Introduction
At present, the development of antitumor drugs on
the basis of natural proteins and peptides that are able to induce
apoptotic death in tumor cells and selectively suppress tumor
growth is one of the most popular strategies in the search for
novel therapeutics (1).
The protein lactaptin (~8.6 kDa), a proteolytic
fragment of human κ-casein, with the ability to induce apoptotic
death in tumor cells in vitro, was previously found in human
breast milk (2,3). A series of recombinant analogs of the
natural peptide has been created (4,5). It has
been demonstrated that the lactaptin analog, RL2, induces apoptosis
in mice and human tumor cells in vitro, and reduces the
growth and metastasis of animal and human tumors in vivo
(6,7).
The preclinical trials of the RL2-based therapeutic drug lactaptin
have been successful (7,8). The safety and antitumor efficacy of this
drug have been demonstrated. However, lactaptin has several major
drawbacks, as with all protein-based therapeutic drugs. Lactaptin
is distributed evenly throughout the organism, which reduces the
concentration of the drug in the tumor and, consequently, reduces
the antitumor efficacy (8).
The present study reports a technique developed to
improve the antitumor efficiency of the drug lactaptin using
conjugates of tumor-specific peptides with RL2. One of the most
common and convenient ways used to isolate tumor-specific peptides
is by screening phage-display peptide libraries. Due to small size,
low immunogenicity and relatively cheap chemical synthesis,
tumor-specific peptides are considered as targeted delivery
vehicles for therapeutic genes, cytokines, imaging agents,
pro-apoptotic peptides and therapeutic drugs. Currently, a number
are in clinical trials, as individual therapeutic agents or as
agents for delivering drugs to the target organs and tissues
(9).
In the present study, tumor-specific peptides were
selected by in vivo screening of a phage-display peptide
library. Recombinant fusion proteins composed of the
apoptosis-inducing protein RL2 and the selected peptides were
synthesized. The cytotoxic properties of the fusion proteins were
assessed using mouse and human tumor cells in culture, and the
targeting properties were investigated using a mouse tumor
model.
Materials and methods
Animals
The animals used in the present were 3–4-month-old
A/Sn, C57Bl/6, CBA, C3H strain mice (22–25 g) bred at the Institute
of Cytology and Genetics, Siberian Branch of the Russian Academy of
Sciences (SB RAS), Novosibirsk, Russia. The mice were kept under
standard conditions and received standard food and water according
to the approved study protocol (EEC directive 86/609/EEC). The
protocol was approved by the Committee on the Ethics of Animal
Experiments of the Administration of SB RAS (permit no. 8–2012),
Novosibirsk, Russia. All the animal experiments were performed in
compliance with the recommendations and requirements of the World
Society for the Protection of Animals, and the European Convention
for the Protection of Vertebrate Animals Used for Experimental and
Other Scientific Purposes (Strasbourg, 1986).
Tumor strains and cell lines
The hepatocarcinoma-1 (HA-1), hepatocarcinoma-29
(HA-29), lung adenocarcinoma (LA), Krebs-2 carcinoma, Lewis lung
carcinoma (LLC) and Ehrlich carcinoma strains were obtained from
the Institute of Cytology and Genetics, SB RAS (Novosibirsk,
Russia) and maintained in the ascitic form by weekly
intraperitoneal transplantations using the following mouse strains:
A/Sn (HA-1, LA), C57 Black (Krebs-2 carcinoma), CBA (LLC, HA-29)
and C3H (Ehrlich carcinoma).
For in vivo experiments, ascites tumor cells
were resuspended in saline to a final concentration of
106 cells/ml and 150 µl of suspension was injected
subcutaneously into the back of each mouse. After the tumor nodes
grew to 0.7–0.9 cm in diameter, the animals were used in the study
experiments.
The HA-1 cells were cultured in vitro in
Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich; Merck
Millipore, Darmstadt, Germany) in the presence of 10% FBS (Gibco;
Thermo Fisher Scientific, Inc., Waltham, MA, USA), 2 mM L-glutamine
(Sigma-Aldrich; Merck Millipore), 250 mg/ml amphotericin В and 100
U/ml penicillin/streptomycin (Gibco; Thermo Fisher Scientific,
Inc.) at 37°С in an atmosphere containing 5% СО2.
The В16 melanoma, human breast adenocarcinoma
MDA-MB-231 and MCF-7 cell lines were obtained from the Russian
Collection of Cell Cultures (Institute of Cytology, RAS, Saint
Petersburg, Russia). В16 melanoma cells were cultured in
vitro in DMEM (Sigma-Aldrich; Merck Millipore) in the presence
of 10% FBS (Gibco; Thermo Fisher Scientific, Inc.), 2 mM
L-glutamine (Sigma-Aldrich; Merck Millipore), 250 mg/ml
amphotericin В and 100 U/ml penicillin/streptomycin (Gibco; Thermo
Fisher Scientific, Inc.) at 37°С in an atmosphere containing 5%
СО2. For the in vivo experiments, В16 melanoma
cells were detached with 0.05% trypsin-EDTA, diluted to a final
concentration of 106 cells/ml, and 150 µl of the
suspension was injected subcutaneously into the back of each mouse.
After the tumor nodes grew to 0.5–0.6 cm in diameter, the animals
were used in the study experiments.
MCF-7 cells were cultured in Iscove's modified
Dulbecco's medium (Sigma-Aldrich; Merck Millipore) in the presence
of 10% FBS, 2 mM L-glutamine, 250 mg/ml amphotericin В and 100 U/ml
penicillin/streptomycin.
MDA-MB-231 cells were cultured in L15 in the
presence of 10% FBS, 2 mM L-glutamine, 250 mg/ml amphotericin В and
100 U/ml penicillin/streptomycin at 37°С in an atmosphere
containing 5% СО2.
In vivo screening of the phage-display
peptide library
A mouse with a subcutaneously transplanted HA-1
tumor was administered, via tail vein injection, 0.5 ml of the
phage-display peptide library (2×109 PFU/ml; New England
Biolabs, Inc., Ipswich, MA, USA) diluted in saline. The phage
particles were allowed to circulate for 5 min, after which the
mouse was sedated with 2.5% Avertin solution at a dose of 0.2 ml
per 20 g of mouse body weight and the heart was perfused with 15 ml
saline to remove the unbound phage particles from the bloodstream.
The tumor was removed, washed with phosphate-buffered saline (PBS)
and homogenized in 1 ml PBS containing 1 mM of the protease
inhibitor phenylmethylsulfonyl fluoride. The homogenate was
centrifuged at 10,000 × g for 10 min at room temperature and
then the supernatant was removed. Pellets were resuspended in 1 ml
blocking buffer and centrifuged at 10,000 × g for 10 min at
room temperature and then the supernatant was removed. Pellets were
resuspended in 1 ml liquid E. сoli ER2738 culture in
lysogeny broth (optical density at 600 nm, 0.2–0.3) to elute the
bacteriophages that had bound to the tumor, and were incubated for
30 min at 37°С. The eluate containing the phage particles was
centrifuged at 10,000 × g for 10 min at room temperature.
The suspension containing the phage particles was then amplified
and the amplified phage particles were used in further rounds of
selection.
Sequencing of phage DNA
After the third and fourth rounds of selection, the
phage particles were titrated to obtain individual phage colonies,
which were used for the isolation of DNA according to the protocol
described by the manufacturer of the phage-display peptide
library.
The single-stranded DNAs were used in Sanger
sequencing (−96 gIII sequencing primer; 5′-CCCTCATAGTTAGCGTAACG-3′;
New England Biolabs, Inc.). The sequencing reaction products were
determined using an ABI 310 Genetic Analyzer (Applied Biosystems;
Thermo Fisher Scientific, Inc.) (located at the Genomics Core
Facility of SB RA, Novosibirsk, Russia). The nucleotide sequences
of the inserts encoding the peptides were analyzed using MEGA 4.0
software (10).
Construction of plasmid DNA-expressing
fusion proteins
The RL2 sequence was amplified from plasmid
pGSDI/RL2 (4) using RL2_F and
c1_RL2_R primers (Table I) with the
following cycling conditions: 96°C for 5 min, followed by 35 cycles
of 96°C for 30 sec, 66°C for 30 sec, 72°C for 30 sec and a final
step of 72°C for 10 min. At the first stage, to produce a fusion
DNA sequence that encodes a selected peptide and RL2, the sequences
that encode the peptides Т1 and Т2 and the region of overlap with
the RL2 sequence were produced. The Т1 sequence was amplified from
ClonN1_target as a template using c1_RL2_F and c1_R primers with
the following cycling conditions: 96°C for 5 min, followed by 35
cycles of 96°C for 30 sec, 65°C for 30 sec, 72°C for 15 sec and a
final step of 72°C for 10 min. The T2 sequence was amplified from
ClonN2_target as a template using c2_RL2_F and c2_R primers with
the following cycling conditions: 96°C for 5 min, followed by 35
cycles of 96°C for 30 sec, 65°C for 30 sec, 72°C for 15 sec and a
final step of 72°C for 10 min. At the second stage, the fusion
T1_RL2 and T2_RL2 DNA sequences encoding the tumor-targeting
peptide and RL2 were produced by polymerase chain reaction using
the primer pairs c1_RL2_F, c1_RL2_R and c2_RL2_F, c1_RL2_R with the
following cycling conditions: 96°C for 5 min, followed by 35 cycles
of 96°C for 30 sec, 65°C for 35 sec, 72°C for 35 sec and a final
step of 72°C for 10 min. The amplicons produced at the first stage
and the RL2 amplicon were used as templates. All the amplicons were
separated on an agarose gel and extracted from the gel according to
the manufacturer's instructions (Qiagen, Inc., Valencia, CA,
USA).
 | Table I.Sequences of the oligonucleotides
used in the construction of plasmid DNA-expressing fusion
proteins. |
Table I.
Sequences of the oligonucleotides
used in the construction of plasmid DNA-expressing fusion
proteins.
|
Oligonucleotide | Sequence
(5′→3′) |
|---|
| RL2_F |
AACCAGAAACAACCAGCATGCCATGAGAATGAT |
| c1_RL2_R |
CATCATGGATCCTTAGTGATGGTGATGGTGATGTGATCCGCCGATGGT |
| c1_RL2_F |
CATCATCCATGGGTTTGCATACTTCGGCT |
| c1_R |
CTGGTTGTTTCTGGTTCGAACCTCACCAGCAA |
| ClonN1_target |
GGTTTGCATACTTCGGCTACTAATCTGTATTTGCATGGTGGAGGTTCG |
| c2_RL2_F |
CATCATCCATGGGCAGTGGTGTGTATAAGGTT |
| c2_R |
TGGTTGTTTCTGGTTCGAACCTCCACCATGCT |
| ClonN2_target |
AGTGGTGTGTATAAGGTTGCGTATGATTGGCAGCATGGTGGAGGTTCG |
DNA fragments encoding the fusion proteins T1_RL2
and T2_RL2 were inserted in plasmid pET-15b (Novagen; Merck
Millipore) using the BamHI and NcoI restriction
sites. The recombinant plasmids were cloned in E. coli TOP10
(Life Technologies; Thermo Fisher Scientific, Inc.).
The sequencing reaction was run using a BigDye
Terminator Cycle Sequencing Ready Reaction kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.) and the reaction products were
purified using a DyeEx Spin kit (Qiagen, Inc.) at the Genomics Core
Facility of SB RAS. For the sequencing reaction, Т7seq_F
5′-TAATACGACTCACTATAGGG-3′ and Т7seq_R 5′-GCTAGTTATTGCTCAGCGGT-3′
were used as primers. The purified products were sequenced using an
ABI 310 Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The nucleotide sequences of interest were
analyzed using MEGA 4.0 software.
The recombinant plasmids pET-15b_Т1_RL and
pET-15b_Т2_RL, which encode the fusion proteins T1_RL and T2_RL,
respectively, were expressed in E. coli BL21 (DE3) (New
England Biolabs, Inc., Ipswich, MA, USA).
MTT assay
The cytotoxic effects of the recombinant lactaptin
analogs RL2, T1_RL and T2_RL on mouse and human tumor cells were
investigated using the МТТ assay (Sigma-Aldrich; Merck Millipore)
according to a protocol described previously (6). The cells that had reached 30% confluence
in a 96-well plate were incubated for 48 h with protein
preparations at various concentrations (0.1–0.4 mg/ml). After
incubation, the supernatant was removed and 200 µl MTT solution in
RPMI 1640 medium (0.5 mg/ml) was added to each well and incubated
for 4 h at 37°С. The formazan crystals were dissolved in 150 µl
dimethyl sulfoxide. The optical density of the formazan solutions
was measured using an Apollo LB912 photometer (Berthold
Technologies, Oak Ridge, TN, USA) at a wavelength of 570 nm. Cell
viability was determined relative to the viability of the control
cells (100%) ± standard deviation in three independent
experiments.
Flow cytometry
The cells were incubated in 6-well culture plates
and used in the experiments upon reaching 80–90% confluence. The
recombinant lactaptin analogs RL2, Т1_RL and T2_RL (0.6 mg/ml) were
added to culture medium and incubated for 18 and 24 h. After
incubation, the cells detached from the support were pooled with
the cells detached by trypsinization (PBS, 0.5 mg/ml trypsin and
0.4 mg/ml EDTA). The pooled cells were precipitated by
centrifugation at 400 × g for 5 min and washed with PBS. The
exposure of phosphatidylserine (PS) at the outer surface of the
cell membrane, and the activation of caspase-3 and −7 were analyzed
by flow cytofluorometry with fluorescein isothiocyanate Annexin V
Apoptosis Detection kit I (BD Pharmingen, San Jose, CA, USA) and
the Vybrant FAM Caspase-3 and −7 assay kit (Life Technologies;
Thermo Fisher Scientific, Inc.) using a FACSCanto II cell analyzer
(BD Biosciences, Franklin Lakes, NJ, USA).
Labeling of the recombinant lactaptin
analogs with sulfo-Cy5
The recombinant lactaptin analogs RL2, Т1_RL and
Т2_RL were isolated from E. coli and purified according to a
protocol for the isolation of RL2 (5).
To label RL2, T1_RL and T2_RL with sulfo-Cy5, 0.2 mM
of each protein was incubated with 1.25 mM of the fluorescent dye
sulfo-Cy5 maleimide (Ex/Em=646/662; Lumiprobe GmbH, Hannover,
Germany) in Tris-HCl buffer (pH 5.5) for 4 h at 25°С. The
purification of the conjugates RL2-Cy5, T1_RL-Cy5 and T2_RL-Cy5 was
performed by reversed-phase high-performance liquid chromatography
on a С18 column using a Milichrom A-02 chromatograph (EcoNova,
Novosibirsk, Russia) with acetonitrile-water as a mobile phase.
Retention of the RL2-Cy5, T1_RL-Cy5
and T2_RL-Cy5 conjugates in tumor tissue
Mice with subcutaneously transplanted HA-1 tumors
were administered with 80 µg of the conjugates RL2-Cy5, T1_RL-Cy5
and T2_RL-Cy5 in 200 µl of PBS via tail vein injection. The
conjugates were allowed to circulate for 10 min, after which the
tumors were removed and analyzed using a Kodak In Vivo FX
Professional Imaging system. The excitation (650 nm) and emission
(700 nm) filters were used to detect the fluorescence of Cy5. The
fluorescence intensity was recorded as total photons per second per
square centimeter (mean ± standard deviation).
Statistical analysis
The data from the mouse experiments were
statistically processed using one-way analysis of variance.
Post-hoc testing was performed using Fisher's least significant
differences (LSD). P<0.05 was considered to indicate a
statistically significant difference. STATISTICA v10.0 was used to
perform the statistical analyses (StatSoft, Inc., Tulsa, OK,
USA).
Results
In vivo screening of the phage-display
peptide library
To isolate tumor-specific peptides, four rounds of
selection were performed in vivo on A/Sn strain mice with
subcutaneously transplanted HA-1 tumors. After the third round of
selection, 37 displayed sequences were identified and analyzed, and
after the fourth round, 38 sequences were identified (Table II).
 | Table II.Sequences and frequencies of
displayed peptides isolated after the third and fourth rounds of
in vivo selection using A/Sn strain mice with subcutaneously
transplanted HA-1 tumors. |
Table II.
Sequences and frequencies of
displayed peptides isolated after the third and fourth rounds of
in vivo selection using A/Sn strain mice with subcutaneously
transplanted HA-1 tumors.
| Peptide
sequence | Frequency, % |
|---|
| III round | 35.1 |
|
GLHTSATNLYLHa | 21.6 |
|
SGVYKVAYDWQHa |
5.4 |
|
GSAPLLTVDTSK |
2.7 |
|
GRIEPHRLFQGA |
2.7 |
|
QFDYMRPANDTH |
2.7 |
|
LGSSHGHGASHQ |
2.7 |
|
DRWVARDPASIFa |
2.7 |
|
YASDLQPLTQFI |
2.7 |
|
STSDYTQWTSYA |
2.7 |
|
YGHGLNQAELRQ |
2.7 |
|
GDGNSVLKPGNW |
2.7 |
|
GTGLVTLPRLTV |
2.7 |
|
DLGRASWNPFFS |
2.7 |
|
ANLTRWPHNVST |
2.7 |
|
DVSTYKTNAQNS |
2.7 |
|
NWSHNVRLNYTY |
2.7 |
|
NTNYVTWSPSSR | 35.1 |
| IV round |
|
GLHTSATNLYLHa | 63.2 |
|
SGVYKVAYDWQHa | 21.1 |
|
MHPNAGHGSLMR |
5.3 |
|
SFKIPYHYDSGQ |
2.6 |
|
DRWVARDPASIFa |
2.6 |
|
ENLMHADKNFRS |
2.6 |
|
LQSTSPAYTHRM |
2.6 |
|
GDGNSVLKPGNW |
2.6 |
After the third round of in vivo selection on
the HA-1 tumor model, the most frequent outputs were two displayed
peptides, GLHTSATNLYLH (35.1%) and SGVYKVAYDWQH (21.6%). After the
fourth round of in vivo selection, the frequency of the
peptide GLHTSATNLYLH increased to 63.2%, while that of the peptide
SGVYKVAYDWQH remained unchanged.
As the displayed peptides GLHTSATNLYLH and
SGVYKVAYDWQH occurred with the highest frequency in the in
vivo experiments, those clones were used in further
experiments.
In vivo specificity of the selected
peptides to various mouse tumors
The ability of the displayed peptides GLHTSATNLYLH
and SGVYKVAYDWQH to specifically bind to tumors in vivo was
studied using HA-1, LA, В16 melanoma, Krebs-2 carcinoma, LLC,
Ehrlich carcinoma and HA-29 tumors.
The individual phage clones displaying the selected
peptides were administered to mice via tail vein injection. At 24 h
post-administration, the tumor and the control organs (lung and
liver) were examined for bacteriophages (PFU/mg tissue) by
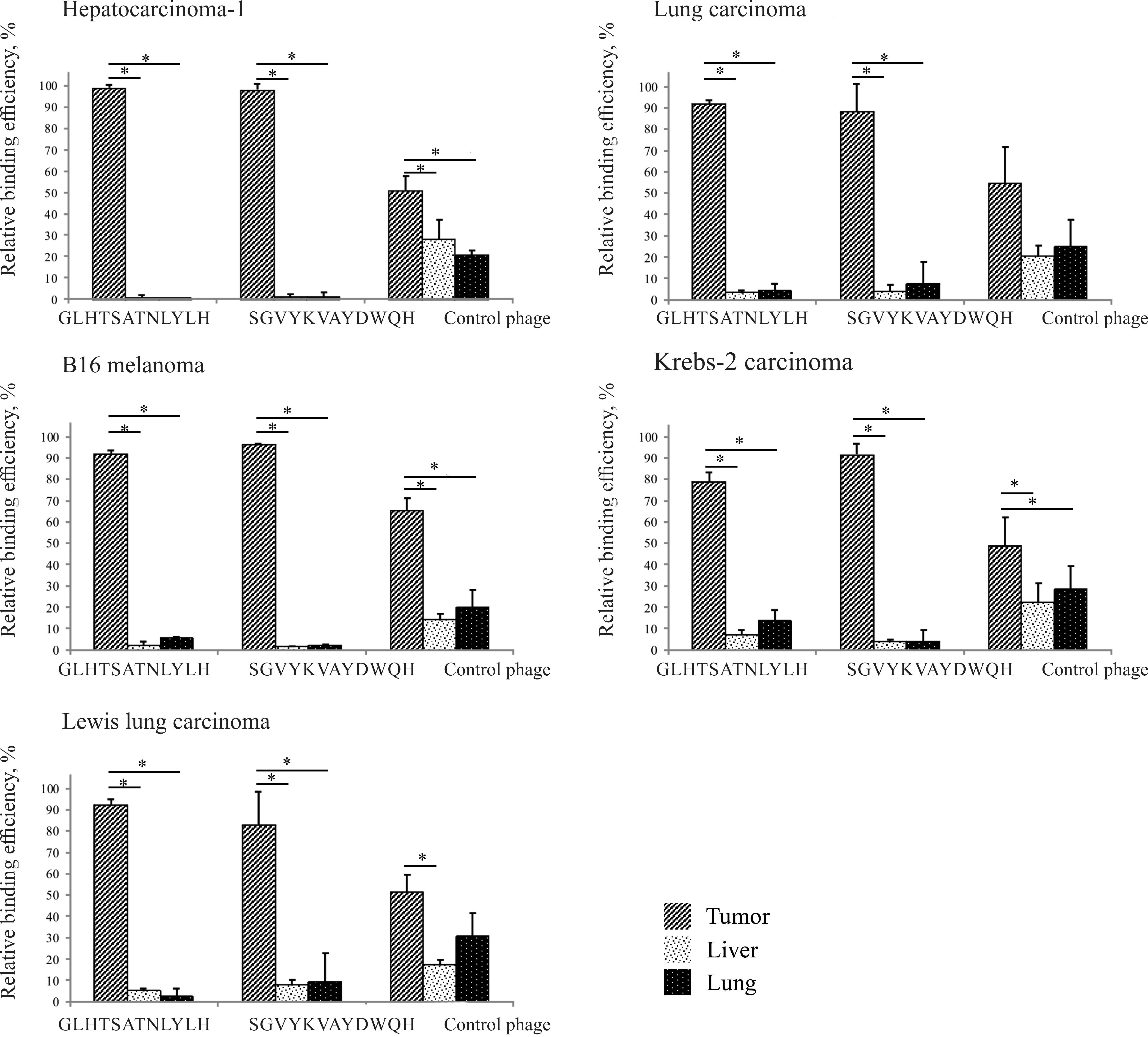
titrating on E. сoli ER2738 culture cells (Fig. 1).
As shown from the data presented in Fig. 1, the selected peptides were
significantly bound not only to the selection-specific tumor
(HA-1), but also to other tumors (LA, В16 melanoma, Krebs-2
carcinoma and LLC) (P<0.05). The highest amount of binding was
for HA-1. At 24 h post-intravenous administration, the
concentration of the control bacteriophage (without any displayed
peptide) in the tumor remained relatively low (just two or three
times the value in the liver and lung). This is consistent with
literature data, according to which the wild-type bacteriophage has
a low capacity for retention and accumulation in tumor tissue, as
the vascular system in tumors is poorly organized (11).
Construction of fusion proteins with
the selected peptides and RL2
Two genetic constructs, pET-15b_T1_RL and
pET-15b_T2_RL, were made for the production of recombinant fusion
proteins composed of RL2 and the selected tumor-targeting peptides
that deliver RL2.
The sequence encoding the fusion protein was
constructed so that the synthesis product appears as a polypeptide
with the tumor-targeting peptide fused to the N-terminus (with
methionine as the first amino acid) and RL2 fused to the
C-terminus. The targeting peptide and RL2 are spaced by a glycine
linker, which provides for the flexibility of the peptide (Fig. 2).
To produce the desired recombinant proteins, two
E. сoli expression systems, BL21 (DE3)/pET-15b_Т1_RL and
BL21 (DE3)/pET-15b_Т2_RL, were developed.
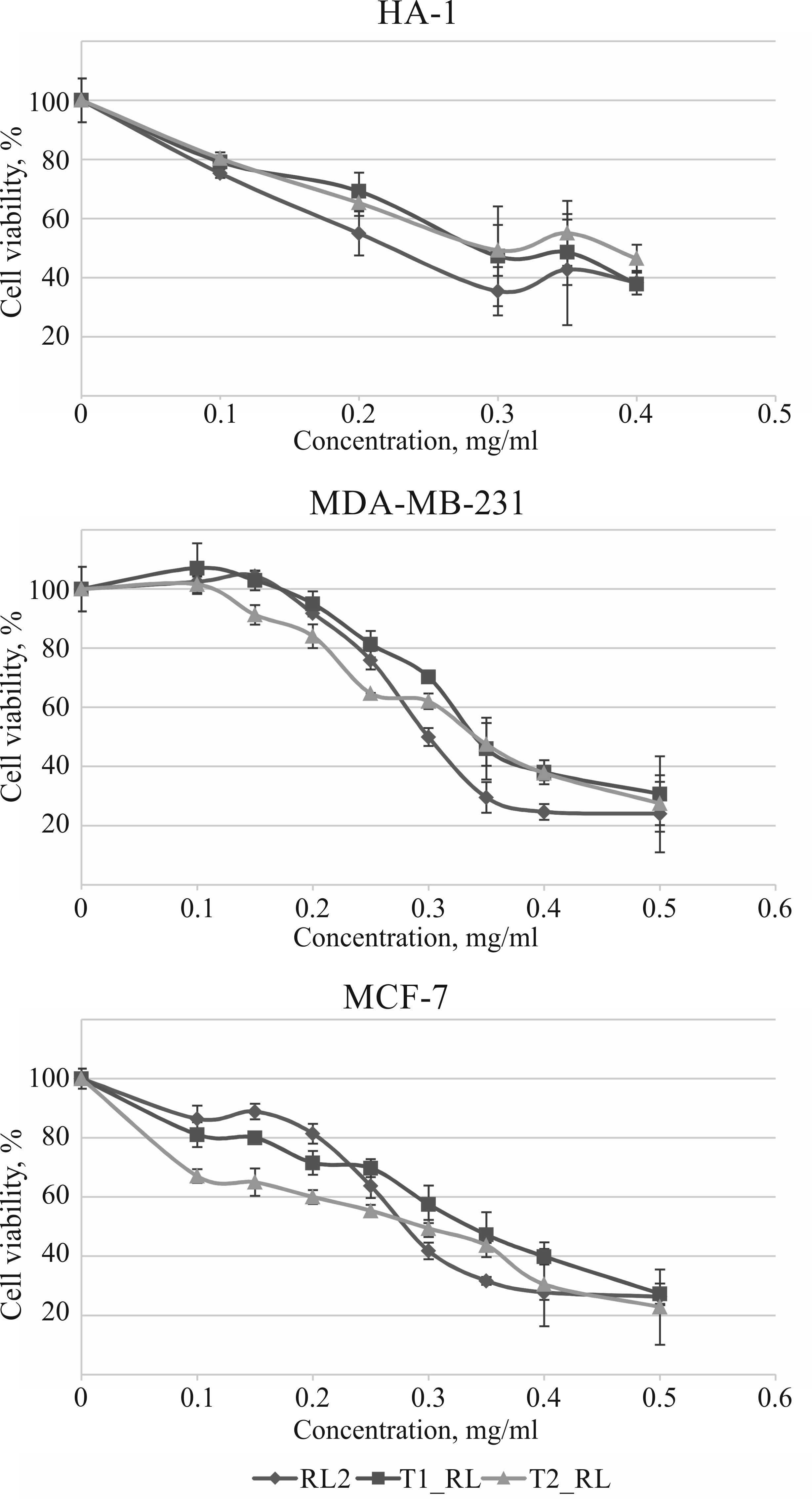
In vitro cytotoxic activity of RL2,
T1_RL and T2_RL
A comparative analysis of the effects of RL2 and the
recombinant fusion proteins T1_RL and T2_RL on tumor cell viability
was performed using HA-1, MDA-MB-231 and MCF-7 cell cultures.
Changes in cell viability were assessed using the МТТ assay.
It was demonstrated that T1_RL and T2_RL reduce the
viability of HA-1, MDA-MB-231 and MCF-7 cells in a dose-dependent
manner, while the cytotoxic activity of the fusion proteins was
practically the same as that of RL2 (Fig.
3).
As is known, no wild-type bacteriophage can infect
eukaryotic cells or produce a cytotoxic effect on them (11). Additionally, the present study
demonstrated that the incubation of MDA-MB-231 and MCF-7 tumor
cells with the bacteriophages that display the peptides
GLHTSATNLYLH and SGVYKVAYDWQH has no effect on cell viability (data
not shown).
Data obtained strongly suggest that the cytotoxic
activity of the fusion proteins is solely due to RL2, while the
tumor-targeting peptide fused to the N-terminus of RL2 has no
effect on its cytotoxic properties.
The cytotoxic activity of RL2 in relation to tumor
cells is due to its ability to induce apoptosis in these cells
(6,7).
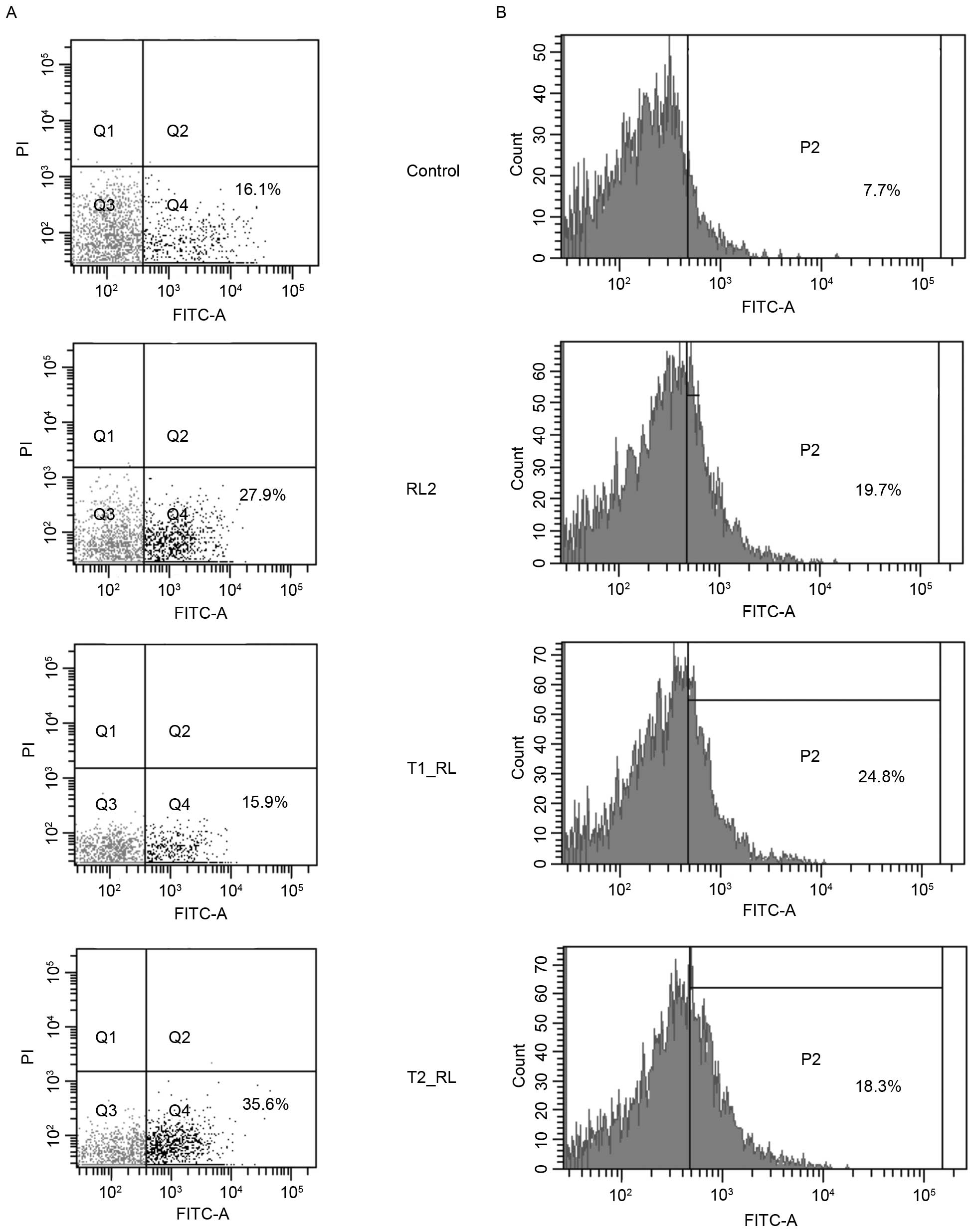
The current study assessed the ability of the fusion proteins T1_RL
and T2_RL to induce apoptosis in MDA-MB-231 cells by cytometry.
After the incubation of MDA-MB-231 cells with RL2 and T2_RL for 18
h, the population of apoptotic MDA-MB-231 cells increased to 27.9
and 35.6%, respectively, compared with the population of control
cells (16.1%). The incubation of MDA-MB-231 cells with T1_RL for 18
h led to no change in the number of apoptotic cells as compared to
that in the control cells (Fig. 4);
however, incubation with T1_RL for a longer time, 24 h, led to an
increase in the number of apoptotic cells (data not shown).
A key role in apoptosis is played by specific
proteins, the caspases (cysteine proteases) (12). As is known, RL2-induced apoptosis is
accompanied by the activation of effector caspase-3 and −7, and
mitochondrial transmembrane potential dissipation (7,13).
Activation of caspase-3 and −7 in MDA-MB-231 cells by the
recombinant proteins RL2, T1_RL and T2_RL was analyzed by
cytofluorometry in the present study. As shown in the data
presented in Fig. 4B, following
incubation with RL2, the percentage of cells expressing active
forms of caspase-3 and −7 increased to 19.7, to 24.8% after
incubation with T1_RL and to 18.3% after incubation with Т2_RL,
while the increase in the control cells was only 7.7%.
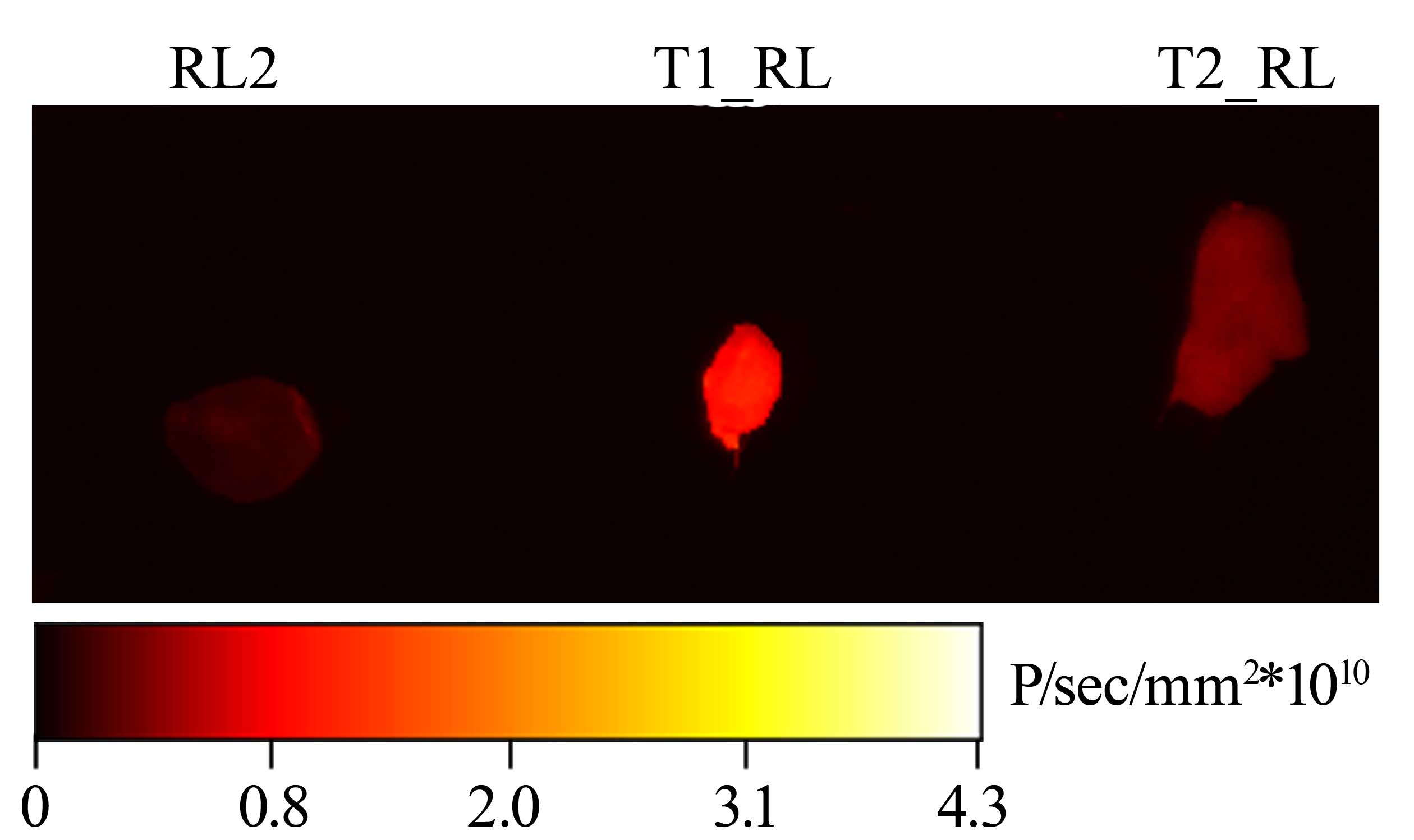
Retention of the RL2-Cy5, T1_RL-Cy5
and T2_RL-Cy5 conjugates in tumor tissue
The accumulation of the recombinant fusion proteins
in tumor tissue was assessed using the mouse HA-1 tumor model. Mice
with subcutaneously transplanted tumors were administered with the
conjugates RL2-Cy5, T1_RL-Cy5 and T2_RL-Cy5 via tail vein
injection. The Cy5-labeled conjugates were allowed to circulate for
10 min, after which the tumors were removed. The intensity of the
fluorescent signal from the tumors was measured using a Kodak In
Vivo FX Professional Imaging system (Fig. 5).
As observed in Fig. 5,
subsequent to circulation of the recombinant proteins in the
bloodstream for 10 min, the strongest fluorescent signal from the
tumor was for T1_RL; its intensity was 5.08±1.15 times as high as
that of the signal from the tumor with RL2 (P<0.05). The
intensity of the fluorescent signal from the tumor with T2_RL was
only 2.81±1.48 (P<0.05) times as high as that the signal from
the tumor with RL2.
Discussion
One of the most promising ways to improve the
efficiency of antitumor drugs is the targeted delivery of these
drugs to tumor cells.
To this end, tumor-targeting peptides of a small
size (not longer than 50 amino acid residues) are often used
(14,15). A quick and convenient way to isolate
peptides such as these is by the screening of phage-display peptide
libraries, which is performing in vitro using tumor cell
cultures and in vivo using animal models (16). The two screening systems each have
their advantages. In vitro screening allows for the
isolation of tumor-targeting peptides that are highly specific for
cell surface receptors, without even knowing what a particular
target may be. In vivo screening allows for the isolation of
peptides that are specific for tumor tissue in its natural
microenvironment. Moreover, peptides that interact with non-target
molecules, such as ubiquitous cell surface protein and plasma
proteins, or those that do not survive the degradative environment
of the vascular system are depleted from the phage population
(17). In addition to those
characteristics that make tumor cells and tumor endothelial cells
distinct from normal cells, physical differences exist between
tumor and normal tissue (temperature change, low oxygen (hypoxia)
and reduction in pH) and they affect in vivo selection
outputs (18–20).
It was previously shown that HA-1 was highly
sensitive to RL2 treatment in vitro and in vivo
(5). Therefore, a targeted delivery
of the drug lactaptin was primarily developed using the mouse HA-1
tumor model in the present study.
To obtain HA-1 specific peptides, a selection method
was applied using phage peptide libraries. Two displayed peptides,
GLHTSATNLYLH and SGVYKVAYDWQH, occurred with the highest frequency
after the fourth round of in vivo selection performed on
A/Sn mice with HA-1 tumors. The selected phage clones were studied
for the ability to bind different types of tumors in vivo at
24 h post-intravenous administration. The affinity of the selected
displayed peptides was shown not only to the selection-specific
tumor (HA-1), but also to other tumors (LA, В16 melanoma, Krebs-2
carcinoma and LLC). It appears that the peptides GLHTSATNLYLH and
SGVYKVAYDWQH are specific for the targets that are typical of
different types of tumor cells.
According to literature data, tumor cells possess
numerous quantitative and/or qualitative properties that make them
distinct from non-tumor cells and that the majority of tumor cell
types have in common. For example, the receptors of growth and
proliferation factors, including epidermal growth factor receptors,
transferrin receptors and folic acid receptors, are often
overexpressed on tumor cells, which provides for uncontrolled cell
proliferation and promotes metastasis (18). Furthermore, markers exist that are
typical of particular groups of cell lines. For example, enhanced
expression of the matrix metalloproteinase-2 and aminopeptidase N
receptors is observed on breast cancer cells (20,21), while
enhanced expression of platelet-derived growth factor receptor-β is
observed on pancreas tumor cells (20).
It is also known that tumor growth is accompanied by
active angiogenesis, which, in a healthy adult, can only be the
case if damaged tissues are undergoing regeneration. Angiogenesis
activation may, for example, occur when the level of the expression
of vascular endothelial growth factors is increasing or when the
level of the expression of receptors for the
αvβ3/5 integrins is increased
(19).
Additionally, incomplete poorly organized
vasculature that associates with tumor growth results in hypoxic
zones and a reduction in nutrient availability within the tumor
tissue. The ability to adapt to hypoxic conditions is crucial to
tumor growth and survival, and occurs through the hypoxia inducible
factor signaling pathway, which affects the expression of >1,000
genes (22).
As there was no significant binding of the displayed
peptides to Ehrlich or HA-29 tumors in the present study (data not
shown), these tumors do not appear to possess the molecular
structures or microenvironment for which selection was run. Thus,
peptides that were able to specifically deliver the associated
cargo to mouse tumors in vivo were selected.
Genetic constructs that provide for the production
of the fusion proteins composed of the selected peptides and RL2
were created and the recombinant fusion proteins, T1_RL and T2_RL,
were prepared. With the genetic constructs that provide for the
synthesis of the fusion proteins in producing strains, such stages
towards end products as the production, isolation and purification
of each component (lactaptin and the tumor-targeting peptide)
separately, as well as the final stage of conjugating two protein
molecules, are unnecessary.
Fusion of a tumor-targeting peptide to RL2 may lead
to a considerable accumulation of this cytotoxic agent in tumor
tissue and improve the antitumor efficiency of the drug.
Admittedly, it should be ensured that fusion of the tumor-targeting
peptide to the N-terminus of RL2 does not affect its cytotoxic
activity in relation to tumor cells. The in vitro
experiments of the present study demonstrated that RL2 and the
fusion proteins T1_RL and T2_RL reduce the viability of HA-1 cancer
cells in the same dose-dependent manner.
The antitumor drug lactaptin, developed on the basis
of the recombinant lactaptin analog RL2, has been successful in
preclinical trials. Lactaptin shows its highest efficiency in
relation to human breast cancers, including triple-negative cancer
[estrogen receptor (ER)−/progesterone receptor
(PR)−/human epidermal growth factor receptor 2
(HER2)−]. Triple-negative cancer is often more
aggressive than any other breast cancer and is unique in that the
therapeutic targets that are typical of this disease, namely ER, PR
and HER2/neu, are not there (23,24).
Therefore, in the present study, the cytotoxic activity was
assessed not only in relation to mouse tumor HA-1 cells, but also
human breast cancers. The data confirmed that RL2 and the fusion
proteins T1_RL and T2_RL have identical cytotoxic properties and
mechanisms of cell death induction.
Based on pharmacokinetic data, lactaptin is quite
evenly distributed throughout the organism, with a certain degree
of preference for the liver and kidneys. The half-life of lactaptin
is 15.6 min (8). The present in
vivo experiments involving the mouse HA-1 tumor model
demonstrated that tumor concentrations of the conjugates T1_RL-Cy5
and T2_RL-Cy5 after 10 min of circulation are higher than that of
RL2-Cy5, with fluorescence intensities of 5.08±1.15 and 2.81±1.48,
respectively.
Several factors may account for the slight
difference in fluorescence intensity that was observed between the
tumors. Each phage particle in the library displays five copies of
a specific peptide as part of coat protein pIII, which provides for
the multivalent binding and a high avidity of the particle, but
reduces affinity of the peptide for the target (25). A single copy of the selected peptide
within the fusion proteins may have not been sufficient for
efficient binding due to a decreased affinity of the molecule for
the receptor as a whole. Additionally, the peptides displayed on
the surface of the phage particle in five copies can interact with
each other to form dimers or multimers, thus providing for specific
binding. With the introduction of a pool of separate peptides,
chances of this happening decrease.
The study by Ahn et al (26) is notable as it demonstrated that the
tumor concentration of fusion proteins with the tumor-targeting RGD
peptide specific for the αvβ3 integrin
increases with increase in circulation time.
Thus, in the present study, a technique to improve
the antitumor efficiency of the drug lactaptin was developed by
conjugating this drug to a tumor-specific targeting peptide. Fusion
proteins composed of lactaptin and peptides specific for the
molecular structures of human tumors (in particular, breast tumors)
are deemed promising.
Acknowledgements
The present study was supported by The Ministry of
Education and Science of the Russian Federation, Federal Targeted
Program ‘R&D in Priority Areas of Russian S&T Development’,
contract 14.607.21.0063 (project unique identifier
RFMEFI60714X0063).
References
|
1
|
Millimouno FM, Dong J, Yang L, Li J and Li
X: Targeting apoptosis pathways in cancer and perspectives with
natural compounds from mother nature. Cancer Prev Res (Phila).
7:1081–1107. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Nekipelaya VV, Semenov DV, Potapenko MO,
Kuligina EV, Kit Yu, Romanova IV and Richter VA: Lactaptin is a
human milk protein inducing apoptosis of MCF-7 adenocarcinoma
cells. Dokl Biochem Biophys. 419:58–61. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Vlassov VV, Richter VA, Semenov DV,
Nekipelaya VV, Kuligina EV and Potapenko MO: Peptide inducing
apoptotic death of human cancer cells. Patent RF N 2317304.
2008.
|
|
4
|
Tikunova NV, Semenov DV, Babkina IN,
Kuligina EV, Koval OA, et al: Recombinant plasmid DNA pFK2,
providing synthesis of the recombinant peptide which is the analog
of human kappa-casein and recombinant peptide-the analog of human
kappa-casein fragment, with the apoptotic activity against human
tumor cells. Patent RF N 2401307. 2010.
|
|
5
|
Semenov DV, Fomin AS, Kuligina EV, Koval
OA, Matveeva VA, Babkina IN, Tikunova NV and Richter VA:
Recombinant analogs of a novel milk pro-apoptotic peptide,
lactaptin, and their effect on cultured human cells. Protein J.
29:174–180. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Koval OA, Fomin AS, Kaledin VI, Semenov
DV, Potapenko MO, Kuligina EV, Nikolin VP, Nikitenko EV and Richter
VA: A novel pro-apoptotic effector lactaptin inhibits tumor growth
in mice models. Biochimie. 94:2467–2474. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Koval OA, Tkachenko AV, Fomin AS, Semenov
DV, Nushtaeva AA, Kuligina EV, Zavjalov EL and Richter VA:
Lactaptin induces p53-independent cell death associated with
features of apoptosis and autophagy and delays growth of breast
cancer cells in mouse xenografts. PLoS One. 9:e939212014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bondarenko DA, Richter VA, Kuligina EV,
Koval OA, Fomin AS, et al: Toxicity studies and pharmacokinetics of
Lactaptin. Russian Journal of Biopharmaceuticals. 7:40–47.
2015.
|
|
9
|
Nemudraya AA, Richter VA and Kuligina EV:
Phage peptide libraries as a source of targeting ligands. Acta
Naturae. 8:48–57. 2016.PubMed/NCBI
|
|
10
|
Tamura K, Dudley J, Nei M and Kumar S:
MEGA4: Molecular evolutionary genetics analysis (MEGA) software
version 4.0. Mol Biol Evol. 24:1596–1599. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bakhshinejad B and Sadeghizadeh M:
Bacteriophages as vehicles for gene delivery into mammalian cells:
Prospects and problems. Expert Opin Drug Deliv. 11:1561–1574. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Galluzzi L, Vitale I, Abrams JM, Alnemri
ES, Baehrecke EH, Blagosklonny MV, Dawson TM, Dawson VL, El-Deiry
WS, Fulda S, et al: Molecular definitions of cell death
subroutines: Recommendations of the Nomenclature Committee on Cell
Death 2012. Cell Death Differ. 19:107–120. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fomin AS, Koval' OA, Semenov DV, Potapenko
MO, Kuligina EV, Kit IuIa and Rikhter VA: The Analysis of
biochemical markers of MCF-7 cells apoptosis induced by recombinant
analog of lactaptin. Bioorg Khim. 38:92–98. 2012.PubMed/NCBI
|
|
14
|
Svensen N, Walton JG and Bradley M:
Peptides for cell-selective drug delivery. Trends Pharmacol Sci.
33:186–192. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Venditto VJ and Szoka FC Jr: Cancer
nanomedicines: So many papers and so few drugs! Adv Drug Deliv Rev.
65:80–88. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hamzeh-Mivehroud M, Alizadeh AA, Morris
MB, Church WB and Dastmalchi S: Phage display as a technology
delivering on the promise of peptide drug discovery. Drug Discov
Today. 18:1144–1157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Krumpe LR and Mori T: The use of
phage-displayed peptide libraries to develop tumor-targeting drugs.
Int J Pept Res Ther. 12:79–91. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wicki A, Witzigmann D, Balasubramanian V
and Huwyler J: Nanomedicine in cancer therapy: Challenges,
opportunities, and clinical applications. J Control Release.
200:138–157. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ruoslahti E: Specialization of tumour
vasculature. Nat Rev Cancer. 2:83–90. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li ZJ and Cho CH: Peptides as targeting
probes against tumor vasculature for diagnosis and drug delivery. J
Transl Med. 10:(Suppl 1). S12012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Brown KC: Peptidic tumor targeting agents:
The road from phage display peptide selections to clinical
applications. Curr Pharm Des. 16:1040–1054. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Parks SK, Cormerais Y, Marchiq I and
Pouyssegur J: Hypoxia optimises tumour growth by controlling
nutrient import and acidic metabolite export. Mol Aspects Med
47–48. 3–14. 2016. View Article : Google Scholar
|
|
23
|
Wahba HA and El-Hadaad HA: Current
approaches in treatment of triple-negative breast cancer. Cancer
Biol Med. 12:106–116. 2015.PubMed/NCBI
|
|
24
|
Bose S: Triple-negative breast carcinoma:
Morphologic and molecular subtypes. Adv Anat Pathol. 22:306–313.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Noren KA and Noren CJ: Construction of
high-complexity combinatorial phage display peptide libraries.
Methods. 23:169–178. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ahn KY, Ko HK, Lee BR, Lee EJ, Lee JH,
Byun Y, Kwon IC, Kim K and Lee J: Engineered protein nanoparticles
for in vivo tumor detection. Biomaterials. 35:6422–6429. 2014.
View Article : Google Scholar : PubMed/NCBI
|