Introduction
Hepatoblastoma (HB) is the most common hepatic
malignancy in children, and accounts for ~1% of all childhood
tumors (1). Usually, the disease is
diagnosed during the first three years of a child's life (2). The etiology of HB is unclear, but has
been associated with Beckwith-Wiedemann syndrome, familial
adenomatosis polypi and low birth weight (3). The primary treatment for HB is surgical
resection, and the use of pre-resection chemotherapy may increase
the likelihood of a resectable tumor (3). Therefore, the pathogenesis of HB
requires urgent understanding.
Differentially expressed genes and microRNAs (miRNAs
or miRs) have been previously indicated to be crucial in HB,
affecting the development, metastasis and treatment of this disease
(4). HB-associated factors have less
effect, compared with differentially expressed factors (5). Despite the large number of genes and
miRNAs that have been reported to affect HB, the association
between them remains unclear.
Transcription factors (TFs), miRNAs and target genes
construct an intricate regulatory network that provides a novel
opportunity to study HB (6). TFs and
miRNAs are prominent regulators of gene expression. Gene regulatory
factors are predominantly comprised of TFs and miRNAs, which
control the expression of genomic information in multicellular
genomes (7). TFs are proteins that
bind to specific DNA sequences, thus controlling the transfer of
genetic information between DNA and messenger (m)RNA (8). In the three networks at various levels
described in the present study, TFs regulate miRNAs and miRNAs
target other genes. As a result, TFs influence the expression of
genes via miRNAs. miRNAs are endogenous RNAs of ~23 nt in length
that are crucial for gene regulation, as they interact with the
mRNAs of protein-coding genes to control their post-transcriptional
repression (9). Magrelli et al
(10) demonstrated that miRNAs
associated with carcinogenesis may play a pivotal role in the onset
of HB. TFs and miRNAs interact with each other (11). Wang et al (12) reported that TFs regulate the
expression of miRNAs, and miRNAs regulate the transcription of TFs.
This complex association is included in the networks of the present
study.
Target genes are targeted by miRNAs (13). Naeem et al (14)indicated that miRNAs may exert a
widespread impact on the regulation of target and non-target genes.
At present, miRNAs are known to target numerous genes (15). These target genes, for which abundant
information may be obtained from available databases and pertinent
literature (16), may provide
essential evidence for the discovery of the biological role of
miRNAs (17).
Host genes are genes where miRNAs are located.
Rodriguez et al (18)
suggested that the transcriptional patterns of all miRNA host genes
were derived from a variety of sources that illustrate the spatial,
temporal and physiological regulation of miRNA expression. This
theory indicates that miRNAs are transcribed in parallel with their
host transcripts, and that the two different transcription classes
of miRNAs identified (‘exonic’ and ‘intronic’) may require slightly
different mechanisms of biogenesis (18). The cooperation of miRNAs with the host
gene may affect the disease process (19). In the present study, the host genes
are considered to be mutated and involved in the progression of
cancer when their corresponding miRNA is differentially
expressed.
From the experimental data obtained in the present
study, the differentially expressed genes and miRNAs were concluded
to have a paramount impact on HB. In addition, the associated genes
and miRNAs also exert certain effect on HB.
In the present study, the underlying networks of HB
were assessed with respect to miRNAs, target genes, TFs, host genes
of miRNAs and the associations between these factors in human HB.
Various data were collected, and the associations between the
aforementioned factors were revealed to be intricate. The
differences and similarities between these factors were compared,
and significant associations were extracted for analysis, in order
to aid the understanding of the pathogenesis of HB.
Materials and methods
Material collection and data
processing
Extraction of the regulatory associations between
miRNAs and target genes
The interactions between human miRNAs and target
genes were extracted from two databases, TarBase 5.0 (20) and miRTarBase (http://mirtarbase.mbc.nctu.edu.tw). The gene symbols
used in the present study were adapted from official symbols in the
National Center for Biotechnology Information (NCBI) database
(http://www.ncbi.nlm.nih.gov/gene). The
miRNA symbols were normalized using miRBase (http://www.mirbase.org). All the information extracted
was termed dataset D1.
Extraction of the regulatory associations between
TFs and miRNAs. The experimentally validated dataset of human TFs
and the miRNAs regulated by these TFs was collected from TransmiR
(12), a manually extracted database
that details the associations between TFs and miRNAs. The TF and
miRNA association data were collected into the dataset D2.
Extraction of the regulatory associations between
miRNAs and host genes. The host genes of human miRNAs were
extracted from the miRBase (21) and
NCBI databases. In order to ensure the accuracy of the present
study, each host gene was manually assigned an official symbol and
an official identity. The miRNA and host gene data were included in
the dataset D3.
Extraction of data regarding genes and miRNAs. The
differentially expressed genes in HB were collected from the Cancer
GeneticsWeb (www.cancerindex.org/geneweb), the NCBI database of
single nucleotide polymorphism (dbSNP) (www.ncbi.nlm.nih.gov/snp) and pertinent literature in
the Science Citation Index (22). The
differentially expressed genes were termed dataset D4. The
associated genes were then extracted from GeneCards®
(www.genecards.org) and pertinent literature.
Predicted TFs were identified from target genes of
the differentially expressed miRNAs. The TFs were extracted using
the P-Match method (http://www.gene-regulation.com/pub/programs.html#pmatch),
an algorithm that operates on the concepts of pattern matching and
weight matrix approaches in order to identify transcription factor
binding sites (TFBSs) in DNA sequences (23). The genes that appeared in TransmiR
were regarded to be HB-associated genes, and were further analyzed.
In total, 1,000 nt promoter region sequences of the targets of
differentially expressed genes were downloaded from the University
of California Santa Cruz database (24). P-Match was used to combine pattern
matching and weight matrix approaches in order to identify the
TFBSs in 1,000 nt promoter region sequences. The TFBSs were mapped
onto the promoter regions of the targets. The P-Match matrix
library consists of a set of known TFBSs that were collected from
the TRANSFAC® database (http://www.gene-regulation.com/pub/databases.html),
thus providing the possibility to search for large numbers of
differentially expressed TFBSs. The differentially expressed genes
that were obtained were also associated genes. The data regarding
differentially expressed genes and associated genes were termed
dataset D5.
The differentially expressed miRNAs were extracted
from pertinent literature and miR2Disease (25,26), which
is a manually curated database regarding differentially expressed
miRNAs in HB. The differentially expressed miRNAs were termed
dataset D6. The associated miRNAs were collected from pertinent
literature. The differentially expressed miRNAs were also
associated miRNAs. The associated miRNAs collected were termed
dataset D7.
Construction of three networks at various levels.
The differentially expressed network, associated network and global
network were constructed. All the regulatory associations depended
upon host genes, target genes, miRNAs and TFs. Specifically, the
associations between host genes, miRNAs, target genes and TFs were
extracted. Through combining all the associations in datasets
D1-D7, the global network was obtained. The differentially
expressed network and associated network were obtained separately
from the global network, and the associated network contained the
differentially expressed network.
The differentially expressed network was identified
as the most crucial network, since genes and miRNAs are the primary
factors to construct this complex network. The factors and pathways
were obtained by mapping the differentially expressed genes and
miRNAs datasets D4 and D6 onto datasets D1, D2 and D3. Then, the
regulatory pathways were used to construct the network.
Differentially expressed genes, associated genes and
the respective miRNAs are all involved in the process of HB. As the
associated genes and miRNAs are also involved in the process of HB,
the datasets D5 and D7 were mapped onto the datasets D1, D2 and D3.
The associated network, including the differentially expressed
network and other factors, demonstrated intricate associations.
The differentially expressed and associated networks
are hypothesized to be important for investigating HB. However,
with the exception of the experimentally validated genes and miRNAs
present in these two networks, certain data that have no direct
association with HB may be involved in the process of HB. All the
TFs and miRNAs that were present in the associated network were
mapped onto the datasets D1, D2 and D3 to obtain the global
network.
Results
Differentially expressed network of HB. A large
number of notable regulatory associations and factors are shown in
Fig. 1. The network consists of 6 TFs
[namely catenin beta 1 (CTNNB1), epidermal growth factor receptor
(EGFR), v-myc avian myelocytomatosis viral oncogene homolog (MYC),
v-myc avian myelocytomatosis viral oncogene neuroblastoma derived
homolog (MYCN), lin-28 homolog B (LIN28B) and cyclin D1 (CCND1)],
25 miRNAs, 9 target genes and 30 host genes.
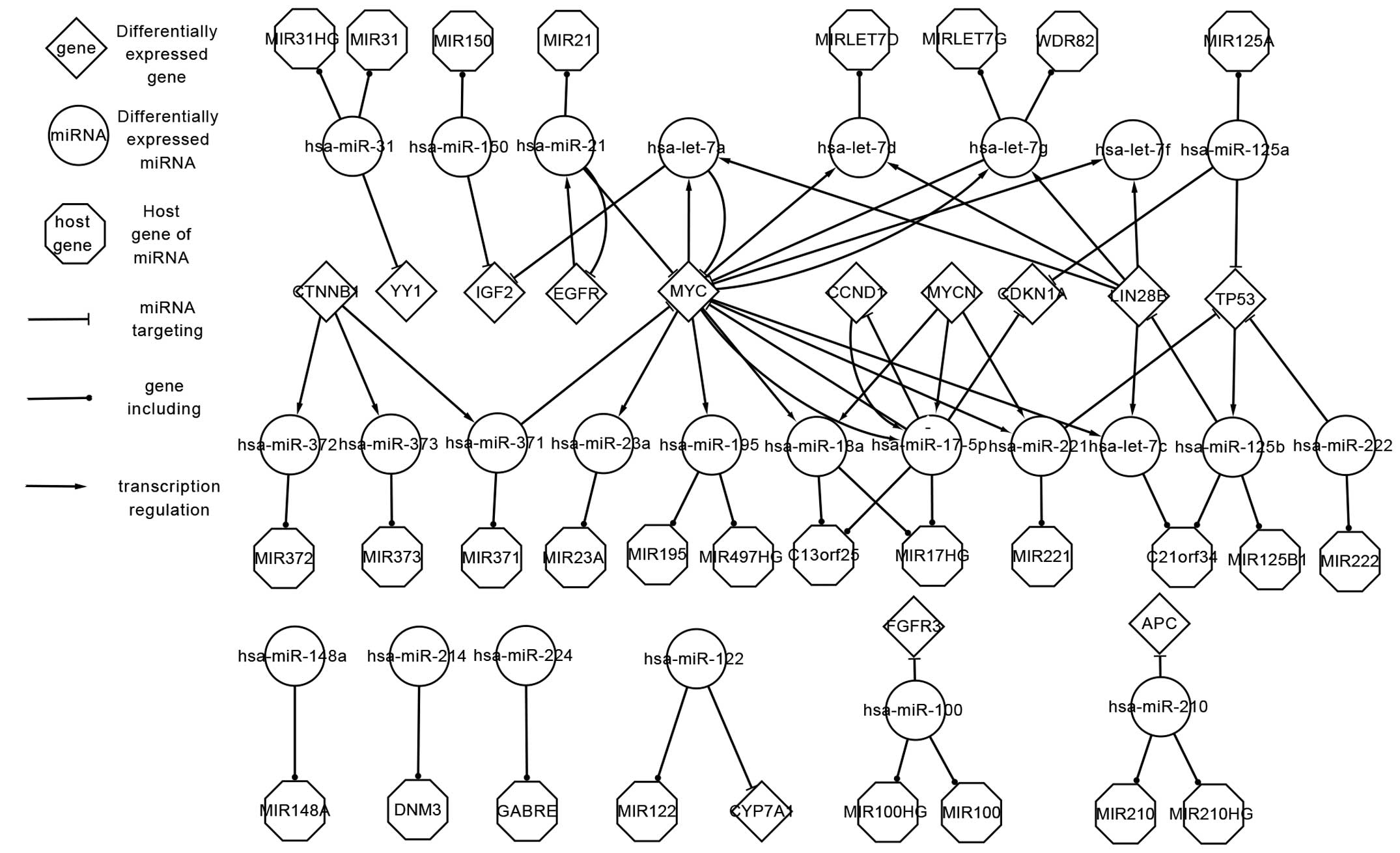 | Figure 1.Differentially expressed network of
genes and microRNAs that are differentially expressed in
hepatoblastoma. miRNA; microRNA; MIR, microRNA; miR, microRNA; HG,
host gene; hsa, Homo sapiens; let, lethal; WDR82, WD
repeat-containing protein 82; CTNNB1, catenin beta 1; YY1, Yin Yang
1; IGF2, insulin-like growth factor 2; EGFR, epidermal growth
factor receptor; MYC; v-myc avian myelocytomatosis viral oncogene
homolog; MYCN, v-myc avian myelocytomatosis viral oncogene
neuroblastoma derived homolog; CCND1, cyclin D1; CDKN1A,
cyclin-dependent kinase inhibitor 1A; LIN28B, lin-28 homolog B;
TP53, tumor protein p53; C13orf25, MIR17HG; C21orf34, MIR99AHG;
FGFR3, fibroblast growth factor receptor 3; APC, adenomatous
polyposis coli; DNM3, dynamin 3; GABRE, gamma-aminobutyric acid
receptor subunit epsilon; CYP7A1, cytochrome P450 family 7
subfamily A member 1. |
The complex associations between genes and miRNAs
may be classified into three classes: i) TFs that regulate miRNAs;
ii) miRNAs that locate at host genes; iii) and miRNAs that interact
with target genes. The reasons behind the intricate associations in
the differentially expressed network may be summarized as follows:
Firstly, a single miRNA may be located at >1 host gene, and
several miRNAs may be located at the same host genes (27). For example, Homo sapiens
(hsa)-miR-210 is located at the genes MIR210 and MIR210 host gene
(HG), while hsa-miR-18a and hsa-miR-17-5p are located at MIR17HG
(also known as C13orf25). Secondly, certain TFs not only regulate
miRNAs, but also act as target genes. For example, LIN28B regulates
hsa-lethal (let)-7a, hsa-let-7c, hsa-let-7d, hsa-let-7g and
hsa-let-7f, and is also targeted by hsa-miR-125b. In addition,
certain feed-forward loops (FFLs) are important to consider. For
example, MYC and CCND1 regulate hsa-miR-17-5p, which in turn
targets MYC and CCND1.
The upstream and downstream information may describe
the association between genes and miRNAs more clearly. For example,
LIN28B and MYC regulate hsa-let-7a, which in turn targets insulin
like growth factor 2 (IGF2) and MYC. Therefore, the upstream genes
LIN28B and MYC impact the downstream genes IGF2 and MYC through
hsa-let-7a.
The differentially expressed network provides
important information regarding the regulatory mechanisms of
HB.
Transcriptional network of HB. The TFs that were
included in the differentially expressed network were further
analyzed. Fig. 2 shows the regulatory
pathways of TFs, differentially expressed miRNAs and host genes
that associate with each other, according to the differentially
expressed network. These factors affect their downstream elements
by regulating or targeting them.
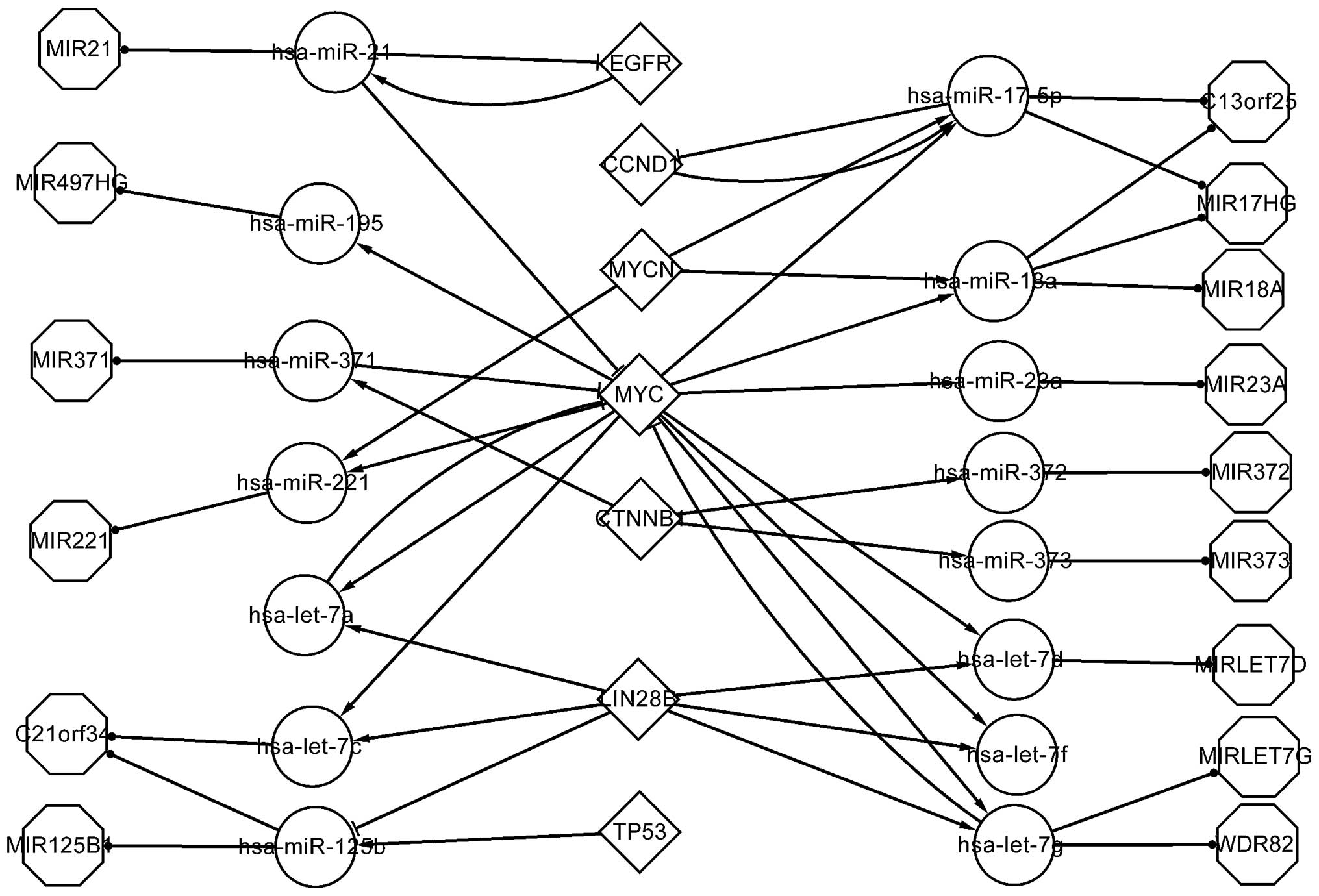 | Figure 2.Transcriptional network of
transcription factors and microRNAs involved in
hepatoblastoma..MIR, microRNA; HG, host gene; C21orf34, MIR99AHG;
hsa, Homo sapiens; let, lethal; EGFR, epidermal growth
factor receptor; CCND1, cyclin D1; MYC, v-myc avian
myelocytomatosis viral oncogene homolog; MYCN, v-myc avian
myelocytomatosis viral oncogene neuroblastoma derived homolog;
CTNNB1, catenin beta 1; LIN28B, lin-28 homolog B; TP53, tumor
protein p53; C13orf25, MIR17HG; WDR82, WD repeat-containing protein
82. |
In total, 7 TFs, namely EGFR, CCND1, MYCN, MYC,
CTNNB1, LIN28B and tumor protein 53 (TP53) are shown in Fig. 2. MYC and MYCN affect CCND1 through
hsa-miR-17-5p, while hsa-miR-21 and EGFR form a FFL.
MYC is a crucial TF in the differentially expressed
network, since it regulates hsa-miR-195, −221, −18a and −17-5p, in
addition to hsa-let-7a, c, d, f and g. Of these miRNAs,
hsa-miR-17-5p targets CCND1, which indicates that MYC affects CCND1
through hsa-miR-17-5p. Cairo et al (28) indicated that hsa-miR-100, hsa-miR-371,
hsa-miR-373 and hsa-let-7a, also known as the four-miR signature,
may be used to classify HB, and reported that MYC activation is
important in the pathogenesis of HB.
Previously, MYCN was indicated to be highly
unregulated in HB (4). However, the
results of the present study indicate that MYCN regulates
hsa-miR-221, hsa-miR-17-5p and hsa-miR-18a. Furthermore, CCND1 and
EGFR were previously reported to exhibit differentially increased
expression in HB (29).
According to the pathways constructed in a previous
study, hsa-let-7a targets MYC, thus exerting tumor suppressive
activity (28). A previous study
indicated that the targeting of the LIN28B gene by MYC contributes
to the regulation of miRNA expression, notably by inhibiting the
biogenesis of hsa-let-7a, c, d, f and g (30). In addition, the expression of LIN28B
was negative correlated with the expression of the aforementioned
miRNAs (31).
The transcriptional network, including genes, miRNAs
and their associations, meaningfully explains the pathogenesis of
HB.
Associated network of HB. Fig. 3 shows the associated network of
associated expressed genes, associated expressed miRNAs and the
host genes of associated expressed miRNAs. The associated network
includes all the factors and pathways that are included in the
differentially expressed network.
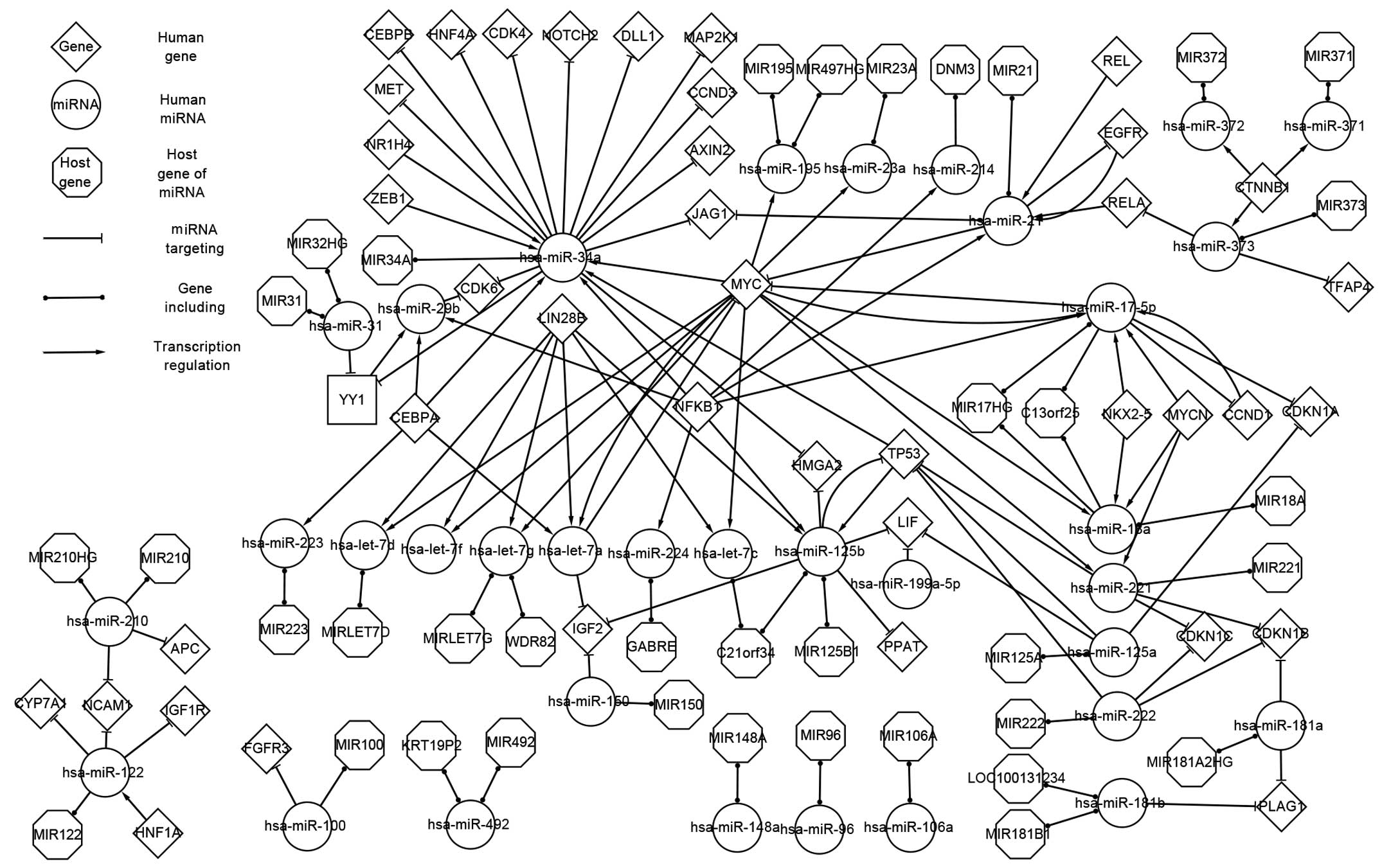 | Figure 3.Associated network describing the
associations between genes and microRNAs involved in
hepatoblastoma. miRNA; microRNA; CEBPB, CCAAT/enhancer-binding
protein beta; HNF, hepatocyte nuclear factor; CDK4,
cyclin-dependent kinase 4; NOTCH2, neurogenic locus notch homolog
protein 2; DLL1, delta-like 1; MAP2K1, mitogen-activated protein
kinase kinase 1; MET, mesenchymal epithelial transition factor;
NR1H4, nuclear receptor subfamily 1, group H, member 4; ZEB1, zinc
finger E-box binding homeobox 1; CCND3, cyclin D3; AXIN2, axis
inhibition protein 2; JAG1, jagged 1; hsa, Homo sapiens;
miR, microRNA; MIR, microRNA; HG, host gene; CDK6, cyclin-dependent
kinase 6; LIN28B, lin-28 homolog B; YY1, Yin Yang 1; CEBPA,
CCAAT/enhancer-binding protein alpha; NFKB1, nuclear factor of
kappa light polypeptide gene enhancer in B-cells 1; MYC, v-myc
avian myelocytomatosis viral oncogene homolog; MYCN, v-myc avian
myelocytomatosis viral oncogene neuroblastoma derived homolog;
DNM3, dynamin 3; REL, v-rel avian reticuloendotheliosis viral
oncogene homolog; EGFR, epidermal growth factor receptor; CTNNB1,
catenin beta 1; TFAP4, transcription factor activating
enhancer-binding protein 4; C13orf25, MIR17HG; NKX2-5, NK2 homeobox
5; CCND1, cyclin D1; CDKN1, cyclin-dependent kinase inhibitor 1;
LOC100131234, MIR181A1HG; PLAG1, pleomorphic adenoma gene 1; HMGA2,
high mobility group AT-hook 2; TP53, tumor protein p53; LIF,
leukemia inhibitory factor; PPAT, phosphoribosyl pyrophosphate
amidotransferase; C21orf34, MIR99AHG; GABRE, gamma-aminobutyric
acid receptor subunit epsilon; IGF2, insulin-like growth factor 2;
WDR82, WD repeat-containing protein 82; KRT19P2, keratin 19
pseudogene 2; FGFR3, fibroblast growth factor receptor 3; APC,
adenomatous polyposis coli; CYP7A1, cytochrome P450 family 7
subfamily A member 1; NCAM1, neural cell adhesion molecule 1;
IGF1R, insulin like growth factor 1 receptor. |
With the exception of the factors included in the
differentially expressed network, the associated network contained
additional 9 TFs, namely CCAAT/enhancer binding protein alpha,
hepatic nuclear factor 4 alpha, nuclear factor of kappa light
polypeptide gene enhancer in B-cells 1 (NFKB1), v-rel avian
reticuloendotheliosis viral oncogene homolog (REL), RELA, CTNNB1,
nuclear receptor subfamily 1, group H, member 4 (NR1H4), zinc
finger E-box binding homeobox 1 (ZEB1) and NK2 homeobox 5. Four of
these TFs, namely ZEB1, REL, NFKB1 and RELA, were obtained using
the P-Match algorithm. With the exception of differentially
expressed miRNAs, 12 associated miRNAs, including hsa-miR-122,
hsa-miR-492 and hsa-miR-34a, are shown in the associated network
(Fig. 3). These associated factors
affect the therapy, prognosis and clinical research of HB (32).
An attractive miRNA (hsa-miR-34a) is shown in
Fig. 3. This miRNA targets 13 genes,
including mesenchymal epithelial transition factor (MET),
cyclin-dependent kinase (CDK)4 and delta-like 1 (DLL1), and is
regulated by 6 genes, including ZEB1, NR1H4 and MYC.
The TF Yin Yang 1 (YY1) was also obtained from a
P-Match algorithm. YY1 is differentially expressed in HB (4), is targeted by hsa-miR-31 and
hsa-miR-29a, and regulates CDK6.
Compared with the differentially expressed network,
the associated network reveals more associations and provides a
wider perspective to aid the understanding of the pathogenesis of
HB.
Global network of HB. The topological associations
derived from the D1, D2 and D3 datasets comprise the global
network, which includes the associated and differentially expressed
networks. The global network provides a more comprehensive
perspective to study the pathogenesis of HB.
Comparison and analysis of the features of
differentially expressed genes in the HB networks. The present
study analyzed the differentially expressed genes and associated
factors obtained from the aforementioned miRNAs and genes pathways.
The genes were classified according to the regulatory associations
of their adjacent nodes with miRNAs derived from the three networks
at various levels, in order to analyze the interacting feature of
each gene.
A total of 30 differentially expressed genes,
including CCND1, CTNNB1 and LIN28B, were analyzed in the present
study. These genes may be classified into 7 classes, according to
their adjacent nodes. The first class of genes consists of genes
that possess 6 types of adjacent nodes. The adjacent nodes include
3 successors (downstream elements) and 3 predecessors (upstream
elements). These genes include CTNNB1, EGFR, MYC, CCND1, MYCN,
LIN28B and TP53. The following section focuses on MYC.
Table I shows the
differentially expressed and regulatory associations of MYC with
miRNAs. In total, 5 differentially expressed miRNAs, including
hsa-miR-21, hsa-miR-17-5p, hsa-let-7a, hsa-let-7g and hsa-miR-371,
target MYC; 13 miRNAs in the global network target MYC; and MYC
regulates 5 miRNAs in the differentially expressed network,
including hsa-miR-18a, hsa-miR-17-5p, hsa-miR-221, hsa-let-7a-2 and
hsa-miR-195. A total of 4 and 13 miRNAs target MYC in the
associated and global networks, respectively; and 7 and 32 miRNAs
are regulated by MYC in the associated and global networks,
respectively. According to these results, it is evident that miRNAs
are able to affect directly the expression of multiple miRNAs. In
addition, hsa-let-7a was observed to target MYC, and MYC regulated
hsa-let-7a at all three network levels. Therefore, hsa-let-7a and
MYC form a feedback loop, and are vital in the process of HB
development. Mutations in MYC may directly influence other genes
via miRNAs. For example, MYC influences the expression of TP53 by
hsa-miR-221. Furthermore, the differential expression of miRNAs
affects other miRNAs through MYC. For example, hsa-miR-17-5p
targets MYC, and MYC regulates hsa-miR-195. Certain additional
miRNAs in the global network are not involved in HB, but aid the
understanding of the pathogenesis of HB.
 | Table I.Regulatory associations between MYC
and miRNAs. |
Table I.
Regulatory associations between MYC
and miRNAs.
| Network | miRNAs that target
MYC | miRNAs that are
regulated by MYC |
|---|
| Differentially
expressed | miR-17-5p, let-7a,
let-7g, miR-21 and miR-371 | miR-18a, miR-17-5p,
miR-221, let-7a and miR-195 |
| Associated | miR-17-5p, let-7a,
miR-21 and miR-34a | miR-18a, miR-17-5p,
miR-221, let-7a, miR-195, miR-29b and miR-34a |
| Global | miR-17-5p, let-7a,
miR-21, miR-34a, let-7g, miR-145, miR-20a, miR-24, miR-26a,
miR-34b, miR-34c-5p, miR-378 and miR-98 | miR-18a, miR-17-5p,
miR-221, let-7a, miR-195, miR-29b, miR-34a, let-7b, let-7c, let-7d,
let-7e, let-7f, let-7g, let-7i, miR-106a, miR-106b, miR-141,
miR-15a, miR-16-1, miR-18a, miR-19a, miR-19b, miR-20b, miR-20b,
miR-22, miR-23a, miR-23b, miR-29a, miR-29b, miR-9, miR-92a and
miR-93 |
The second class of genes consists of genes that
possess 4 types of adjacent nodes. MYCN regulates 3 miRNAs in the
differentially expressed network, namely hsa-miR-18a, hsa-miR-17-5p
and hsa-miR-221. However, hsa-miR-18a does not target any gene,
indicating that hsa-miR-18a may directly act on HB.
The third class of genes consists of genes that
possess 3 types of adjacent nodes. This class includes IGF2, YY1,
cyclin-dependent kinase inhibitor 1A (CDKN1A), adenomatous
polyposis coli, cytochrome P450 family 7 subfamily A member 1 and
fibroblast growth factor receptor 3 (FGFR3), which do not regulate
any miRNAs in the three network levels. Therefore, these genes may
be last actors in HB.
Neurogenic locus notch homolog protein 2 possesses 2
types of adjacent nodes, thus constituting the fourth class of
genes, and is not suspected to be important in the pathogenesis of
HB.
Comparison and analysis of the features of
differentially expressed miRNAs in the HB networks. The same method
was used to compare the differentially expressed miRNAs and genes.
From the extracted data, 34 differentially expressed miRNAs,
including hsa-miR-17-5p, hsa-miR-18a and hsa-miR-125a, were
identified. To facilitate the understanding of the pathogenesis of
HB, the following section focuses on the miRNAs that are associated
with other genes and, extracts the information regarding these
associations.
The aforementioned 34 miRNAs were classified into 5
classes: The first class of miRNAs consists of miRNAs that possess
6 types of adjacent nodes. A total of 7 miRNAs, including
hsa-miR-21, hsa-let-7a, hsa-let-7g, hsa-miR-125b, hsa-miR-221,
hsa-miR-371 and hsa-miR-17-5p, were observed to have 3 successors
and 3 predecessors. To use hsa-miR-17-5p as an example, Table II shows that 3 differentially
expressed genes, namely CCND1, MYC and MYCN, regulate
hsa-miR-17-5p; and 3 differentially expressed genes, namely CCND1,
MYC and CDKN1A, are targeted by hsa-miR-17-5p. Therefore, CCND1,
MYC and MYCN directly influence CCND1, MYC and CDKN1A through
hsa-miR-17-5p. In total, 4 and 11 genes regulate hsa-miR-17-5p in
the associated and global networks, respectively; and hsa-miR-17-5p
targets 3 and 17 genes in the associated and global networks,
respectively. In addition, certain miRNAs are affected by
hsa-miR-17-5p through certain genes. For example, hsa-miR-17-5p
targets MYC, which regulates miRNAs, including hsa-miR-18a and
hsa-miR-195.
 | Table II.Regulatory associations between
hsa-miR-17-5p and various genes. |
Table II.
Regulatory associations between
hsa-miR-17-5p and various genes.
| Network | Genes that regulate
hsa-miR-17-5p | Target genes of
hsa-miR-17-5p |
|---|
| Differentially
expressed | MYCN, CCND1 and
MYC | CCND1, MYC and
CDKN1A |
| Associated | MYCN, CCND1, MYC
and NFKB1 | CCND1, MYC and
CDKN1A |
| Global | MYCN, CCND1, MYC,
E2F1, ESR1, NFKB1, SPI1, STAT5, TLX1, TLX3 and TNF | CCND1, MYC, CDKN1A,
APP, CCL1, BCL2L11, BMPR2, CCND2, FBXO31, GPR137B, DNAJC27, E2F3,
ICAM1, E2F1, JAK1, MAP3K12 and BCL2 |
The second and third classes of miRNAs consist of
miRNAs that possess 4 types of adjacent nodes. The miRNAs in the
second class, specifically, have 1 predecessor and 3 successors.
For example, hsa-miR-125a has no regulatory gene in the
differentially expressed network or the associated network, but 2
genes share a regulatory pathway with hsa-miR-125a in the global
network. In addition, TP53 and CDKN1A target hsa-miR-125a in the
differentially expressed network, while a total of 3 and 19 genes
target hsa-miR-18a in the associated and global networks,
respectively.
In contrast, the third class of miRNAs has 3
predecessors and 1 successor. For example, 2 genes, namely MYC and
MYCN, regulate hsa-miR-18a in the differentially expressed network,
and 2 and 9 genes regulate hsa-miR-18a in the associated and global
networks, respectively. However in the global network, only 2 genes
target hsa-miR-18a.
The fourth class consists of miRNAs that only
possess 3 successors, such as hsa-miR-100, which is targeted by
FGFR3 in the differentially expressed and associated networks,
while 6 genes target hsa-miR-100 in the global network. The fifth
class has <3 adjacent nodes. For example, hsa-miR-148a is only
regulated by 3 genes, and targets 7 genes in the global
network.
Comparison and analysis of the features of predicted
TFs in the HB networks. The analysis of predicted TFs was conducted
following the aforementioned method. A total of 5 TFs, namely ZEB1,
YY1, REL, RELA and NFKB1, regulate miRNAs in the associated
network. The first class of TFs consists of TFs that possess 6
types of adjacent nodes, specifically 3 successors and 3
predecessors. NFKB1, an example of a TF from the first class,
possesses 6 types of adjacent nodes. Table III shows that the differentially
expressed miRNA hsa-let-7a targets NFKB1, which regulates the
differentially expressed miRNAs hsa-miR-214, hsa-miR-224,
hsa-miR-125b and hsa-miR-17-5p. In the associated and global
networks, respectively, 1 and 5 miRNAs target NFKB1, which in turn
regulates 7 and 19 miRNAs in the above networks. In the present
study, NFKB1 was not observed to be a differentially expressed TF,
but exhibited a close association with differentially expressed
miRNAs. Since NFKB1 is only targeted by hsa-let-7a-2 in the
associated network, it may be possible to conclude that
hsa-let-7a-2 directly results in expression errors involving other
miRNAs through NFKB1. The miRNAs that are not differentially
expressed possess certain associations with HB.
 | Table III.Regulatory associations between NFKB1
and miRNAs. |
Table III.
Regulatory associations between NFKB1
and miRNAs.
| Network | miRNAs that target
NFKB1 | miRNAs that are
regulated by NFKB1 |
|---|
| Differentially
expressed | let-7a | miR-17-5p, miR-214,
miR-224, miR-125b and miR-21 |
| Associated | let-7a | miR-17-5p, miR-214,
miR-224, miR-125b, miR-21, miR-34a and miR-29b |
| Global | let-7a, miR-146a,
miR-146b-1, miR-15a and miR-9 | miR-17-5p, miR-214,
miR-224, miR-125b, miR-21, miR-34a, miR-29b, miR-199a-5p, let-7a-3,
let-7b, miR-10b, miR-125b-2, miR-146a, miR-155, miR-16, miR-9,
miR-29a, miR-365 and miR-448 |
The second class of TFs, which includes RELA, REL,
ZEB1 and YY1, consists of TFs that possess 4 types of adjacent
nodes. RELA regulates the differentially expressed miRNA
hsa-miR-21. However, none of the differentially expressed miRNAs
target RELA, while hsa-miR-373 targets RELA in the associated
network. As for YY1, none of the differentially expressed miRNAs
regulate or target YY1. In the associated network, hsa-miR-34a
targets YY1, which regulates hsa-miR-29b. ZEB1 and REL are similar,
in that no miRNAs target them. Therefore, RELA may be the first
actor in the pathogenesis of HB. Future studies are recommended to
account for these important factors.
Discussion
In the present study, certain data were collected
from authoritative websites and pertinent literature, while certain
data were obtained using P-Match. A total of three networks at
various levels, including differentially expressed, associated and
global networks, were constructed in order to analyze all the
experimentally validated data. The pathways involving TFs, target
genes, miRNAs and host genes were analyzed. Since these genes and
miRNAs are not isolated, their development or mutation may affect
other genes or miRNAs that are associated with HB.
In the present study, numerous important genes and
miRNAs were identified. In total, 3 differentially expressed TFs,
namely CCND1, MYC and EGFR, were observed to form FFLs that
correspond to hsa-miR-17-5p, hsa-let-7a and hsa-miR-21,
respectively. MYCN regulates 3 miRNAs (hsa-miR-17-5p, hsa-miR-18a
and hsa-miR-221) in the differentially expressed and associated
networks, and notably, no miRNAs target MYCN. The associated miRNA
hsa-miR-34a targets 13 genes, and is regulated by 6 TFs. Following
the P-Match method, 6 TFs were obtained, which may be also
important, as they suggest potential associations between miRNAs
and TFs. The associations between miRNAs and TFs are complex, and
lead to the interaction between miRNAs and TFs and the regulation
of HB.
Core genes and miRNAs are unlikely to only affect
one type of cancer, and may play a significant role in other types
of cancer. A previous study indicated that MYCN is not only
differentially expressed in HB, but also in Wilms' tumor (33). Furthermore, hsa-miR-373 is
overexpressed in HB, and is also differentially expressed in
retinoblastoma (34). Future studies
should focus on the genes and miRNAs that are common in various
types of cancer. The authoritative data and pathways discussed in
the present study provide a novel perspective for the understanding
of HB.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (Beijing, China; grant no.
60905022).
Glossary
Abbreviations
Abbreviations:
|
HB
|
hepatoblastoma
|
|
miRNA
|
microRNA
|
|
TF
|
transcription factor
|
|
TFBs
|
transcription factors binding
sites
|
|
NCBI
|
National Center for Biotechnology
Information
|
|
FFL
|
feed-forward loop
|
References
|
1
|
De Ioris M, Brugieres L, Zimmermann A,
Keeling J, Brock P, Maibach R, Pritchard J, Shafford L, Zsiros J,
Czaudzerna P and Perilongo G: Hepatoblastoma with a low serum
alpha-fetoprotein level at diagnosis: The SIOPEL group experience.
Eur J Cancer. 44:545–550. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lack EE, Neave C and Vawter GF:
Hepatoblastoma. A clinical and pathologic study of 54 cases. Am J
Surg Pathol. 6:693–705. 1982. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Herzog CE, Andrassy RJ and Eftekhari F:
Childhood cancers: Hepatoblastoma. Oncologist. 5:445–453. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shin E, Lee KB, Park SY, Kim SH, Ryu HS,
Park YN, Yu E and Jang JJ: Gene expression profiling of human
hepatoblastoma using archived formalin-fixed and paraffin-embedded
tissues. Virchows Arch. 458:453–465. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Baynam GS and Goldblatt J: A child with an
FGFR3 mutation, a laterality disorder and an hepatoblastoma: Novel
associations and possible gene-environment interactions. Twin Res
Hum Genet. 13:297–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zimmermann A: The emerging family of
hepatoblastoma tumours: From ontogenesis to oncogenesis. Eur J
Cancer. 41:1503–1514. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hobert O: Gene regulation by transcription
factors and microRNAs. Science. 319:1785–1786. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Latchman DS: Transcription factors: An
overview. Int J Biochem Cell Biol. 29:1305–1312. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Magrelli A, Azzalin G, Salvatore M,
Viganotti M, Tosto F, Colombo T, Devito R, Di Masi A, Antoccia A,
Lorenzetti S, et al: Altered microRNA expression patterns in
hepatoblastoma patients. Transl Oncol. 2:157–163. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hobert O: Common logic of transcription
factor and microRNA action. Trends Biochem Sci. 29:462–468. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang J, Lu M, Qiu C and Cui Q: TransmiR: A
transcription factor-microRNA regulation database. Nucleic Acids
Res. 38:D119–D122. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Griffiths-Jones S, Grocock RJ, van Dongen
S, Bateman A and Enright AJ: miRBase: MicroRNA sequences, targets
and gene nomenclature. Nucleic Acids Res. 34:D140–D144. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Naeem H, Küffner R and Zimmer R: MIRTFnet:
Analysis of miRNA regulated transcription factors. PLoS One.
6:e225192011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Volinia S, Calin GA, Liu CG, Ambs S,
Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et
al: A microRNA expression signature of human solid tumors defines
cancer gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishimoto K, Nakamura H, Tachibana K,
Yamasaki D, Ota A, Hirano K, Tanaka T, Hamakubo T, Sakai J, Kodama
T and Doi T: Sterol-mediated regulation of human lipin 1 gene
expression in hepatoblastoma cells. J Biol Chem. 284:22195–22205.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brand AH and Perrimon N: Targeted gene
expression as a means of altering cell fates and generating
dominant phenotypes. Development. 118:401–415. 1993.PubMed/NCBI
|
|
18
|
Rodriguez A, Griffiths-Jones S, Ashurst JL
and Bradley A: Identification of mammalian microRNA host genes and
transcription units. Genome Res. 14:1902–1910. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gennarino VA, Sardiello M, Avellino R,
Meola N, Maselli V, Anand S, Cutillo L, Ballabio A and Banfi S:
MicroRNA target prediction by expression analysis of host genes.
Genome Res. 19:481–490. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Papadopoulos GL, Reczko M, Simossis VA,
Sethupathy P and Hatzigeorgiou AG: The database of experimentally
supported targets: A functional update of TarBase. Nucleic Acids
Res. 37:D155–D158. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kozomara A and Griffiths-Jones S: miRBase:
Integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Garfield E: “Science Citation Index” - A
new dimension in indexing. Science. 144:649–654. 1964. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chekmenev DS, Haid C and Kel AE: P-Match:
Transcription factor binding site search by combining patterns and
weight matrices. Nucleic Acids Res. 33:(Web Server). W432–W437.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fujita PA, Rhead B, Zweig AS, Hinrichs AS,
Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A,
et al: The UCSC Genome Browser database: Update 2011. Nucleic Acids
Res. 39:D876–D882. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bao J, Li D, Wang L, Wu J, Hu Y, Wang Z,
Chen Y, Cao X, Jiang C, Yan W and Xu C: MicroRNA-449 and
microRNA-34b/c function redundantly in murine testes by targeting
E2F transcription factor-retinoblastoma protein (E2F-pRb) pathway.
J Biol Chem. 287:21686–21698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Jiang Q, Wang Y, Hao Y, Juan L, Teng M,
Zhang X, Li M, Wang G and Liu Y: miR2Disease: A manually curated
database for microRNA deregulation in human disease. Nucleic Acids
Res. 37:D98–104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Golan D, Levy C, Friedman B and Shomron N:
Biased hosting of intronic microRNA genes. Bioinformatics.
26:992–995. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Cairo S, Wang Y, de Reyniès A, Duroure K,
Dahan J, Redon MJ, Fabre M, McClelland M, Wang XW, Croce CM and
Buendia MA: Stem cell-like micro-RNA signature driven by Myc in
aggressive liver cancer. Proc Natl Acad Sci USA. 107:20471–20476.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
López-Terrada D, Gunaratne PH, Adesina AM,
Pulliam J, Hoang DM, Nguyen Y, Mistretta TA, Margolin J and
Finegold MJ: Histologic subtypes of hepatoblastoma are
characterized by differential canonical Wnt and Notch pathway
activation in DLK+ precursors. Hum Pathol. 40:783–794.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
King CE, Wang L, Winograd R, Madison BB,
Mongroo PS, Johnstone CN and Rustgi AK: LIN28B fosters colon cancer
migration, invasion and transformation through let-7-dependent and
-independent mechanisms. Oncogene. 30:4185–4193. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Viswanathan SR and Daley GQ: Lin28: A
microRNA regulator with a macro role. Cell. 140:445–449. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Giardiello FM, Petersen GM, Brensinger JD,
Luce MC, Cayouette MC, Bacon J, Booker SV and Hamilton SR:
Hepatoblastoma and APC gene mutation in familial adenomatous
polyposis. Gut. 39:867–9. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zirn B, Hartmann O, Samans B, Krause M,
Wittmann S, Mertens F, Graf N, Eilers M and Gessler M: Expression
profiling of Wilms tumors reveals new candidate genes for different
clinical parameters. Int J Cancer. 118:1954–1962. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li J, Xu ZW, Wang KH, Wang N, Li DQ and
Wang S: Networks of microRNAs and genes in retinoblastomas. Asian
Pac J Cancer Prev. 14:6631–6636. 2014. View Article : Google Scholar : PubMed/NCBI
|

















