | 9T | G | C | C | G | G | 0.014 | 0.494 | 0.059 |
Pairwise LD analysis was performed with the detected
variations having a MAF >10%. LD blocks were defined by the
‘solid spine of LD’ algorithm, and LD structure was exhibited by
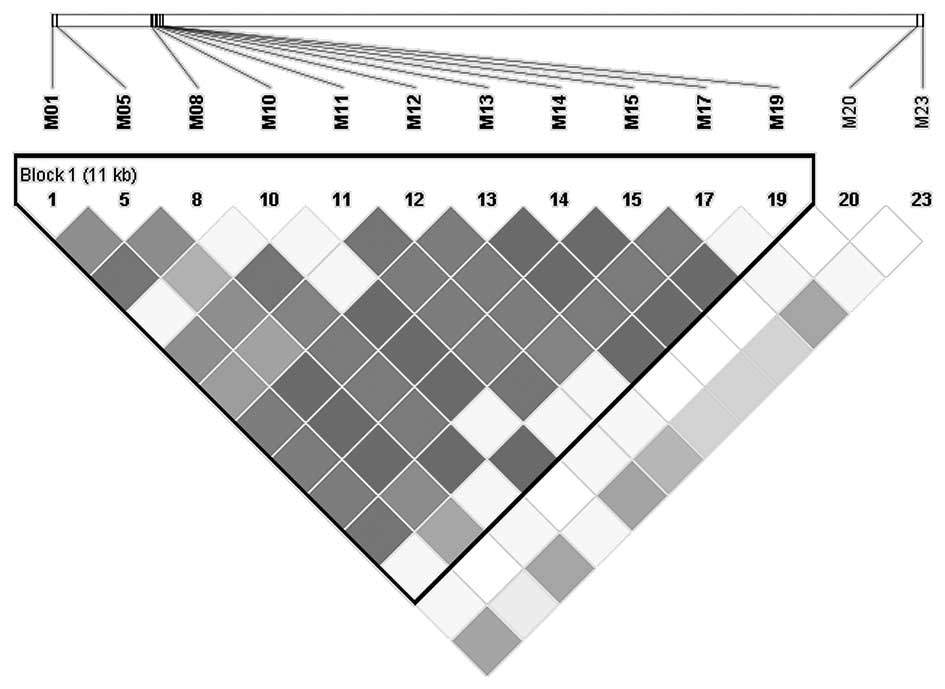
the GOLD heatmap color scheme. As shown in Fig. 1, a linkage block was observed across
the sequenced region in the present metastatic gastrointestinal
cancer patients. The variants detected in the UGT1A7 and UGT1A9
regions were closely associated (block 1). Out of the 6 SNPs that
exhibited an association with grade 3–4 neutropenia, 5 SNPs were
located in the UGT1A7-UGT1A9 block (block 1). In addition, M17
(UGT1A7*3) was the only functional variant among the 5 SNPs. It was
hypothesized that the significant association with grade 3–4
neutropenia of the other 4 SNPs was attributed to UGT1A7*3.
In total, 2 functional mutations exhibited a
significant association with grade 3–4 neutropenia, including
UGT1A7*3 (P=0.011; OR, 3.391; 95% CI, 1.384–8.312) and UGT1A1*6
(P=0.016; OR, 3.373; 95% CI, 1.350–8.431). The risk of severe
neutropenia associated with UGT1A7*3 and UGT1A1*6 genotypes
(Table VI), and the combined effects
of those two alleles, was analyzed (Table VII). In the present study, patients
with one UGT1A7*3 allele had an OR of 4.444 (95% CI, 1.060–18.627)
for the risk of severe neutropenia. Compared with patients
heterozygous for UGT1A7*3, patients homozygous for UGT1A7*3
demonstrated a 5-fold increase in risk, with an OR of 23.333 (95%
CI, 1.609–338.419). The OR for patients homozygous for UGT1A1*6
(OR, 12.333; 95% CI, 0.607–250.508) was two times higher than the
OR of patients heterozygous for UGT1A1*6 (OR, 6.167; 95% CI,
1.486–25.586). Combined effects analysis of those 2 alleles
revealed that patients carrying the UGT1A7*3 and UGT1A1*6 alleles
had a higher OR (OR, 7.333; 95% CI, 1.485–36.209) than the OR of
patients carrying either UGT1A7*3 (OR, 5.556; 95% CI; 1.371–22.507)
or UGT1A1*6 (OR, 6.491; 95% CI, 1.595–26.423). However, UGT1A9*1b
was not significantly associated with the risk of severe toxicity
in the heterozygous (P=0.617) or homozygous (P=0.065) state.
 | Table VI.Risk of severe toxicity associated
with UGT1A7*3, UGT1A1*6 and UGT1A9*1b. |
Table VI.
Risk of severe toxicity associated
with UGT1A7*3, UGT1A1*6 and UGT1A9*1b.
| SNP ID | Allele |
Genotypesa | Severe toxicity,
n | Overall, n | P-value | OR | 95% CI |
|---|
| M17 | UGT1A7*3 | 0 | 3b | 38 |
|
|
|
|
|
| 1 | 8b | 29 | 0.046 |
4.444 | 1.060–18.627 |
|
|
| 2 | 2b | 3 | 0.035 | 23.333 | 1.609–338.419 |
| M23 | UGT1A1*6 | 0 | 3b | 40 |
|
|
|
|
|
| 1 | 9b | 27 | 0.010 |
6.167 | 1.486–25.586 |
|
|
| 2 | 1b | 2 | 0.184 | 12.333 | 0.607–250.508 |
| M05 | UGT1A9*1b | 0 | 1c | 27 |
|
|
|
|
|
| 1 | 3c | 31 | 0.617 |
2.786 | 0.272–28.496 |
|
|
| 2 | 3c | 11 | 0.065 |
9.750 | 0.886–107.249 |
 | Table VII.Risk of severe neutropenia associated
with combined effects of UGT1A7*3 and UGT1A1*6. |
Table VII.
Risk of severe neutropenia associated
with combined effects of UGT1A7*3 and UGT1A1*6.
| SNP ID | Allele | Genotypes | Severe neutropenia,
n | Overall, n | P-value | OR | 95% CI |
|---|
| M17 | UGT1A7*3 | 0 |
3a | 38b | 0.014 | 5.556 | 1.371–22.507 |
|
|
| 1+2 | 10a | 31b |
| M23 | UGT1A1*6 | 0 |
3a | 40b | 0.010 | 6.491 | 1.595–26.423 |
|
|
| 1+2 | 10a | 29b |
| M17&M23 |
| 0 |
2a | 34b | 0.012 | 7.333 | 1.485–36.209 |
|
|
| Others | 11a | 35b |
Discussion
Polymorphisms in the UGT1A family have been
demonstrated to repeatedly affect the variability of
irinotecan-induced toxicity, and the results are dependent on
ethnicity. The toxicity of irinotecan remains unpredictable and
requires additional investigation (11,12,20).
Direct sequencing was adopted to avoid ethnic
heterogeneity and to identify novel variations. Subsequent to
sequencing of the promoter (−1000 bp) and exon 1 regions of UGT1A1,
UGT1A7 and UGT1A9, comprehensive analysis of genetic polymorphisms
in these alleles was performed in Chinese patients with metastatic
gastrointestinal cancer treated using irinotecan-based regimens. A
total of 23 different genetic variants were detected, including 2
novel polymorphisms. In total, 7 SNPs exhibited an association with
grade 3–4 toxicity (P<0.05). Finally, 3 polymorphisms were
focused on, 2 of which were the functional mutations UGT1A7*3 and
UGT1A1*6 that exhibited a significant association with grade 3–4
neutropenia (UGT1A7*3: P=0.011, OR=3.391, 95% CI=1.384–8.312; and
UGT1A1*6: P=0.016, OR=3.373, 95% CI=1.350–8.431). Patients that
carried the UGT1A7*3 and UGT1A1*6 alleles had an increased risk of
grade 3–4 neutropenia (OR, 7.333; 95% CI, 1.485–36.209). Another
key mutation is UGT1A9*1b (−118 9T>10T), which is found
predominantly in the Asian population, and leads to increased
enzyme expression and glucuronidation rates (16,19). This
was the only mutation significantly associated with grade 3–4
diarrhea (P=0.045; OR, 3.273; 95% CI, 1.033–10.37) in the present
study.
UGT1A7*3 is located on UGT1A7 exon 1, and a
transition of T>C results in the 208th amino acid, tryptophan,
changing to arginine. The UGT1A7*3 allele has been reported to
generate an enzyme with 50% reduced catalytic activity (27). It has also been reported that UGT1A7*3
affects the pharmacokinetics of SN-38 in studies investigating both
Caucasian and Asian patients (11,16,18,20).
In the present study, patients homozygous for UGT1A7*3 exhibited a
23.333-fold increased risk of grade 3–4 neutropenia (OR, 23.333;
95% CI, 1.609–338.419), while patients heterozygous for UGT1A7*3
had an OR of 4.444 (95% CI, 1.060–18.627).
UGT1A1*6, a unique allele in the Asian population,
is located on UGT1A1 exon 1 (11,12). A
transition of G>A results in a change of the 71st amino acid
from glycine to arginine. Previous studies in the Asian population
found that UGT1A1*28/*6 is a risk factor for irinotecan-induced
toxicity (18,28). The OR for UGT1A1*6-homozygous patients
(OR, 12.333; 95% CI, 0.607–250.508) was increased two-fold compared
with the OR for UGT1A1*6-heterozygous patients (OR, 6.167; 95% CI,
1.486–25.586).
In conclusion, the UGT1A1*6, UGT1A7*3 and UGT1A9*1b
polymorphisms are predictive markers for severe toxicity in Chinese
metastatic gastrointestinal cancer patients treated with
irinotecan-based regimens.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (grant no. 81402999).
References
|
1
|
Mathijssen RH, van Alphen RJ, Verweij J,
Loos WJ, Nooter K, Stoter G and Sparreboom A: Clinical
pharmacokinetics and metabolism of irinotecan (CPT-11). Clin Cancer
Res. 7:2182–2194. 2001.PubMed/NCBI
|
|
2
|
Guillemette C: Pharmacogenomics of human
UDP-glucuronosyltransferase enzymes. Pharmacogenomics J. 3:136–158.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cunningham D, Pyrhönen S, James RD, Punt
CJ, Hickish TF, Heikkila R, Johannesen TB, Starkhammar H, Topham
CA, Awad L, et al: Randomized trial of irinotecan plus supportive
care versus supportive care alone after fluorouracil failure for
patients with metastatic colorectal cancer. Lancet. 352:1413–1418.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saltz LB, Cox JV, Blanke C, Rosen LS,
Fehrenbacher L, Moore MJ, Maroun JA, Ackland SP, Locker PK, Pirotta
N, et al: Irinotecan plus fluorouracil and leucovorin for
metastatic colorectal cancer. N Engl J Med. 343:905–914. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Fuchs CS, Moore MR, Harker G, Villa L,
Rinaldi D and Hecht JR: Phase III comparison of two irinotecan
dosing regimens in second-line therapy of metastatic colorectal
cancer. J Clin Oncol. 21:807–814. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Zhao ZY, Wu Q, Li J, Fu Q, Cheng
J, Xu N, Wu CP and Xu LG: Multicenter phase II study of modified
FOLFIRI in patients in a Chinese population with advanced
colorectal cancer (CRC) refractory to fluoropyrimidine and
oxaliplatin. J Clin Oncol. 23:292s. 2005.
|
|
7
|
Maitland ML, Grimsley C, Kuttab-Boulos H,
Witonsky D, Kasza KE, Yang L, Roe BA and Di Rienzo A: Comparative
genomics analysis of human sequence variation in the UGT1A gene
cluster. Pharmacogenomics J. 6:52–62. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gong QH, Cho JW, Huang T, Potter C,
Gholami N, Basu NK, Kubota S, Carvalho S, Pennington MW, Owens IS
and Popescu NC: Thirteen UDPglucuronosyltransferase genes are
encoded at the human UGT1 gene complex locus. Pharmacogenetics.
11:357–368. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Iyer L, King CD, Whitington PF, Green MD,
Roy SK, Tephly TR, Coffman BL and Ratain MJ: Genetic predisposition
to the metabolism of irinotecan (CPT-11). Role of uridine
diphosphate glucuronosyltransferase isoform 1A1 in the
glucuronidation of its active metabolite (SN-38) in human liver
microsomes. J Clin Invest. 101:847–854. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gagné JF, Montminy V, Belanger P,
Journault K, Gaucher G and Guillemette C: Common human UGT1A
polymorphisms and the altered metabolism of irinotecan active
metabolite 7-ethyl-10-hydroxycamptothecin (SN-38). Mol Pharmacol.
62:608–617. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Han JY, Lim HS, Shin ES, Yoo YK, Park YH,
Lee JE, Jang IJ, Lee DH and Lee JS: Comprehensive analysis of UGT1A
polymorphisms predictive for pharmacokinetics and treatment outcome
in patients with non-small-cell lung cancer treated with irinotecan
and cisplatin. J Clin Oncol. 24:2237–2244. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Minami H, Sai K, Saeki M, Saito Y, Ozawa
S, Suzuki K, Kaniwa N, Sawada J, Hamaguchi T, Yamamoto N, et al:
Irinotecan pharmacokinetics/pharmacodynamics and UGT1A genetic
polymorphisms in Japanese: Roles of UGT1A1*6 and *28. Pharmacogenet
Genomics. 17:497–504. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pharmacia and Upjohn Company LLC, .
Camptosar - irinotecan hydrochloride injection, solution.
http://labeling.pfizer.com/Show
Labeling.aspx?id=533
|
|
14
|
Sai K, Saeki M, Saito Y, Ozawa S, Katori
N, Jinno H, Hasegawa R, Kaniwa N, Sawada J, Komamura K, et al:
UGT1A1 haplotypes associated with reduced glucuronidation and
increased serum bilirubin in irinotecan-administered Japanese
patients with cancer. Clin Pharmacol Ther. 75:501–515. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bowcock AM, Anderson LA, Friedman LS,
Black DM, Osborne-Lawrence S, Rowell SE, Hall JM, Solomon E and
King MC: THRA1 and D17S183 flank an interval of <4 cM for the
breast-ovarian cancer gene (BRCA1) on chromosome 17q21. Am J Hum
Genet. 52:718–722. 1993.PubMed/NCBI
|
|
16
|
Fujita K, Ando Y, Nagashima F, Yamamoto W,
Eodo H, Araki K, Kodama K, Miya T, Narabayashi M and Sasaki Y:
Genetic linkage of UGT1A7 and UGT1A9 polymorphisms to UGT1A1*6 is
associated with reduced activity for SN-38 in Japanese patients
with cancer. Cancer Chemother Pharmacol. 60:515–522. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lankisch TO, Schulz C, Zwingers T,
Erichsen TJ, Manns MP, Heinemann V and Strassburg CP: Gilbert's
syndrome and irinotecan toxicity: Combination with
UDP-glucuronosyltransferase 1A7 variants increases. Cancer
Epidemiol Biomarkers Prev. 17:695–701. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hazama S, Mishima H, Tsunedomi R, Okuyama
Y, Kato T, Takahashi K, Nozawa H, Ando H, Kobayashi M, Takemoto H,
et al: UGT1A1*6, 1A7*3, and 1A9*22 genotypes predict severe
neutropenia in FOLFIRI-treated metastatic colorectal cancer in two
prospective studies in Japan. Cancer Sci. 104:1662–1669. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yamanaka H, Nakajima M, Katoh M, Hara Y,
Tachibana O, Yamashita J, McLeod HL and Yokoi T: A novel
polymorphism in the promoter region of human UGT1A9 gene
(UGT1A9*22) and its effects on the transcriptional activity.
Pharmacogenetics. 14:329–332. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cecchin E, Innocenti F, D'Andrea M, Corona
G, De Mattia E, Biason P, Buonadonna A and Toffoli G: Predictive
role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their
haplotypes on the outcome of metastatic colorectal cancer patients
treated with fluorouracil, leucovorin, and irinotecan. J Clin
Oncol. 27:2457–2465. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Cui C, Shu C, Yang Y, Liu J, Shi S, Shao
Z, Wang N, Yang T and Hu S: XELIRI compared with FOLFIRI as a
second-line treatment in patients with metastatic colorectal
cancer. Oncol Lett. 8:1864–1872. 2014.PubMed/NCBI
|
|
22
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Barrett JC, Fry B, Maller J and Daly MJ:
Haploview: Analysis and visualization of LD and haplotype maps.
Bioinformatics. 21:263–265. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Abecasis GR and Cookson WO: GOLD-graphical
overview of linkage disequilibrium. Bioinformatics. 16:182–183.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uhlén M, Fagerberg L, Hallström BM,
Lindskog C, Oksvold P, Mardinoglu A, Sivertsson Å, Kampf C,
Sjöstedt E, Asplund A, et al: Proteomics. Tissue-based map of the
human proteome. Science. 347:12604192015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Purcell S, Neale B, Todd-Brown K, Thomas
L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly
MJ and Sham PC: PLINK: A toolset for whole-genome association and
population-based linkage analysis. Am J Hum Genet. 81:559–575.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Guillemette C, Ritter JK, Auyeung DJ,
Kessler FK and Housman DE: Structural heterogeneity at the
UDP-glucuronosyltransferase 1 locus: Functional consequences of
three novel missense mutations in the human UGT1A7 gene.
Pharmacogenetics. 10:629–644. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang H, Bian T, Jin T, Chen Y, Lin A and
Chen C: Association analysis of UGT1A genotype and haplotype with
SN-38 glucuronidation in human livers. Pharmacogenomics.
15:785–798. 2014. View Article : Google Scholar : PubMed/NCBI
|