Introduction
Wilms tumor (WT) is the most common cancer that
primarily develops in abdominal solid organ of children, with an
incidence rate of 2.5/1 million in China (1,2). It has no
incipient symptom, and the most frequent symptoms are a painless,
palpable abdominal mass. Bilateral WT (BWT) accounts for 5–7% and
is more complex than unilateral WT (UWT) in diagnosis and treatment
(1,2).
However, at present no tumor marker has been identified to diagnose
BWT accurately. With the continuous development of proteomics
technologies in recent years, new ideas and methods for detecting
differential expression protein of WB especially BWT have been
adopted (3,4).
From January, 2009 to December, 2013, we collected
serum samples of BWT, UWT and healthy children, and screened BWT
differential expression protein with surface-enhanced laser
desorption/ionisation time-of-flight mass spectrometry (SELDI-TOF
MS), to determine the differential expression protein associated
with BWT.
Materials and methods
Serum analysis
Serum samples were selected from 10 cases of
children with BWT (BWT group), 20 cases of children with UWT (UWT
group) and 20 cases of healthy children (control group). All of the
children were sampled after overnight fasting. The 3 males and 2
females in the BWT group, aged 7–48 months (average 34.20±12.41
months), were confirmed as BWT pathologically. Six males and 4
females in the UWT group, aged 4–56 months (average 41.20±10.12
months), were confirmed as UWT pathologically. The 5 males and 5
females in the control group were aged 4–74 months (average of
34.53±15.52 months). According to WT staging criteria, BWT was in
stage V and UWT in stages I–IV. The serum samples standing at 25°C
for 1–2 h were centrifuged for 20 min at 2,500 × g, and the
supernatant was placed in Eppendorf tubes and preserved at
−80°C.
The present study was approved by the Ethics
Committee of Zhengzhou University (Zhejiang, China). Written
informed consent was obtained from the patient's relatives and/or
guardians.
Reagents and instruments
Urea, NaNC, dithiothreitol, acetonitrile,
3-[(3-cholamidopropyl) dimethylammonio)-1-propanesulfonate,
trifluoroacetic acid, sinapinic acid and trypsin were purchased
from Sigma (St. Louis, MO, USA). Spectra Multicolor Low Range
Protein Ladder and SPD SpeedVac were purchased from Thermo Fisher
Scientific (Waltham, MA, USA). PBSII SELDI-TOF MS and the WCX2
protein chip were purchased from Ciphergen Biosystems Inc.
(Fremont, CA, USA). Ammonium persulfate, 2X/4X Laemmli Sample
Buffer, 10X Tris/Glycine/SDS, TEMED, Powerpac Universal and
Mini-PROTEAN Tetra vertical electrophoresis system were purchased
from Bio-Rad (Berkeley, CA, USA). Matrix-assisted laser desorption
ionization/time-of-flight mass spectrometry (MALDI-TOF MS) was
purchased from Kratos Analytica Inc. (Spring Valley, NY, USA).
Uhraflex III MALDI-TOF/TOF MS were purchased from Bruker Corp.
(Bremen, Germany).
Screening serum differential protein
markers with SELDI-TOF MS
Sera taken from the three groups were thawed on
ice-bath for 30–60 min and centrifuged at 8,000 × g. A total of 5
µl of serum was taken and added to 10 µl U9 serum conditioning
fluid, mixed fully at 600 times/min for total of 30 min on shaker
at 4°C. The serum conditioned with U9 was diluted with binding
buffer to 200 µl, and incubated on a shaker at 4°C 600 times/min
for 2 min and WCX2 protein chip was applied (1). All-in-One standard protein chip with
known molecular weight was used to correct the error of SELDI MS
system to <0.1%, and then the protein chip well connected with
protein was put into the mass spectrometer for detection (2,3). Ciphergen
Biosystems Inc. (Version 3.1) was used to correct the data
collected, to homogenize the total ionic strength and molecular
weight, and Biomarker wizard and ZUCI protein chip data analysis
system were used for de-noising of the signal of original data with
non-sampling discrete wavelet transform approach. A mark of signal
strength was adjusted according to the three peak values marked and
appeared in all selected spectra. The baseline of spectrum was
corrected based on the monotonous and aligned minimum curve and
quality, and peaks with signal-to-noise ratio >2 were filtered.
In the cluster analysis, the minimum threshold was set to 10%, and
peaks with proton number/charge number (m/z) difference <0.3% in
each sample were clustered as a category (4).
Separation of the target protein with
SDS-PAGE
Gene electrophoresis was performed using a protein
marker (1.7–40 kDa) in the first lane, and serum samples of the
tumor and control groups were added in other lanes (5 µl in each
lane) at 90 V. The spacer gel was removed and dyed using Coomassie
blue coloring agent overnight, and then de-colored for 1.5 h. The
de-coloring liquid was discarded to continue decolorizing for 2.5
h, by comparing with a protein ladder. The corresponding target
protein bands were incised in aseptic condition, chopped and
preserved for protein identification (5).
Determination of candidate protein
markers by MALDI- TOF MS
The purified protein fluid at different periods of
time were, respectively, pulled into an integrated centrifuging
enrichment system (SPD SpeedVac) and freeze-dried to 20 µl.
Freeze-dried proteins (1.5 µl) were mixed with 1.5 µl CHCA,
respectively, and cytochrome C (molecular weight of 12,361.96) +
CHCA and insulin (molecular weight of 5,734.51) + CHCA were used to
correct the samples. The target board was placed in MALDI-TOF MS
for detection and peaks of the target protein were tracked
again.
Enzymolysis of the target
proteins
The protein containing the target protein was added
with ultrapure water (40 µl) and 4 µl 0.1 mol DTT solution of
certain concentration was placed in 37°C warm water for 1 h. The
1.6 µl IAM was added into the mixture and placed in the dark for 1
h, and 1.6 µl of 1 mol DTT solution, 150 µl of 0.1 mol
NH4HCO3 (ammonium bicarbonate) and 2 µl
trypsin were added successively, and placed in 37°C warm water
overnight (6–8 h).
Identification of the target
protein
Trypsin samples were digested and the substrate CCA
was mixed and applied to the target sample for natural drying. The
protein chip was placed into MALDI-TOF/TOF MS to detect the peptide
fragment of enzymolysis, and Mascot search software (Matrix
Science, Ltd., London, UK) was used to retrieve the proteins that
may match with the identified peptides and amino acids in the
SwissProt database (6).
Confirmation of candidate protein
biomarkers by western blotting
First, SDS-PAGE was used to extract and separate
total protein from the serum sample. Antigen-antibody immunological
reaction was carried out after semi-dry trarsmembrane, GAPDH was
used as internal reference. The chemiluminescence, developing and
photographic fixing operation were carried out, and negative film
was retained. After scanning negative film, Image pro plus 6.0
software (Media Cybernetics, Silver Spring, MD, USA) was used to
measure the gray value of each negative protein band of the film,
and the gray values were divided into the BWT, UWT and healthy
control groups, and the gray values were statistically
assessed.
Statistical analysis
After noise filtering and clustering analysis, the
Wilcoxon rank sum test was conducted against the m/z peaks
selected, and the t-test was conducted against the data of each
group. P<0.01 was considered statistically significant.
Results
Screening serum differential protein
markers with SELDI-TOF MS
SELDI-TOF MS was applied to screen the serum samples
of each group, to obtain a series of data of protein expression
intensity, and then the Wilcoxon rank sum test (α=0.01) was
conducted against data of the BWT, UWT and healthy control groups,
to screen the m/z peaks of differential expression protein.
Fourteen differential protein peaks were selected from the BWT and
healthy control groups, 8 of which were of high level expression in
the BWT group and 4 were high level expression in the healthy
control group. Ten differential protein peaks were selected from
the BWT and UWT groups, 8 of which were of high level expression in
the BWT group and 2 of high level expression in the UWT group. A
total of 13 differential protein peaks were selected from the UWT
and healthy control group, 11 of which were of high level
expression in the BWT group and 2 of high level expression in the
UWT group. By comparing the differential protein peaks of the above
groups, the difference of the 5,648 kDa m/z differential expression
proteins selected between the BWT group and healthy control group
was of statistical significance (P<0.01) (Table I), and the difference between the BWT
and UWT groups was of statistical significance (P<0.01)
(Table II), and the difference
between UWT group and the normal group was of statistical
significance (P<0.01) (Table
III). The protein expression in the BWT, UWT and healthy
control group were 100, 200 and 300, respectively.
 | Table I.Expression of m/z in 5,648 kDa peptide
fragment and protein in BWT group and control group (mean ±
SD). |
Table I.
Expression of m/z in 5,648 kDa peptide
fragment and protein in BWT group and control group (mean ±
SD).
| Group | Expression
intensity |
|---|
| BWT |
3,889.36±1,796.83a |
| Normal control | 432.21±730.42 |
 | Table II.Expression of m/z in 5,648 kDa peptide
fragment and protein in the BWT and UWT groups (mean ± SD). |
Table II.
Expression of m/z in 5,648 kDa peptide
fragment and protein in the BWT and UWT groups (mean ± SD).
| Group | Expression
intensity |
|---|
| BWT |
3,889.36±1,796.83a |
| UWT |
2,886.81±1,404.65 |
 | Table III.Expression of m/z in 5,648 kDa peptide
fragment and protein in the UWT and control groups (mean ± SD). |
Table III.
Expression of m/z in 5,648 kDa peptide
fragment and protein in the UWT and control groups (mean ± SD).
| Group | Expression
intensity |
|---|
| UWT |
2,886.81±1,404.65a |
| Normal control | 432.21±730.42 |
Identification of the target
proteins
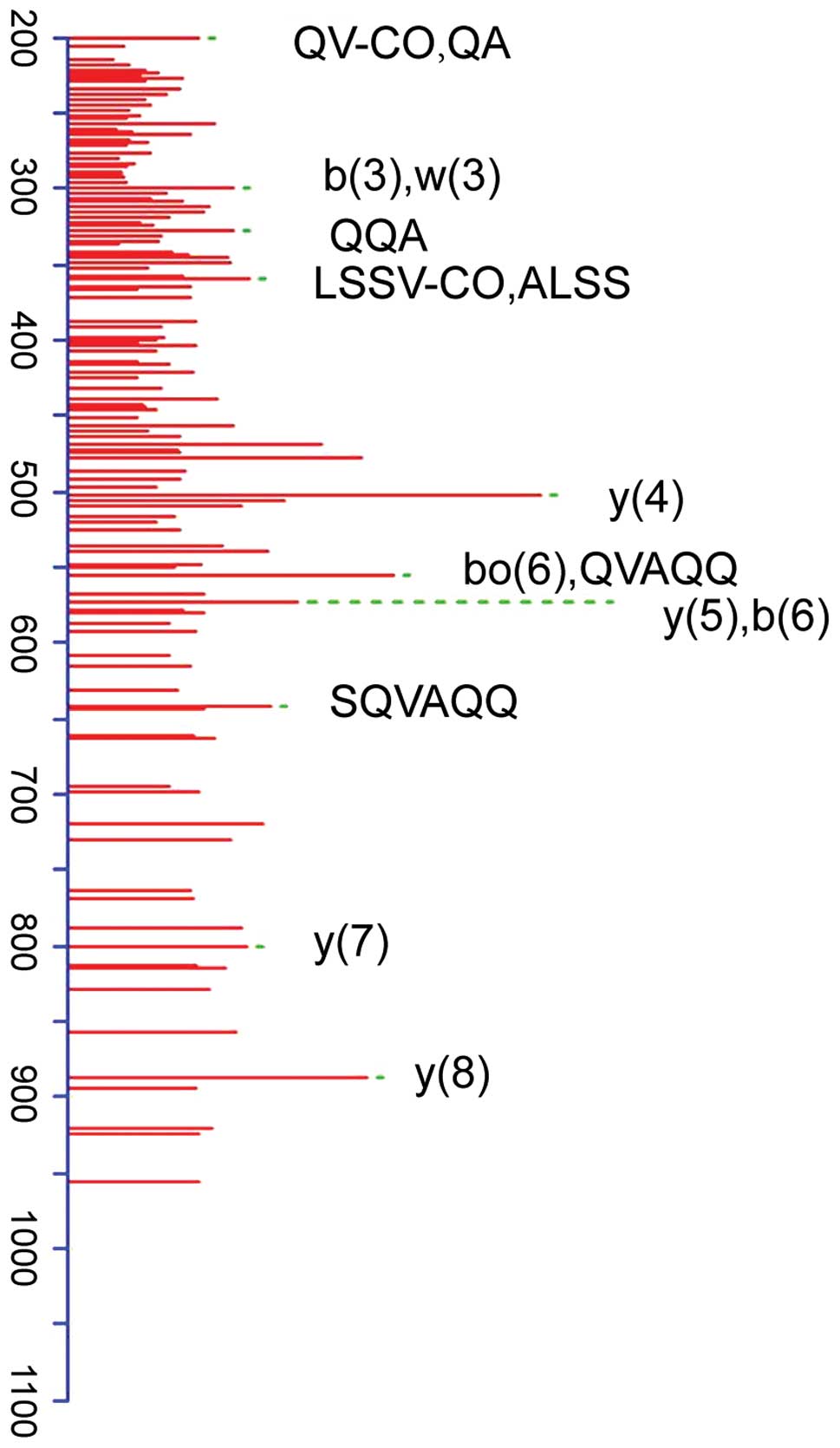
Peptide fragments of m/z of the 5,648 kDa or protein
samples were digested by enzymolysis, respectively, and peptide
fragments were detected with the 2D-LC-LTQ-MS system (Fig. 1). According to the results of
importing peptide fragments to SEQUEST retrieval program and
retrieving in Bioworks database, the peptides were apolipoprotein
C-III (APO C-III) with a coverage rate of 57.22% (Table IV).
 | Table IV.Protein or peptide segments with m/z
values of 5,648 kDa in the target protein. |
Table IV.
Protein or peptide segments with m/z
values of 5,648 kDa in the target protein.
| m/z, kDa | Protein name | Sequence
identified | Sequence coverage
(%) | Score |
|---|
| 5,648 | Apolipoprotein
C-III |
TAKDALSSVQESQVAQQARGWVTDG | 57.22% | 52.11 |
|
|
|
FSSLKDYWSTVKDKFSEFWDLDPE |
|
|
Confirmation of candidate protein
biomarkers
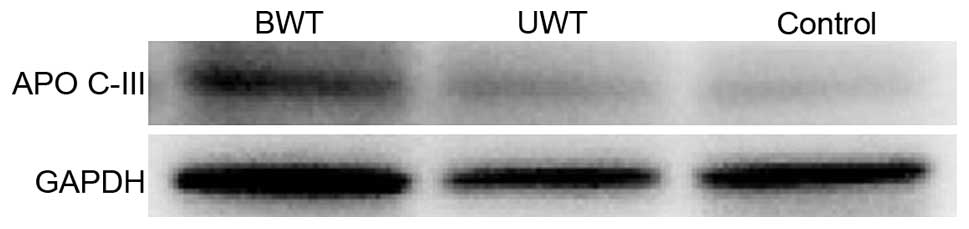
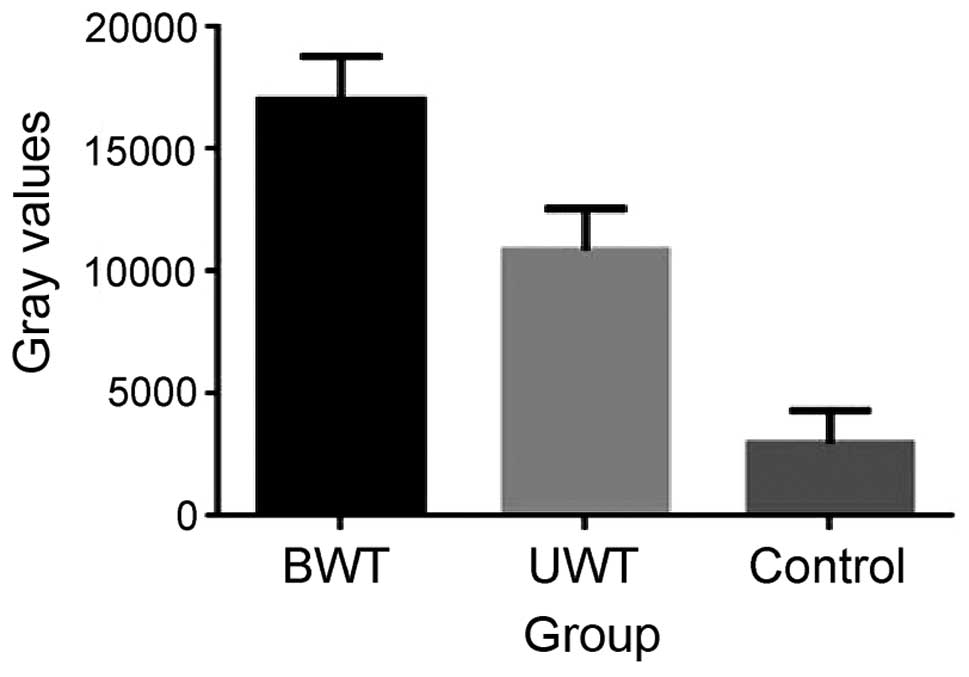
A total of 18 patients were randomly selected to
carry out western blot analysis, including 6 samples from the BWT
group, 6 samples from the UWT group, and 6 samples from the control
group. Target protein APO C-III appeared in all the bands,
statistical analysis on the band gray value of target protein in
the 3 groups of sera. The result indicated that the differences
between the APO C-III gray values of the APO and control groups,
and the differences between the APO C-III gray values of the BWT
and UWT groups have statistical significance (Figs. 2 and 3).
Discussion
WT is the most common abdominal solid cancer of
children, mostly discovered by parents during bathing or while
dressing the children (7). BWT is
rare, accounting for ~5% of all patients with WT, and is more
complex in its diagnosis and treatment compared with UWT (7). In addition, according to WT staging
criteria formulated by the Children Tumor Institute based on
surgery and histopathology, BWT was confirmed as stage V and was
worse than stages I–IV of WT in prognosis (8). There is currently no report available on
the significant improvement of the prognosis by early WBT diagnosis
and treatment or WBT tumor marker. Therefore, to improve the early
diagnostic rate and improve the prognostic effect of children with
BWT, detecting the differential protein expression of BWT is
particularly important (8).
Proteomics refers to all the proteins and existing
ways of genome expression. SELDI-TOF MS technology can detect the
molecular weight and expression level of protein of all types in
the samples under test, and MALDI-TOF/TOF MS technology can detect
the protein molecular weight, structure and other information of
the samples under test. These technologies have wide prospect of
application in detecting tumor markers, and with the emergence and
development of the technologies, and new ideas and methods of
detecting differential expression proteins of WB especially of BWT
were developed (9).
Protein of m/z of 5,648 kDa obtained was APO C-III,
and APO C-III with intensities of the protein expression of
(3,889.36±1,796.83), (2,886.81±1,404.65) and (432.21±730.42),
respectively, in BWT, UWT and the control groups. Pairwise
comparison was conducted against the relative expression levels of
the three groups of proteins stated above, and the difference was
statistically significant (P<0.01). Currently, it is believed
that histopathological features and staging are the most important
factors determining prognosis effect of children with WT (10). According to WT staging criteria
formulated by the Children Tumor Institute based on surgery and
histopathology, BWT was in stage V and UWT in stages I–IV. In BWT
group (stage V), intensity of protein expression of m/z of 5,648
kDa in the BWT group was significantly higher than that in the UWT
group (stages I–IV), and intensity of protein expression of UWT and
BWT groups was significantly higher than that in the healthy
control group, indicating that the protein may be positively
associated with WT development. According to previous studies, APO
C-III has low level of expression in the serum of healthy children
and a high level of expression in the serum of children in stages
I–IV, and the intensity of protein expression increases gradually.
Serum of the children in teh BWT group (stage V) was studied,
showing that the intensity of APO C-III expression was
significantly higher than that of the UWT group (stages I–IV),
confirming that the protein is a type of WT protein marker and its
protein expression intensity is positively correlated with WT
staging (11). In addition, we
verified the diverse expression of APO C-III in BWT, UWT and
healthy control groups to show that APO C-III can be used as a
serum protein marker of WT, especially BWT.
APO C-III is a type of protein in lipoprotein and
can be divided into A, B, C, D, E and other major categories
(11,12). It acts as a lipid carrier in plasma
and can identify APO C-III receptors and regulate plasma
lipoprotein metabolism enzyme activity (12). APO C-III is a protein produced mainly
by liver and can inhibit the combination between APO E and liver
APO E receptor, thus affecting the intake of lipoprotein with very
low density and chylomicron by the liver. Additionally, protein can
inhibit the activity of hepatic lipase, thus inhibiting the
conversion and metabolism of triglyceride lipoprotein remnants
(13).
An increase in the level of APO C-III expression in
cancer patients is likely to be caused by the inhibited activity of
immune cells in the body and functional decline, resulting in
excessive high concentrations of cholesterol and insulin in blood
not excluding raised level of APO C-III expression due to stronger
exuberance of metabolic activities of tumor tissues than other
tissues, and it has no clear causality with BWT development
(14). In addition, use of APO C-III
expression in diagnosis initially requires identification of
metabolic syndrome and the disease affecting homergy of the human
body (15).
In conclusions, in the present study, a differential
expression of APO C-III of 5,648 kDa m/z of BWT was identified
using proteomic technology as APO C-III. APO C-III has good value
and prospect of application in the diagnosis and prognosis of WT,
especially BWT, and is expected to become a new marker for early
diagnosis and prognosis (16).
However, because of the low incidence of BWT, relatively few
samples were involved in the present study, and the unclear
associations between APO C-III and BWT pathogenesis, and
reliability of the differential expression APO C-III still remain
to be studied further.
Acknowledgements
The present study was supported by the National
Natural Science Foundation of China (no. 81071782), and the Medical
Science and Technique Foundation of Henan Province (no.
201503055).
References
|
1
|
Wilde JC, Aronson DC, Sznajder B, Van
Tinteren H, Powis M, Okoye B, Cecchetto G, Audry G, Fuchs J,
Schweinitz DV, et al: Nephron sparing surgery (NSS) for unilateral
Wilms tumor (UWT): The SIOP 2001 experience. Pediatr Blood Cancer.
61:2175–2179. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Venkatramani R, Malogolowkin MH and
Mascarenhas L: Treatment of multiply relapsed Wilms tumor with
vincristine, irinotecan, temozolomide and bevacizumab. Pediatr
Blood Cancer. 61:756–759. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamai T, Tomosugi N, Abe H, Kaji Y, Oyama
T and Yoshida K: Protein profiling of blood samples from patients
with hereditary leiomyomatosis and renal cell cancer by
surface-enhanced laser desorption/ionization time-of-flight mass
spectrometry. Int J Mol Sci. 13:14518–14532. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Q, Wang J, Dong R, Yang S and Zheng
S: Identification of novel serum biomarkers in child nephroblastoma
using proteomics technology. Mol Biol Rep. 38:631–638. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kahar UM, Chan KG, Salleh MM, Hii SM and
Goh KM: A high molecular-mass Anoxybacillus sp. SK3-4
amylopullulanase: Characterization and its relationship in
carbohydrate utilization. Int J Mol Sci. 14:11302–11318. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Wang L, Zhang D, Fan Y, Jia Z, Qin
P, Yu J, Zheng S and Yang F: Identification of potential serum
biomarkers for Wilms tumor after excluding confounding effects of
common systemic inflammatory factors. Mol Biol Rep. 39:5095–5104.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Al-Hussain T, Ali A and Akhtar M: Wilms
tumor: An update. Adv Anat Pathol. 21:166–173. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ehrlich PF: Bilateral Wilms tumor: The
need to improve outcomes. Expert Rev Anticancer Ther. 9:963–973.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Datta J and Vollmer CM Jr: Investigational
biomarkers for pancreatic adenocarcinoma: Where do we stand? South
Med J. 107:256–263. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Breslow NE, Palmer NF, Hill LR, Buring J
and d'Angio GJ: Wilms' tumor: Prognostic factors for patients
without metastases at diagnosis. Results of the national Wilms'
tumor study. Cancer. 41:1577–1589. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yousuf FA and Iqbal MP: Review:
Apolipoprotein E (Apo E) gene polymorphism and coronary heart
disease in Asian populations. Pak J Pharm Sci. 28:1439–1444.
2015.PubMed/NCBI
|
|
12
|
Gaudet D, Brisson D, Tremblay K, Alexander
VJ, Singleton W, Hughes SG, Geary RS, Baker BF, Graham MJ, Crooke
RM, et al: Targeting APOC3 in the familial chylomicronemia
syndrome. N Engl J Med. 371:2200–2206. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chan DC, Wong AT, Pang J, Barrett PH and
Watts GF: Inter-relationships between proprotein convertase
subtilisin/kexin type 9, apolipoprotein C-III and plasma
apolipoprotein B-48 transport in obese subjects: A stable isotope
study in the postprandial state. Clin Sci (Lond). 128:379–385.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ooi EM, Barrett PH, Chan DC and Watts GF:
Apolipoprotein C-III: Understanding an emerging cardiovascular risk
factor. Clin Sci (Lond). 114:611–624. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Geach T: Genetics. APOC3 mutations lower
CVD risk. Nat Rev Cardiol. 11:4962014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ji H, Ai N, Li Q, Zhang K and Di W:
Clinical pathologies of breast cancer in the elderly and youths and
their prognosis. Pak J Med Sci. 30:535–538. 2014.PubMed/NCBI
|