Introduction
Ovarian cancer is the second most common
gynecological cancer worldwide, accounting for ~3% of all female
cancer cases, with the typical age of diagnosis being 63-years-old.
It is challenging to diagnose ovarian cancer at an early stage, and
this cancer has a poor prognosis, with a five-year survival rate of
~47% (1). In order to provide a
foundation for detecting and treating ovarian cancer, studies
investigating the etiology of the disease are required. Previously,
a study by the London Research Institute (Cancer Research UK,
London, UK) found helicase, POLQ-like (HELQ) to be a novel gene
that prevents ovarian cancer (2). In
this study, the possibility of ovarian cancer in mice was increased
two-fold when a copy of the HELQ gene was missing. Even the
deficiency of one copy could lead to the formation of an increased
number of tumors in mice. According to this study, detection of
HELQ gene deficiency may be adopted to screen for ovarian cancer
patients among women in the future, if HELQ helicase performs the
same role in humans and mice (2).
HELQ is a superfamily II DNA helicase that was first
identified in the human and mouse genomes through its homology to
mutagen-sensitive 308 (Mus308) (3), a
DNA repair enzyme required for DNA interstrand crosslink (ICL)
resistance in Drosophila melanogaster (4,5). DNA ICLs
are particularly toxic as they disrupt genetic information on
strands, potently inhibiting DNA replication and transcription. In
normal cells, DNA ICLs and DNA damage often lead to the occurrence
of cancer (6). However, DNA repair is
an important reaction following DNA damage, which may cause the
damaged DNA to revert to the original appearance and perform the
original function (7). DNA helicases
play an important role in this process. HELQ, as a DNA helicase,
has been studied from the perspective of association with cancer.
Previous genome-wide association studies have identified single
nucleotide polymorphisms at loci within or near the HELQ gene that
are associated with an increased risk of several different cancers,
including upper aerodigestive tract cancers and head and neck
cancers (8–11).
To assess the association between the structure of
HELQ and the carcinogenesis of ovarian cancer, bioinformatics
methods were used to analyze this association from a theoretical
angle in the present study. It was expected that such a study may
provide direction and basis for the clinical treatment of ovarian
cancer.
Materials and methods
Gene sequence
The HELQ gene sequence was obtained from the
Nucleotide resource of the National Center for Biotechnology
Information (NCBI; http://www.ncbi.nlm.nih.gov/nucleotide), using the
GenBank accession number AF436845.1 and Gene ID 113510. The HELQ
gene is also termed HEL308.
Bioinformatics analysis
The BioEdit software (http://www.mbio.ncsu.edu/bioedit/bioedit.html) was
used for the analysis of open reading frames. The ExPASy tools
ProtParam (http://web.expasy.org/protparam/) and ProtScale
(http://web.expasy.org/protscale/) were
used to compute gene features, including the amino acid
composition, molecular mass, isoelectric point,
hydrophobicity/hydrophilicity, instability index and aliphatic
index. The SignalP-HMM Server was used to predict the signal
peptides (12). The transmembrane
region was analyzed using the TMHMM Server V.2.0 system (http://www.cbs.dtu.dk/services/TMHMM/).
Subcellular localization was predicted using the Protein
Subcellular Localization Prediction Tool (PSORT) database
(http://www.psort.org/). The Simple Modular
Architecture Research Tool (SMART; http://smart.embl-heidelberg.de/) was used for
analysis of protein domains. Hopfield Neural Network (HNN) was used
to make the secondary structure predictions (13). PHYRE2 Protein Fold Recognition Server
online software (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index)
was employed to obtain the three-dimensional structure of HELQ and
the 3DLigandSite Server (http://www.sbg.bio.ic.ac.uk/3dligandsite/) was used to
construct the three-dimensional ligand binding model. STRING9.0
interactive database (http://string-db.org/) was utilized to find the
associated proteins. Gene ontology analysis was performed using
PredictProtein software (https://www.predictprotein.org/).
Results
Analysis of physicochemical properties
of HELQ
Through NCBI database retrieval, the whole
nucleotide sequence and amino acid sequence of HELQ were obtained,
with a total sequence of 3,591 bp encoding 1,101 amino acid
residues. Open reading frame (ORF) prediction and BioEdit analysis
showed that the HELQ gene contained 15 ORFs. ProtParam predicted
that the molecular weight of the HELQ gene was 124,175.3 Da, the
theoretical isoelectric point was 6.12, the content of leucine
(Leu) was 16.2% of the total components, acidic amino acids were
more common than basic amino acids, and the instability index was
45.55. Therefore, it was inferred that HELQ was an unstable and
acidic protein. In addition, the value of grand average of
hydropathicity was −0.317 and the aliphatic index was 92.34, which
indicated that HELQ is liposoluble.
Subcellular localization and motif
prediction
The location of proteins in cells is closely
associated with the function of proteins. Subcellular localization
analysis of PSORT prediction showed that HELQ was mainly
distributed in the nucleus, mitochondrial matrix space, microbody
(peroxisome) and mitochondrial inner membrane (Table I). HELQ was located in areas with the
presence of DNA.
 | Table I.Subcellular localization of helicase,
POLQ-like encoding product. |
Table I.
Subcellular localization of helicase,
POLQ-like encoding product.
| Subcellular
localization | Certainty |
|---|
| Nucleus | 0.600 |
| Mitochondrial matrix
space | 0.510 |
| Microbody
(peroxisome) | 0.300 |
| Mitochondrial inner
membrane | 0.234 |
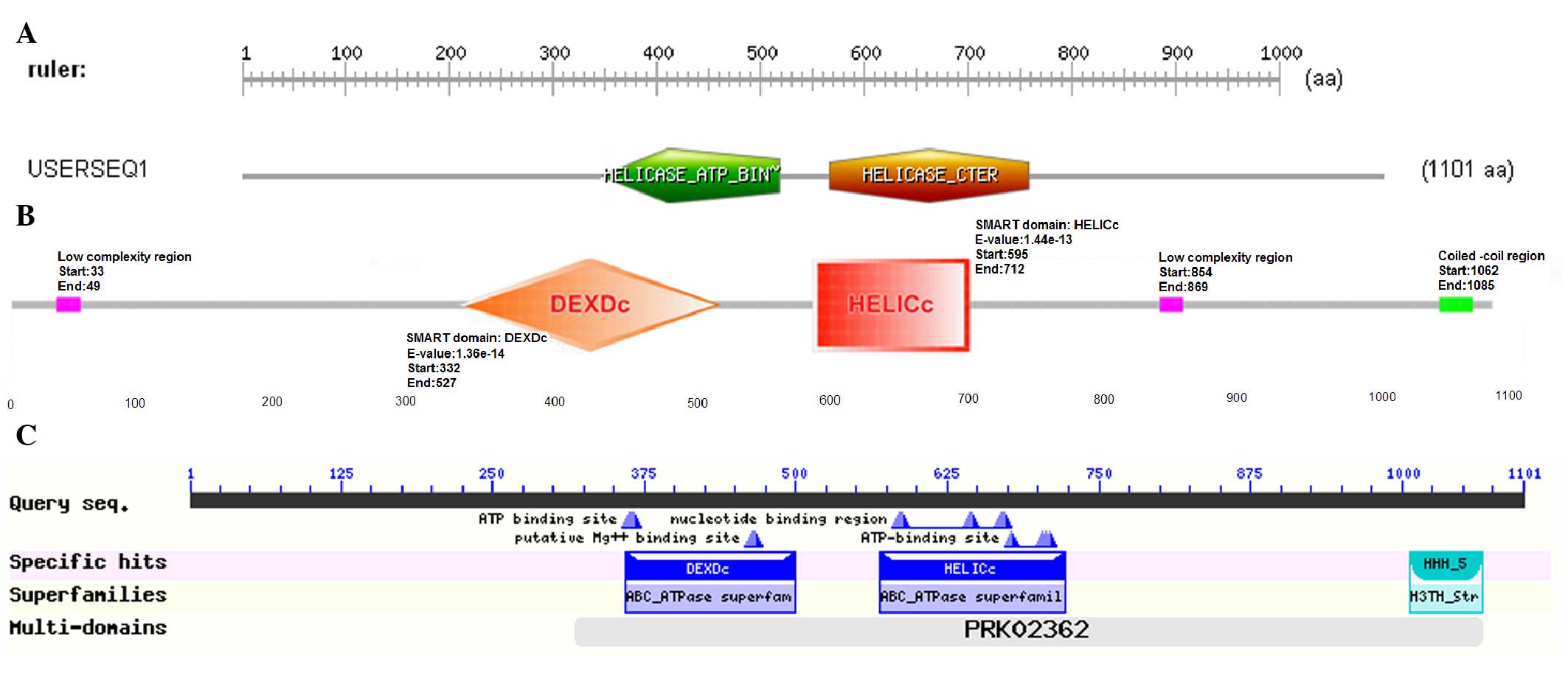
As for motifs, the functional structure domain
database of ScanProsite predicted that the HELQ protein contained
two functional domains, consisting of Helicase_ATP-Bind_1 and
Helicase_Cter, and these domains were responsible for binding and
hydrolyzing ATP, respectively, which conformed to the
characteristic of DNA helicase depending on ATP hydrolysis
(Fig. 1A). Domain architecture
analysis using SMART showed that 5 main domains could be found in
the HELQ sequence, with other features not shown in the diagram
(Fig. 1B) due to overlap. By
searching the NCBI Conserved Domains Database, 4 domains were
found, which were DEXDc, HELICc, HHH-5 and PRK02362 (Fig. 1C). According to the prediction of
SignalP-HMM and TMHMM, the HELQ protein has no evident signal
peptide and transmembrane domain.
Structural analysis of the HELQ
protein
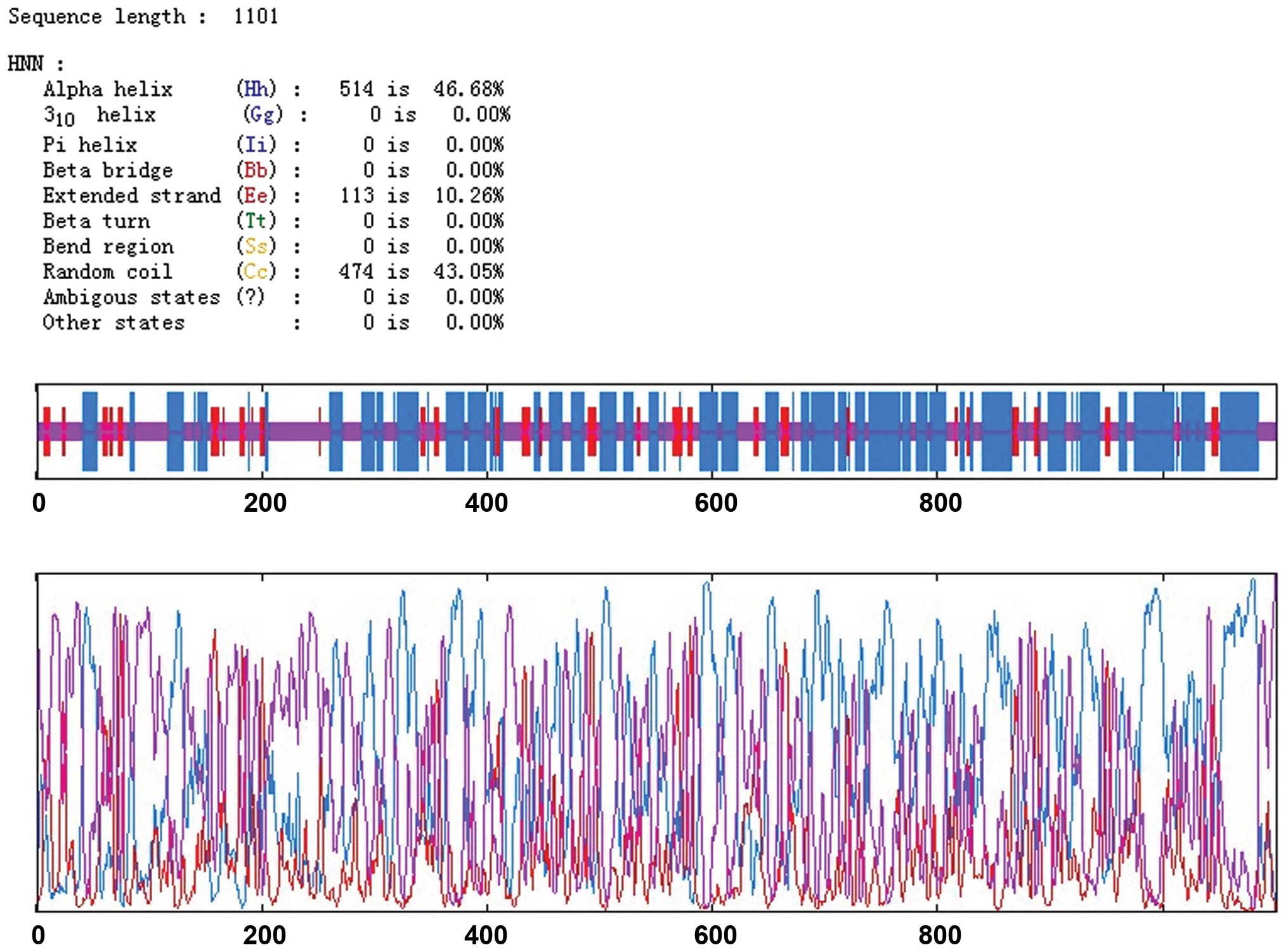
HNN was used to analyze the secondary structure of
the HELQ protein. Results showed that HELQ was mainly composed of
α-helixes (46.48%) and random coils (43.05%), with extended strands
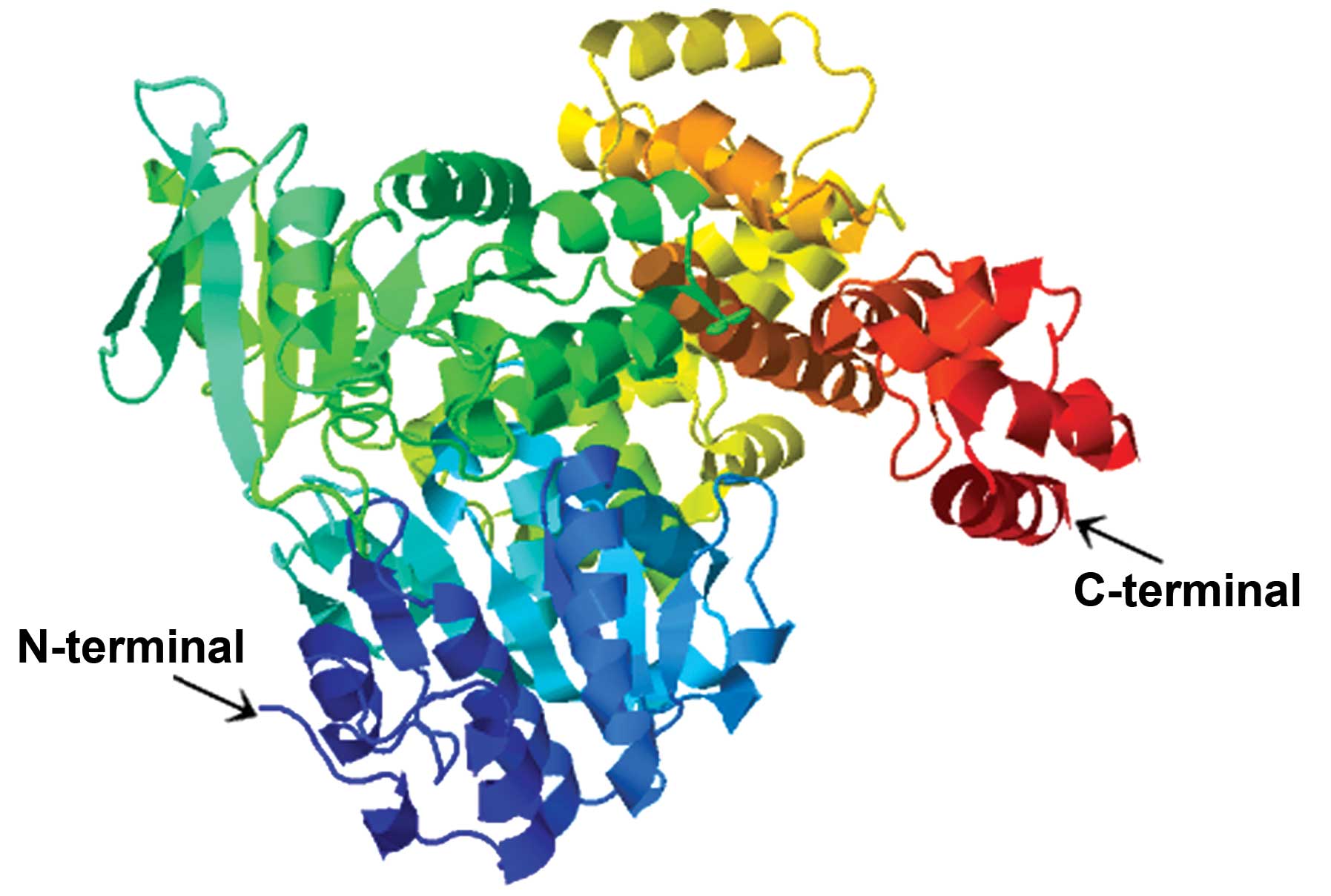
accounting for only 10.26% of the structure (Fig. 2). According to the prediction of the
tertiary structure using the threading method in the PHYRE2 Protein
Fold Recognition Server, the best template protein was c2va8A
(Protein Data Bank code), which had high homology with HELQ
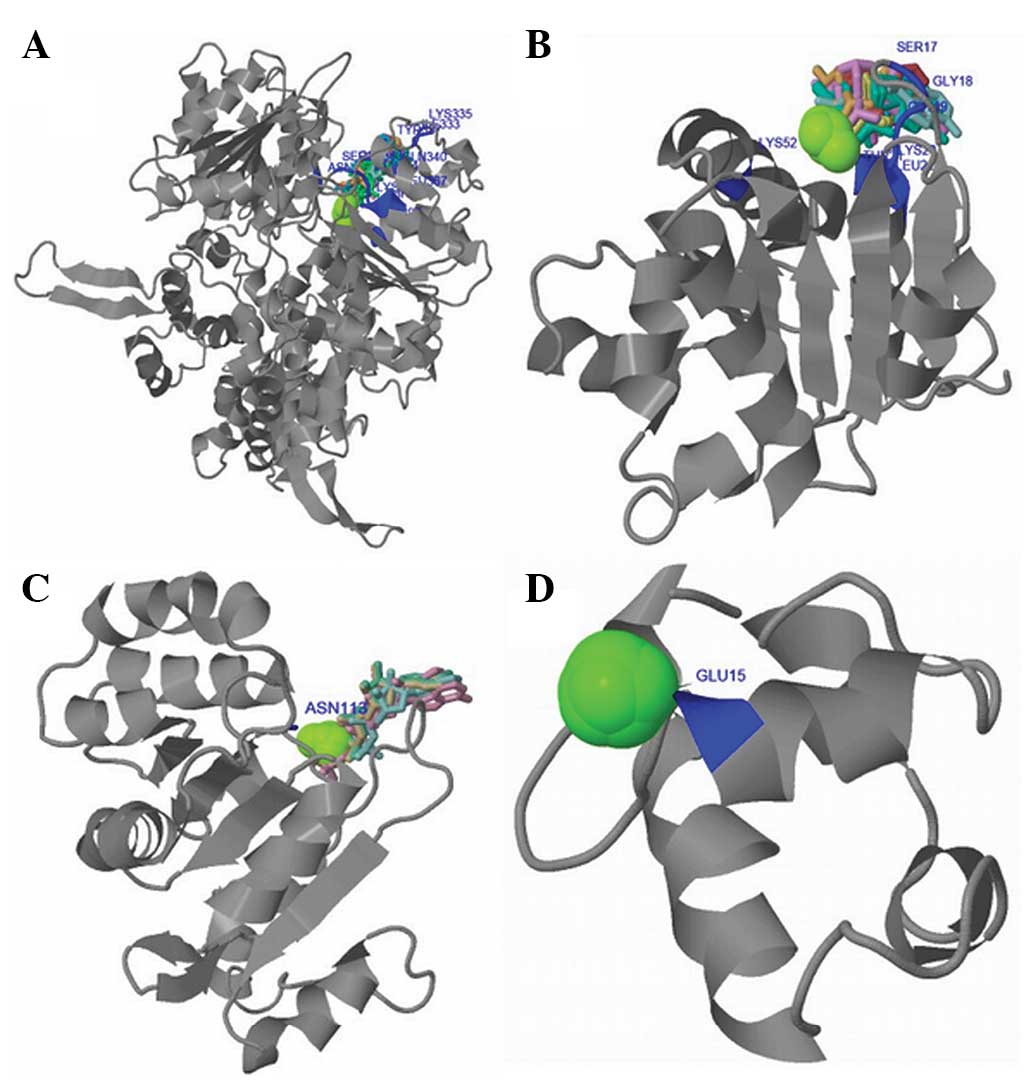
(Fig. 3). The 3DLigandSite Server was
used to construct the three-dimensional ligand binding model
(Fig. 4). It was found that the
ligand binding sites were distributed over ILE333, LYS335, TYR337,
GLN340, SER362, GLY363, GLY364, LYS365, THR366, LEU367, LYS397 and
ASN678. Heterogens present in the predicted binding sites consisted
of 10 adenosine diphosphate (ADP), 14 Mg2+ (MG) and 3
adenosine monophosphate (AMP). The functional domains
Helicase_ATP-Bind_1, Helicase_Cter and HHH_5 were separated from
the HELQ sequence and were respectively analyzed by building ligand
binding models. The results suggested that ligand binding sites in
the Helicase_ATP-Bind_1 domain distributed over SER17, GLY18,
GLY19, LYS20, THR21, LEU22, LYS52 and the heterogens present in the
predicted binding sites contained 12 ADP, 14 MG and 3 AMP. The
binding site present in the Helicase_Cter domain was ASN113, and
the heterogens contained 4 ADP, 12 MG and 1 ATP. The binding site
in the HHH_5 domain was GLU15 and the heterogens contained 7 MG and
33 Ca2+. The activity of HELQ is dependent on the
binding of ATP by the Helicase_Cter domain to supply energy. When
functional domains of HELQ are not complete, DNA replication cannot
progress normally.
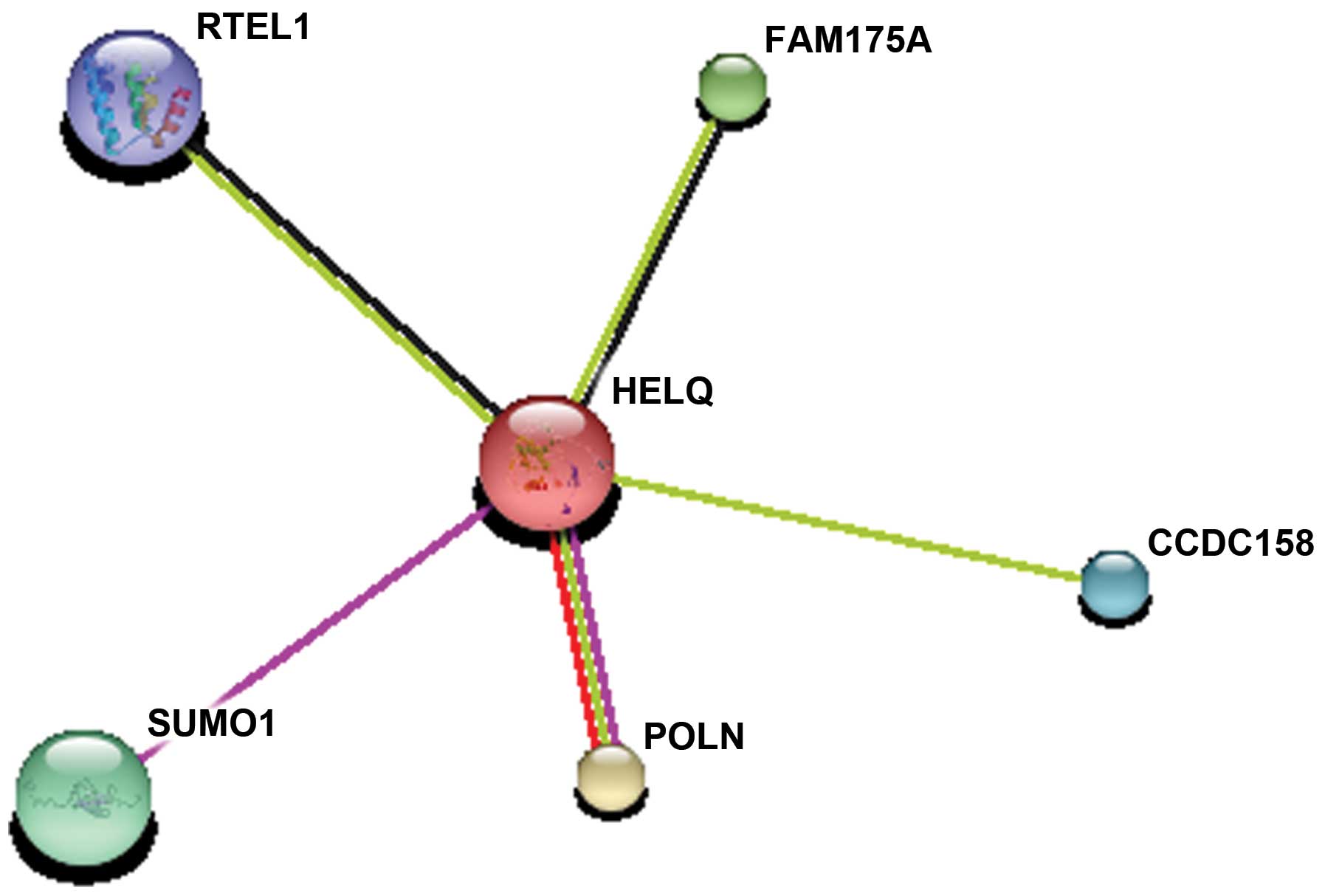
Prediction of protein-protein
interactions
STRING9.0 interactive database was utilized to
determine the functional protein association networks. Proteins
that interact with HELQ mainly included regulator of telomere
elongation helicase 1 (RTEL1), family with sequence similarity 175
member A (FAM175A), small ubiquitin-like modifier 1 (SUMO1), DNA
polymerase ν (POLN) and coiled-coil domain containing 158 (Fig. 5). They were coexpressed and had
similar functions in repairing the DNA lesions appearing in the
process of DNA replication during cell proliferation.
Gene ontology analysis
Gene ontology analysis was performed using
PredictProtein software. Molecular function ontology showed that
the activity of HELQ mainly consisted of protein binding, and
helicase, ATP-dependent RNA helicase, DNA strand annealing, ATPase,
ATP-dependent helicase, RNA helicase and RNA-dependent ATPase
activity (Table II). In addition,
biological process ontology analysis indicated that HELQ mainly
participated in the process of DNA repair (Table III).
 | Table II.Molecular function ontology. |
Table II.
Molecular function ontology.
| GO ID | GO term | Reliability, % |
|---|
| GO:0005515 | Protein binding | 26 |
| GO:0004386 | Helicase
activity | 17 |
| GO:0004004 | ATP-dependent RNA
helicase activity | 14 |
| GO:0000739 | DNA strand annealing
activity | 14 |
| GO:0016887 | ATPase activity | 14 |
| GO:0008026 | ATP-dependent
helicase activity | 12 |
| GO:0003724 | RNA helicase
activity | 11 |
| GO:0008186 | RNA-dependent ATPase
activity | 10 |
| GO:0042802 | identical protein
binding | 6 |
| GO:0043140 | ATP-dependent 3′-5′
DNA helicase activity | 4 |
| GO:0043621 | Protein
self-association | 4 |
| GO:0005524 | ATP binding | 4 |
| GO:0043008 | ATP-dependent protein
binding | 4 |
| GO:0008432 | JUN kinase
binding | 3 |
| GO:0017151 | DEAD/H-box RNA
helicase binding | 3 |
| GO:0003678 | DNA helicase
activity | 3 |
| GO:0030621 | U4 snRNA binding | 2 |
| GO:0003712 | Transcription
cofactor activity | 2 |
| GO:0017070 | U6 snRNA binding | 2 |
| GO:0003743 | Translation
initiation factor activity | 2 |
| GO:0003730 | mRNA 3′-UTR
binding | 2 |
| GO:0003677 | DNA binding | 2 |
| GO:0000339 | RNA cap binding | 2 |
| GO:0003723 | RNA binding | 2 |
| GO:0008143 | Poly(A) RNA
binding | 2 |
| GO:0008094 | DNA-dependent
ATPase activity | 1 |
| GO:0033592 | RNA strand
annealing activity | 1 |
| GO:0000403 | Y-form DNA
binding | 1 |
 | Table III.Biological process ontology. |
Table III.
Biological process ontology.
| GO ID | GO term | Reliability, % |
|---|
| GO:0006281 | DNA repair | 28 |
| GO:0006310 | DNA
recombination | 9 |
| GO:0006397 | mRNA
processing | 9 |
| GO:0006364 | rRNA
processing | 9 |
| GO:0009615 | Response to
virus | 8 |
| GO:0045449 | Regulation of
transcription | 8 |
| GO:0006314 | Intron homing | 8 |
| GO:0006268 | DNA unwinding
involved in replication | 8 |
| GO:0000398 | Nuclear mRNA
splicing, via spliceosome | 8 |
| GO:0008380 | RNA splicing | 8 |
| GO:0040007 | Growth | 8 |
| GO:0006260 | DNA
replication | 8 |
| GO:0007126 | Meiosis | 8 |
| GO:0006350 | Transcription | 8 |
| GO:0009792 | Embryonic
development ending in birth or egg hatching | 8 |
| GO:0006417 | Regulation of
translation | 8 |
| GO:0000393 | Spliceosomal
conformational changes to generate catalytic conformation | 8 |
Discussion
DNA helicases have crucial roles in maintaining
genome stability and stable DNA replication in all organisms. They
have been implicated in nucleotide excision repair, mismatch
repair, base excision repair, double strand break repair and
cross-link repair (14). The
helicases PriA, RecG, RuvAB, RecBCD, UurD, Srs2, Rep and RecQ are
well known for roles in promoting DNA repair and recombination by
several possible mechanisms (15–17).
Similar to RecQ, the Mus308 family of helicases supports genome
stability. The Mus308 locus was identified in Drosophila
melanogaster, being required for resistance to DNA crosslinking
agents (4). Moldovan et al
(18) found that the deletion of
HEL308 in human cells could induce sensitivity to
replication-blocking lesions, namely the ICLs induced by mitomycin
C and irinotecan hydrochloride. Biochemical studies have
established that human HEL308 is an ATP-dependent enzyme that
unwinds DNA with a 3′-5′ polarity (3,19), which
is consistent with the structure of HELQ containing ATP-binding
sites found in the present study. In the functional domains of
HELQ, Helicase_ATP-Bind_1 and Helicase_Cter locate in the center
position and they mainly perform helicase activity. At the
C-terminal, the helix-hairpin-helix configuration may be important
for binding to DNA strands. The results of the functional protein
association networks found that HELQ was involved in ubiquitination
in the process of DNA repair. RTEL1 is responsible for the
maintenance of chromosome ends and has a synergistic effect with
proliferating cell nuclear antigen (PCNA) in the process of DNA
replication to ensure cell growth and division, and to avoid
genetic mistakes. When RTEL1 cannot combine with PCNA, DNA
replication cannot continue and the errors produced lead to cancer
(20). FAM175A mediates the formation
of the BRCA1-RAP80 complex, which adds or modifies existing
ubiquitin chains to promote DNA damage repair (21). SUMO1 is important in the control
genomic integrity. Hu et al (22) found that SUMO1 repaired DNA lesions
through an ubiquitin-like modification path. POLN works as a DNA
polymerase and participates in DNA repair to make DNA replication
proceed normally (18). However, the
exact mechanism of HELQ remains unclear.
An increasing number of studies focus on the
function exploration of HELQ. Ward et al (23) found that HELQ-1 played a role in
meiotic double-strand break repair by promoting postsynaptic RAD-51
filament disassembly in Caenorhabditis elegans. Another
study identified that in humans, HELQ was expressed in the ovaries,
testes, heart and skeletal muscle (24). Luebben et al (25) reported that mammalian HELQ contributed
to genome stability in unchallenged conditions through a mechanism
distinct from the function of Fanconi anemia complementation group
C. HELQ in humans requires additional investigation, particularly
the association with cancer. The occurrence of tumors is usually
closely associated with abnormal DNA replication and cell
proliferation. Appropriate readjustment in the structure of HELQ to
change its primary function may be used to alleviate DNA damage or
promote DNA variation.
The genetic complexity of cancer has posed a
challenge for devising successful therapeutic treatments. Tumor
resistance to cytotoxic chemotherapy drugs and radiation, which
induce DNA damage, has limited their effectiveness (26). Targeting the DNA damage response is
one strategy for combating cancer. The prospect for success of
chemotherapy treatment may be improved by the selective
inactivation of a DNA repair pathway.
In conclusion, the structure of HELQ protein was
predicted and analyzed in this study. Its unique structural
characteristics will have an important role in future
investigations of HELQ gene deletion, as well as the etiological
analysis and targeted therapy of ovarian cancer.
References
|
1
|
Thomson CA, E Crane T, Wertheim BC,
Neuhouser ML, Li W, Snetselaar LG, Basen-Engquist KM, Zhou Y and
Irwin ML: Diet quality and survival after ovarian cancer: Results
from the Women's health initiative. J Natl Cancer Inst.
106:dju3142014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Adelman CA, Lolo RL, Birkbak NJ, Murina O,
Matsuzaki K, Horejsi Z, Parmar K, Borel V, Skehel JM, Stamp G, et
al: HELQ promotes RAD51 paralogue-dependent repair to avert germ
cell loss and tumorigenesis. Nature. 502:381–384. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Marini F and Wood RD: A human DNA helicase
homologous to the DNA cross-link sensitivity protein Mus308. J Biol
Chem. 277:8716–8723. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Boyd JB, Sakaguchi K and Harris PV: mus308
mutants of Drosophila exhibit hypersensitivity to DNA cross-linking
agents and are defective in a deoxyribonuclease. Genetics.
125:813–819. 1990.PubMed/NCBI
|
|
5
|
Leonhardt EA, Henderson DS, Rinehart JE
and Boyd JB: Characterization of the mus308 gene in Drosophila
melanogaster. Genetics. 133:87–96. 1993.PubMed/NCBI
|
|
6
|
Dolgova EV, Alyamkina EA, Efremov YR,
Nikolin VP, Popova NA, Tyrinova TV, Kozel AV, Minkevich AM,
Andrushkevich OM, Zavyalov EL, et al: Identification of cancer stem
cells and a strategy for their elimination. Cancer Biol Ther.
15:1378–1394. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Roy S: Maintenance of genome stability in
plants: Repairing DNA double strand breaks and chromatin structure
stability. Front Plant Sci. 5:4872014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li WQ, Hu N, Hyland PL, Gao Y, Wang ZM, Yu
K, Su H, Wang CY, Wang LM, Chanock SJ, et al: Genetic variants in
DNA repair pathway genes and risk of esophageal squamous cell
carcinoma and gastric adenocarcinoma in a Chinese population.
Carcinogenesis. 34:1536–1542. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gao Y, He Y, Xu J, Xu L, Du J, Zhu C, Gu
H, Ma H, Hu Z, Jin G, et al: Genetic variants at 4q21, 4q23 and
12q24 are associated with esophageal squamous cell carcinoma risk
in a Chinese population. Human Genet. 132:649–656. 2013. View Article : Google Scholar
|
|
10
|
Liang C, Marsit CJ, Houseman EA, Butler R,
Nelson HH, McClean MD and Kelsey KT: Gene-environment interactions
of novel variants associated with head and neck cancer. Head Neck.
34:1111–1118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
McKay JD, Truong T, Gaborieau V, Chabrier
A, Chuang SC, Byrnes G, Zaridze D, Shangina O, Szeszenia-Dabrowska
N, Lissowska J, et al: A genome-wide association study of upper
aerodigestive tract cancers conducted within the INHANCE
consortium. PLoS Genet. 7:e10013332011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Käll L, Krogh A and Sonnhammer EL: A
combined transmembrane topology and signal peptide prediction
method. J Mol Biol. 338:1027–1036. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Scott L, Chahine J and Ruggiero J:
Prediction of Protein Structures using a Hopfield NetworkSixth
Brazilian Symposium on Neural Networks 2000. IEEE Computer Society
Press; Washington, DC: pp. 284. 2000, View Article : Google Scholar
|
|
14
|
McGlynn P: Helicases at the replication
fork. Adv Exp Med Biol. 767:97–121. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsang E and Carr AM: Replication fork
arrest, recombination and the maintenance of ribosomal DNA
stability. DNA repair (Amst). 7:1613–1623. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
McGlynn P and Lloyd RG: Recombinational
repair and restart of damaged replication forks. Nat Rev Mol Cell
Biol. 3:859–870. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Singleton MR and Wigley DB: Modularity and
specialization in superfamily 1 and 2 helicases. J Bacteriol.
184:1819–1826. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Moldovan GL, Madhavan MV, Mirchandani KD,
McCaffrey RM, Vinciguerra P and D'Andrea AD: DNA polymerase POLN
participates in cross-link repair and homologous recombination. Mol
Cell Biol. 30:1088–1096. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tafel AA, Wu L and McHugh PJ: Human HEL308
localizes to damaged replication forks and unwinds lagging strand
structures. J Biol Chem. 286:15832–15840. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Vannier JB, Sandhu S, Petalcorin MI, Wu X,
Nabi Z, Ding H and Boulton SJ: RTEL1 is a replisome-associated
helicase that promotes telomere and genome-wide replication.
Science. 342:239–242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Coleman KA and Greenberg RA: The
BRCA1-RAP80 complex regulates DNA repair mechanism utilization by
restricting end resection. J Biol Chem. 286:13669–13680. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hu Y and Parvin JD: Small ubiquitin-like
modifier (SUMO) isoforms and conjugation-independent function in
DNA double-strand break repair pathways. J Biol Chem.
289:21289–21295. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ward JD, Muzzini DM, Petalcorin MI,
Martinez-Perez E, Martin JS, Plevani P, Cassata G, Marini F and
Boulton SJ: Overlapping mechanisms promote postsynaptic RAD-51
filament disassembly during meiotic double-strand break repair. Mol
Cell. 37:259–272. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Marini F, Kim N, Schuffert A and Wood RD:
POLN, a nuclear PolA family DNA polymerase homologous to the DNA
cross-link sensitivity protein Mus308. J Biol Chem.
278:32014–32019. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Luebben SW, Kawabata T, Akre MK, Lee WL,
Johnson CS, O'Sullivan MG and Shima N: Helq acts in parallel to
Fancc to suppress replication-associated genome instability.
Nucleic Acids Res. 41:10283–10297. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kelley MR and Fishel ML: DNA repair
proteins as molecular targets for cancer therapeutics. Anticancer
Agents Med Chem. 8:417–425. 2008. View Article : Google Scholar : PubMed/NCBI
|